Comparative Analysis of the Polymorphism of the Casein Genes in Camels Bred in Kazakhstan
Abstract
1. Introduction
2. Materials and Methods
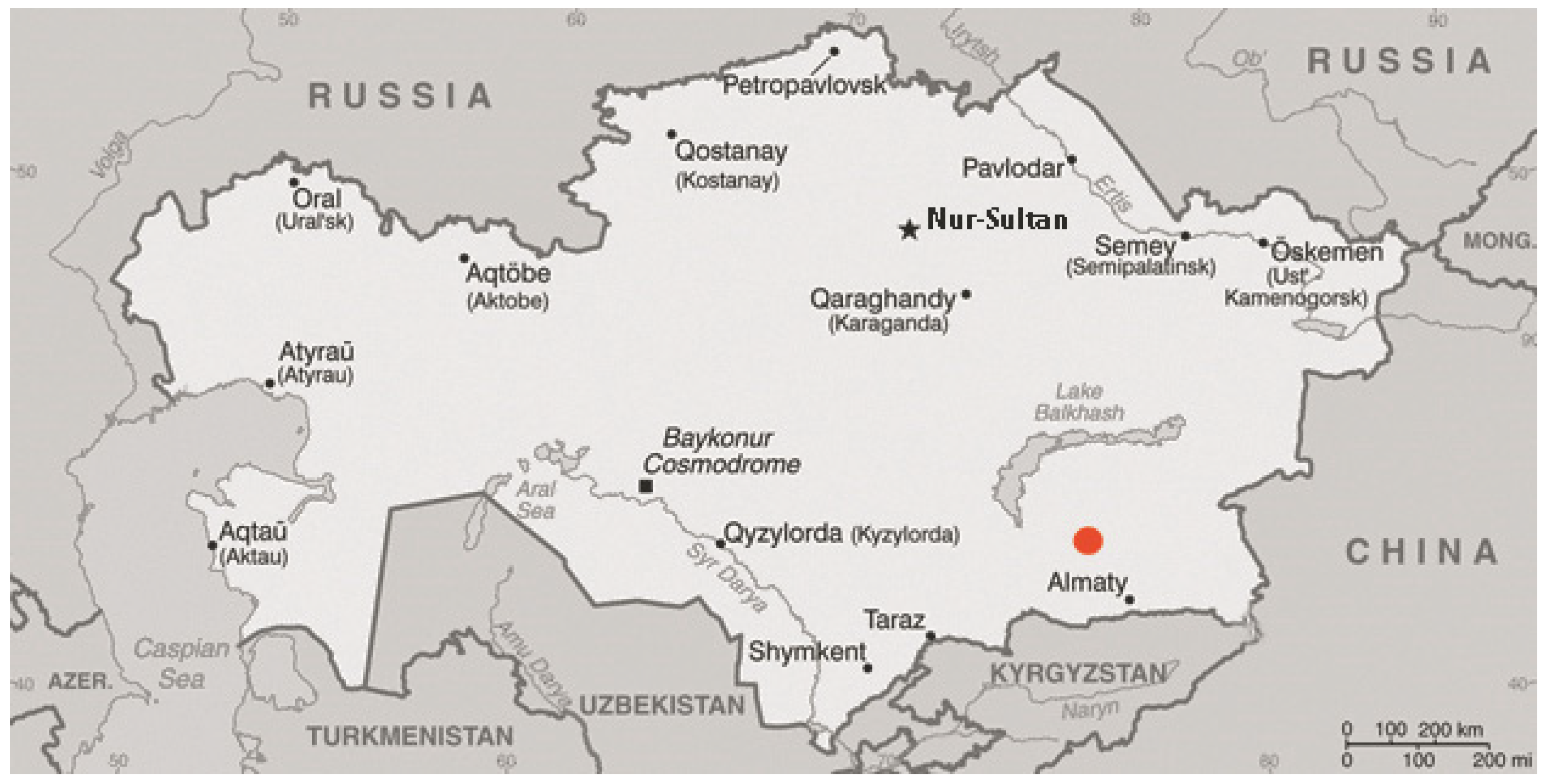
2.1. Collection of Materials
2.2. Genomic DNA Extraction and PCR-RFLP Analysis
2.3. Statistical Data Processing
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dioli, M. Dromedary (Camelus dromedarius) and Bactrian camel (Camelus bactrianus) crossbreeding husbandry practices in Turkey and Kazakhstan: An in-depth review. Pastor. Res. Policy Pract. 2020, 10, 6. [Google Scholar] [CrossRef]
- Ming, L.; Yuan, L.; Yi, L.; Ding, G.; Hasi, S.; Chen, G.; Jambl, T.; Hedayat-Evright, N.; Batmunkh, M.; Badmaevna, G.K.; et al. Whole-genome sequencing of 128 camels across Asia reveals origin and migration of domestic Bactrian camels. Commun. Biol. 2020, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Almathen, F.; Charruau, P.; Mohandesan, E.; Mwacharo, J.M.; Orozco-Terwengel, P.; Pitt, D.; Abdussamad, A.M.; Uerpmann, M.; Uerpmann, H.-P.; De Cupere, B.; et al. Ancient and modern DNA reveal dynamics of domestication and cross-continental dispersal of the dromedary. Proc. Natl. Acad. Sci. USA 2016, 113, 6707–6712. [Google Scholar] [CrossRef] [PubMed]
- Farah, Z. Composition and characteristics of camel milk. J. Dairy Res. 1993, 60, 603–626. [Google Scholar] [CrossRef] [PubMed]
- Mal, G.; Sena, D.S.; Jain, V.K.; Sahani, M.S. Therapeutic Value of Camel Milk as a Nutritional Supplement for Multiple Drug Resistant (MDR) Tuberculosis Patients. Isr. J. Vet. Med. 2006, 61, 88–91. [Google Scholar]
- Pauciullo, A.; Shuiep, E.; Cosenza, G.; Ramunno, L.; Erhardt, G. Molecular characterization and genetic variability at κ-casein gene (CSN3) in camels. Gene 2012, 15, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Shori, A.B. Camel milk as a potential therapy for controlling diabetes and its complications: A review of in vivo studies. J. Food Drug Anal. 2015, 23, 609–618. [Google Scholar] [CrossRef]
- Rachagani, S.; Gupta, I.D. Bovine kappa-casein gene polymorphism and its association with milk production traits. Genet. Mol. Biol. 2008, 31, 893–897. [Google Scholar] [CrossRef]
- Agrawal, R.P.; Beniwal, R.; Kochar, D.K.; Tuteja, F.C.; Ghorui, S.K.; Sahani, M.S.; Sharma, S. Camel milk as an adjunct to insulin therapy improves long-term glycemic control and reduction in doses of insulin in patients with type-1 diabetes: A 1 year randomized controlled trial. Diabetes Res. Clin. Pract. 2005, 68, 176–177. [Google Scholar] [CrossRef]
- Beg, O.U.; von Bahr-Lindström, H.; Zaidi, Z.H.; Jörnvall, H. Characterization of a camel milk protein rich in proline identifies a new β-casein fragment. Regul. Pept. 1986, 15, 55–61. [Google Scholar] [CrossRef]
- Shamsia, S.M. Nutritional and therapeutic properties of camel and human milks. Int. J. Genet. Mol. Biol. 2009, 1, 52–58. [Google Scholar]
- Faye, B. The enthusiasm for camel production. Editor. Emir. J. Food Agric. 2018, 30, 249–250. [Google Scholar]
- Nagy, P.; Fabri, Z.N.; Varga, L.; Reiczigel, J.; Juhasz, J. Effect of genetic and nongenetic factors on chemical composition of individual milk samples from dromedary camels (Camelus dromedarius) under intensive management. J. Dairy Sci. 2017, 100, 8680–8693. [Google Scholar] [CrossRef] [PubMed]
- Burger, P.A.; Clani, E.; Faye, B. Old World camels in a modern world—A balancing act between conservation and genetic improvement. Anim. Genet. 2019, 50, 598–612. [Google Scholar] [CrossRef]
- Hipp, N.J.; Groves, M.L.; Custer, J.N.; McMeekin, T.L. Separation of α-, β- and γ-casein. J. Dairy Sci. 1952, 35, 272–281. [Google Scholar] [CrossRef]
- Alhaj, O.A.; Al-kanhal, H.A. Compositional, technological and nutritional aspects of dromedary camel milk. Int. Dairy J. 2010, 20, 811–821. [Google Scholar] [CrossRef]
- Erhardt, G.; Shuiep, E.S.; Lisson, M.; Weimann, C.; Wang, Z.; Ibtisam, E.M.; Zubeir, E.I.; Pauciullo, A. Alpha S1-casein polymorphisms in camel (Camelus dromedarius) and descriptions of biological active peptides and allergenic epitopes. Trop. Anim. Health Prod. 2016, 48, 879–887. [Google Scholar] [CrossRef]
- Ryskaliyeva, A.; Henry, C.; Miranda, G.; Faye, B.; Konuspayeva, G.; Martin, P. Combining different proteomic approaches to resolve complexity of the milk protein fraction of dromedary, Bactrian camels and hybrids, from different regions of Kazakhstan. PLoS ONE 2018, 13, e0197026. [Google Scholar] [CrossRef]
- Singh, K.; Jayakumar, S.; Dixit, S.P.; Malik, S.Z. Molecular characterization and genetic variability of Alpha Casein gene, CSN1S1 in Bikaneri camel (Camelus dromedarius) milk. Indian J. Anim. Res. 2018, 53, 67–70. [Google Scholar] [CrossRef]
- Mati, A.; Senoussi-Ghezali, C.; Si Ahmed Zennia, S.; Almi-Sebbane, D.; El-Hatmi, H.; Girardet, J.M. Dromedary camel milk proteins, a source of peptides having biological activities—A review. Int. Dairy J. 2017, 73, 25–37. [Google Scholar] [CrossRef]
- Pauciullo, A.; Ogah, D.M.; Iannaccone, M.; Erhardt, G.; Stasio, L.D.; Cosenza, G. Genetic characterization of the oxytocinneurophysin I gene (OXT) and its regulatory regions analysis in domestic Old and New World camelids. PLoS ONE 2018, 13, e0195407. [Google Scholar] [CrossRef] [PubMed]
- Alim, N.; Fondrini, F.; Ionizzi, I.; Feligini, M.; Enne, G. Characterization of casein fractions from Algerian dromedary (Camelus dromedarius) milk. Pak. J. Nutr. 2005, 4, 112–116. [Google Scholar] [CrossRef]
- Yelubayeva, M.E.; Buralkhiyev, B.A.; Tyshchenko, V.I.; Terletskiy, V.P.; Usenbekov, Y.S. Results of Camelus dromedarius and Camelus bactrianus Genotyping by Alpha-S1-Casein, Kappa-Casein Loci, and DNA Fingerprinting. Cytol. Genet. 2018, 52, 179–185. [Google Scholar] [CrossRef]
- Shild, T.A.; Geldermann, H. Variants within the 5’-flanking regions of bovine milk-protein-encoding genes. III. Genes encoding the Ca-sensitive Caseins alpha-s1, alpha-s2 and beta. Theor. Appl. Genet. 1996, 93, 887–893. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, S.A.; Umrikar, U.D.; Sawane, M.P.; Pawar, V.D.; Deshmukh, R.S.; Dahiya, S.S.; Mehta, S.C. Genetic polymorphism at k-casein gene in Indian camel breeds (Camelus dromedarius). J. Camel Pract. Res. 2020, 27, 201–206. [Google Scholar] [CrossRef]
- Pauciullo, A.; Giambra, I.; Iannuzzi, L.; Erhardt, G. The β-casein in camels: Molecular characterization of the CSN2 gene, promoter analysis and genetic variability. Gene 2014, 547, 159–168. [Google Scholar] [CrossRef]
- Amigo, L.; Recio, I.; Ramos, M. Genetic polymorphism of ovine milk proteins: Its influence on technological properties of milk—A review. Int. Dairy J. 2000, 10, 135–149. [Google Scholar] [CrossRef]
- Soyudal, B.; Ardicli, S.; Samli, H.; Dincel, D.; Balci, F. Association of polymorphisms in the CSN2, CSN3, LGB and LALBA genes with milk production traits in Holstein cows raised in Turkey. J. Hellenic Vet. Med. Soc. 2018, 69, 1271–1282. [Google Scholar] [CrossRef]
- Nowier, A.M.; Ramadan, S.I. Association of β-casein gene polymorphism with milk composition traits of Egyptian Maghrebi camels (Camelus dromedarius). Arch. Anim. Breed. 2020, 63, 493–500. [Google Scholar] [CrossRef]
- Giambra, I.J.; Chianese, L.; Ferranti, P.; Erhardt, G. Short communication: Molecular genetic characterization of ovine αS1-casein allele H caused by alternative splicing. J. Dairy Sci. 2010, 93, 792–795. [Google Scholar] [CrossRef]
- Ramunno, L.; Cosenza, G.; Rando, A.; Pauciullo, A.; Illario, R.; Gallo, D.; Di Berardino, D.; Masina, P. Comparative analysis of gene sequence of goat CSN1S1 F and N alleles and characterization of CSN1S1 transcript variants in mammary gland. Gene 2005, 345, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Kappeler, S.; Farah, Z.; Puhan, Z. Sequence analysis of Camelus dromedarius milk caseins. J. Dairy Res. 1998, 65, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Shuiep, E.; Giambra, I.J.; El Zubeir, I.E.M.; Erhardt, G. Biochemical and molecular characterization of polymorphisms of αs1-casein in Sudanese camel (Camelus dromedarius) milk. Int. Dairy J. 2013, 28, 88–93. [Google Scholar] [CrossRef]
- Kim, J.; Yu, J.; Bag, J.; Bakovic, M.; Cant, J.P. Translation attenuation via 3′ terminal codon usage in bovine csn1s2 is responsible for the difference in αs2- and β-casein profile in milk. RNA Biol. 2015, 12, 354–367. [Google Scholar] [CrossRef][Green Version]
- Kharrati, H.K.; Esmailizadeh, A.K. SNPs genotyping technologies and their applications in farm animals breeding programs: Review. Braz. Arch. Biol. Technol. 2014, 57, 87–95. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlex 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Yeh, F.C.; Yang, R.C.; Boyle, T. POPGENE Version 1.32: Microsoft Window-Based Freeware for Population Genetics Analysis; University of Alberta: Edmonton, AB, Canada, 1999. [Google Scholar]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987; pp. 176–187. [Google Scholar] [CrossRef]
- Kimura, M.; Crow, F.J. The number of alleles that can be maintained in a finite population. Genetics 1964, 49, 725–738. [Google Scholar] [CrossRef]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef] [PubMed]
- Lewontin, R.C. The Apportionment of Human Diversity. Evol. Biol. 1972, 6, 381–398. [Google Scholar]
- Smekenov, I.T.; Akishev, Z.D.; Altybayeva, N.A.; Mukhitdinov, N.M.; Bisenbayev, A.K. Evaluation of genetic polymorphism of the Berberis iliensis population in the Ili-Balkhash region based on ISSR markers. Rep. Natl. Acad. Sci. Repub. Kazakhstan 2012, 4, 49–57. [Google Scholar]
- Reiner, A.P.; Gross, M.D.; Carlson, C.S.; Bielinski, S.J.; Lange, L.A.; Fornage, M.; Jenny, N.S.; Walston, J.; Tracy, R.P.; Williams, O.D.; et al. Common coding variants of the HNF1A gene are associated with multiple cardiovascular risk phenotypes in community-based samples of younger and older European American adults: The coronary artery risk development in young adults study and the cardiovascular health study. Circ. Cardiovasc. Genet. 2009, 2, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Othman, E.O.; Nowier, A.M.; Medhat, E.E. Genetic Variations in Two Casein Genes Among Maghrabi Camels Reared in Egypt. Biosci. Biotechnol. Res. Asia 2016, 13, 473–480. [Google Scholar] [CrossRef][Green Version]
- Giambra, I.J.; Brandt, H.; Erhardt, G. Milk protein variants are highly associated with milk performance traits in East Friesian Dairy and Lacaune sheep. Small Rumin. Res. 2014, 121, 382–394. [Google Scholar] [CrossRef][Green Version]
- Caroli, A.M.; Chessa, S.; Erhardt, G.J. Invited review: Milk protein genetic variation in cattle: Impact on animal breeding and human nutrition. J. Dairy Sci. 2009, 92, 5335–5352. [Google Scholar] [CrossRef]
- Imamura, K. Camel Production in Kazakhstan. J. Nagoya Gakuin Univ. 2015, 52, 1–13. [Google Scholar]
- Tanana, L.A.; Karaba, V.I.; Peshko, V.V. Breeding of Farm Animals and the Basics of Selection: A Tutorial; Republican Institute of Professional Education: Minsk, Belarus, 2017; pp. 236–237. [Google Scholar]
| № | SNP | Fragment Length | Primers | Annealing Temperature | Restriction Enzymes | References |
|---|---|---|---|---|---|---|
| 1 | CSN3 g.1029T > C | 488 bp | 5′-CACAAAGATGACTCTGCTATCG-3′ 5′-GCCCTCCACATATGTCTG-3′. | 60 °C | AluI | [6] |
| 2 | CSN2 g.2126A > G | 659 bp | 5′-GTTTCTCCATTACAGCATC-3′ 5′-TCAAATCTATACAGGCACTT-3′. | 53 °C | HphI | [26] |
| 3 | CSN1S1 c.150G > T | 930 bp | 5′-TGAACCAGACAGCATAGAG-3′ 5′-CTAAACTGAATGGGTGAAAC-3′ | 55 °C | SmoI | [17] |
| Population | Overall | CSN3 | CSN2 | CSN1S1 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identified Genotypes | Allele Frequency | Identified Genotypes | Allele Frequency | Identified Genotypes | Allele Frequency | |||||||||||
| CC | CT | TT | C | T | AA | AG | GG | A | G | GG | GT | TT | G | T | ||
| 1 | 8 | 1 | 3 | 4 | 0.30 | 0.70 | 5 | 2 | 1 | 0.75 | 0.25 | 6 | 2 | 0 | 0.88 | 0.12 |
| 2 | 9 | 1 | 2 | 6 | 0.18 | 0.82 | 4 | 2 | 3 | 0.60 | 0.40 | 8 | 1 | 0 | 0.94 | 0.06 |
| 3 | 20 | 2 | 10 | 8 | 0.37 | 0.63 | 18 | 2 | 0 | 0.95 | 0.05 | 20 | 0 | 0 | 1.00 | 0 |
| 4 | 16 | 8 | 3 | 5 | 0.44 | 0.56 | 1 | 6 | 9 | 0.25 | 0.75 | 14 | 2 | 0 | 0.94 | 0.06 |
| Overall | 53 | 12 | 18 | 23 | 0.40 | 0.60 | 28 | 12 | 13 | 0.64 | 0.36 | 48 | 5 | 0 | 0.94 | 0.06 |
| χ2 = 12.1 | χ2 = 8.6 | χ2 = 14.5 | ||||||||||||||
| Populations | Overall | CSN3 | CSN2 | CSN1S1 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identified Genotypes | Allele Frequency | Identified Genotypes | Allele Frequency | Identified Genotypes | Allele Frequency | |||||||||||
| CC | CT | TT | C | T | AA | AG | GG | A | G | GG | GT | TT | G | T | ||
| One-humped camels (Camelus dromedarius) | 9 | 1 | 4 | 4 | 0.33 | 0.67 | 5 | 2 | 2 | 0.66 | 0.33 | 7 | 2 | 0 | 0.89 | 0.11 |
| Two-humped camels (Camelus bactrianus) | 22 | 8 | 7 | 7 | 0.52 | 0.48 | 7 | 7 | 8 | 0.48 | 0.52 | 20 | 2 | 0 | 0.95 | 0.05 |
| Hybrids | 22 | 3 | 7 | 12 | 0.30 | 0.70 | 16 | 3 | 3 | 0.80 | 0.20 | 21 | 1 | 0 | 0.98 | 0.02 |
| Overall | 53 | 12 | 18 | 23 | 0.40 | 0.60 | 28 | 12 | 13 | 0.64 | 0.36 | 48 | 5 | 0 | 0.94 | 0.06 |
| χ2 = 0.40 | χ2 = 0.98 | χ2 = 36.32 | ||||||||||||||
| Locus | Na | Ne | h | I | |
|---|---|---|---|---|---|
| CSN3 | CC | 2.0000 | 1.5392 | 0.3503 | 0.5349 |
| CT | 2.0000 | 1.8134 | 0.4486 | 0.6408 | |
| TT | 2.0000 | 1.9657 | 0.4913 | 0.6844 | |
| CSN2 | AA | 2.0000 | 1.9824 | 0.4956 | 0.6887 |
| AG | 2.0000 | 1.5879 | 0.3702 | 0.5571 | |
| GG | 2.0000 | 1.4902 | 0.3289 | 0.5107 | |
| CSN1S1 | GG | 2.0000 | 1.2061 | 0.1709 | 0.3125 |
| GT | 2.0000 | 1.2061 | 0.1709 | 0.3125 | |
| TT | 1.0000 | 1.0000 | 0.0000 | 0.0000 | |
| Mean | 1.8889 | 1.5323 | 0.3141 | 0.4713 | |
| Statistical error | 0.3333 | 0.3483 | 0.1684 | 0.2255 | |
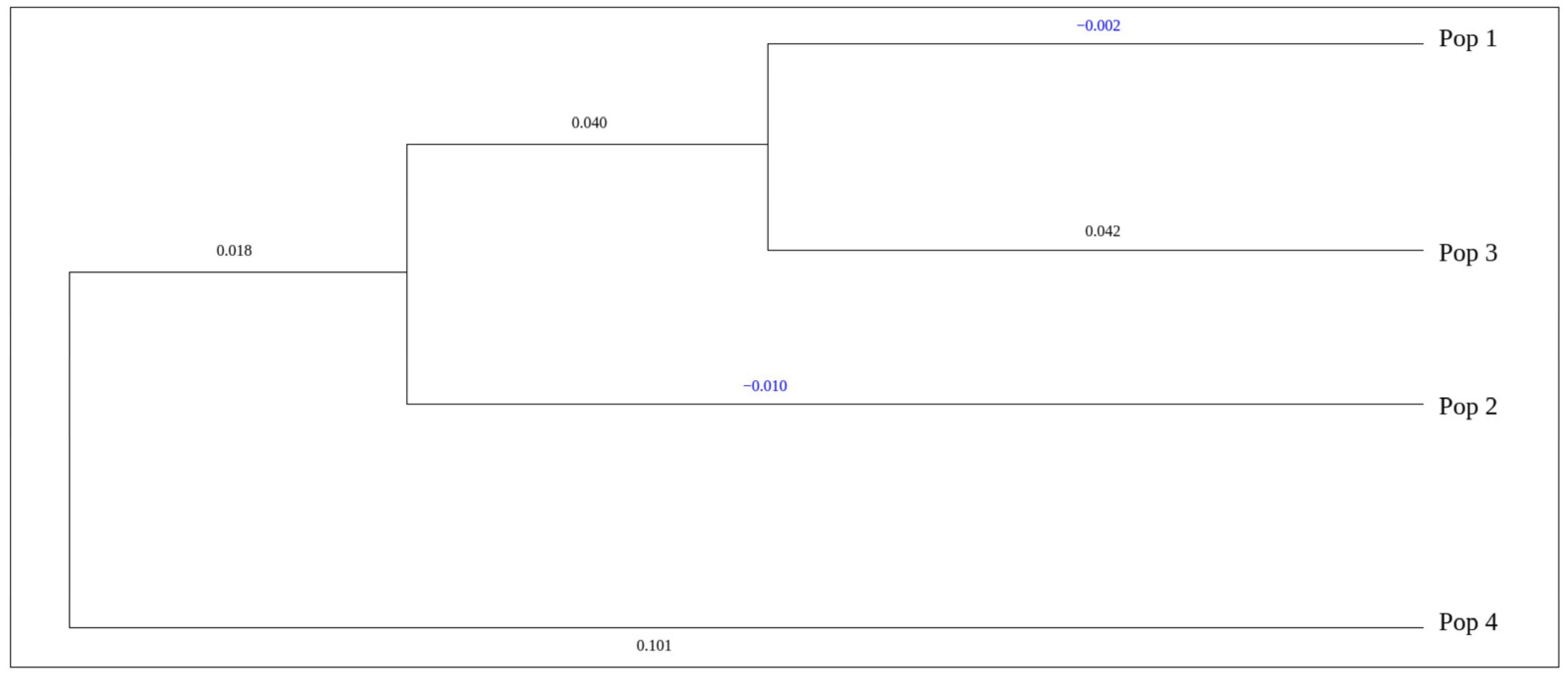
| Population | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| 1 | – | 0.9567 | 0.9603 | 0.8684 |
| 2 | 0.0443 | – | 0.9447 | 0.8969 |
| 3 | 0.0405 | 0.0569 | – | 0.8046 |
| 4 | 0.1411 | 0.1088 | 0.2174 | – |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amandykova, M.; Dossybayev, K.; Mussayeva, A.; Bekmanov, B.; Saitou, N. Comparative Analysis of the Polymorphism of the Casein Genes in Camels Bred in Kazakhstan. Diversity 2022, 14, 285. https://doi.org/10.3390/d14040285
Amandykova M, Dossybayev K, Mussayeva A, Bekmanov B, Saitou N. Comparative Analysis of the Polymorphism of the Casein Genes in Camels Bred in Kazakhstan. Diversity. 2022; 14(4):285. https://doi.org/10.3390/d14040285
Chicago/Turabian StyleAmandykova, Makpal, Kairat Dossybayev, Aizhan Mussayeva, Bakytzhan Bekmanov, and Naruya Saitou. 2022. "Comparative Analysis of the Polymorphism of the Casein Genes in Camels Bred in Kazakhstan" Diversity 14, no. 4: 285. https://doi.org/10.3390/d14040285
APA StyleAmandykova, M., Dossybayev, K., Mussayeva, A., Bekmanov, B., & Saitou, N. (2022). Comparative Analysis of the Polymorphism of the Casein Genes in Camels Bred in Kazakhstan. Diversity, 14(4), 285. https://doi.org/10.3390/d14040285






