Distribution of Homozygosity Regions in the Genome of Kazakh Cattle Breeds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sample Collection and Genotyping
2.3. Quality Control and Data Analysis
3. Results
4. Discussion
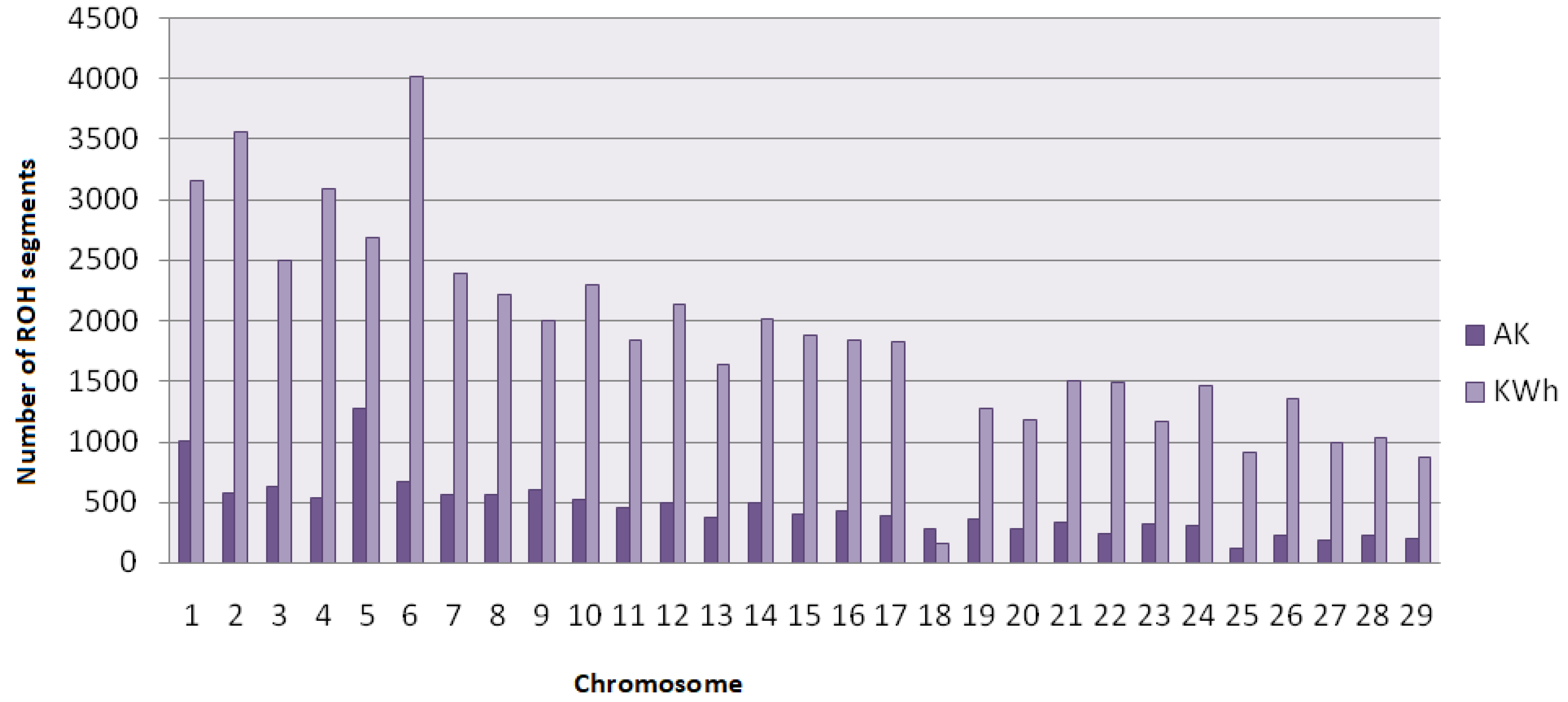
4.1. Distribution of ROH
4.2. ROH-Based Genomic Inbreeding Coefficients
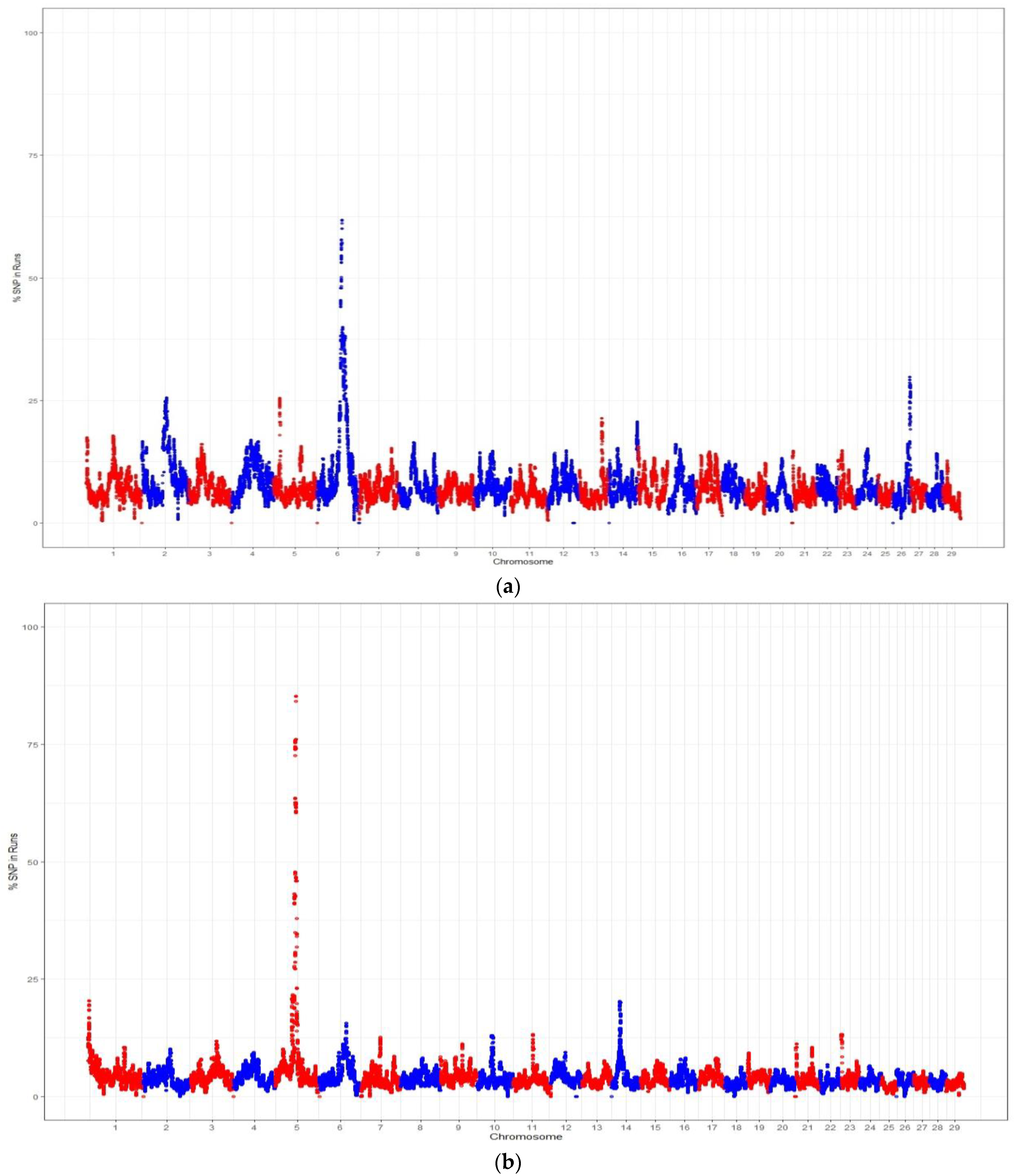
4.3. Genomic Regions within ROH
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Danilenko, O.V. Scientific and Practical Substantiation of Improving the Breeding and Productive Qualities of Auliekol Cattle in the Northern Region of Kazakhstan. Ph.D. Thesis, Mosscow, Russia, 2019. [Google Scholar]
- Makaev, S.H.A.; Nurzhanov, B.S.; Fomin, A.V. Domestic meat breed—Kazakh white-headed. Bull. Meat Cattle Breed. 2015, 4, 57–62. [Google Scholar]
- Tyngozieva, A.T. Development of Methods for Assessing the Exterior of Pedigree Bulls of the Kazakh White-Headed Breed and Their Classification by Body Type. Ph.D. Thesis, Almaty, Kazakhstan, 2018. [Google Scholar]
- Bagriy, B.A.; Dorotyuk, E.N.; Kryuchkov, V.D.; Isabekov, K.I.; Zhakipov, A.P. On the use of combined breeds in increasing meat productivity and improving the Kazakh white-headed breed. Bull. Agric. Sci. 2000, 3, 31–37. [Google Scholar]
- Weber, J.L.; Broman, K.W. 7 Genotyping for human whole-genome scans: Past, present, and future. Adv. Genet. 2001, 42, 77–96. [Google Scholar] [CrossRef] [PubMed]
- Gibson, J.; Morton, N.E.; Collins, A. Extended tracts of homozygosity in outbred human populations. Hum. Mol. Genet. 2006, 15, 789–795. [Google Scholar] [CrossRef] [Green Version]
- Kirin, M.; McQuillan, R.; Franklin, C.S.; Campbell, H.; McKeigue, P.M.; Wilson, J.F. Genomic Runs of Homozygosity Record Population History and Consanguinity. PLoS ONE 2010, 5, e13996. [Google Scholar] [CrossRef] [Green Version]
- Goszczynski, D.; Molina, A.; Terán, E.; Morales-Durand, H.; Ross, P.; Cheng, H.; Giovambattista, G.; Demyda-Peyrás, S. Runs of homozygosity in a selected cattle population with extremely inbred bulls: Descriptive and functional analyses revealed highly variable patterns. PLoS ONE 2018, 13, e0200069. [Google Scholar] [CrossRef] [Green Version]
- Williams, J.; Hall, S.; Del Corvo, M.; Ballingall, K.T.; Colli, L.; Marsan, P.A.; Biscarini, F. Inbreeding and purging at the genomic Level: The Chillingham cattle reveal extensive, non-random SNP heterozygosity. Anim. Genet. 2015, 47, 19–27. [Google Scholar] [CrossRef]
- Shi, L.; Wang, L.; Liu, J.; Deng, T.; Yan, H.; Zhang, L.; Liu, X.; Gao, H.; Hou, X.; Wang, L.; et al. Estimation of inbreeding and identification of regions under heavy selection based on runs of homozygosity in a Large White pig population. J. Anim. Sci. Biotechnol. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Szmatoła, T.; Gurgul, A.; Ropka-Molik, K.; Jasielczuk, I.; Ząbek, T.; Bugno-Poniewierska, M. Characteristics of runs of homozygosity in selected cattle breeds maintained in Poland. Livest. Sci. 2016, 188, 72–80. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Biscarini, F.; Cozzi, P.; Gaspa, G.; Marras, G. DetectRUNS: Detect Runs of Homozygosity and Runs of Heterozygosity in Diploid Genomes. R Package Version 0.9.5. Available online: https://cran.r-project.org/web/packages/detectRUNS/vignettes/detectRUNS.vignette.html (accessed on 3 April 2022).
- Ferenčaković, M.; Hamzic, E.; Gredler, B.; Solberg, T.R.; Klemetsdal, G.; Curik, I.; Sölkner, J. Estimates of autozygosity derived from runs of homozygosity: Empirical evidence from selected cattle populations. J. Anim. Breed. Genet. 2013, 130, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Ferenčaković, M.; Sölkner, J.; Curik, I. Estimating autozygosity from high-throughput information: Effects of SNP density and genotyping errors. Genet. Sel. Evol. 2013, 45, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McQuillan, R.; Leutenegger, A.-L.; Abdel-Rahman, R.; Franklin, C.S.; Pericic, M.; Barac-Lauc, L.; Smolej-Narancic, N.; Janicijevic, B.; Polasek, O.; Tenesa, A.; et al. Runs of Homozygosity in European Populations. Am. J. Hum. Genet. 2008, 83, 359–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, P.D.; Campbell, M.J.; Kejariwal, A.; Mi, H.; Karlak, B.; Daverman, R.; Diemer, K.; Muruganujan, A.; Narechania, A. PANTHER: A Library of Protein Families and Subfamilies Indexed by Function. Genome Res. 2003, 13, 2129–2141. [Google Scholar] [CrossRef] [Green Version]
- Purfield, D.C.; Berry, D.P.; McParland, S.; Bradley, D.G. Runs of homozygosity and population history in cattle. BMC Genet. 2012, 13, 70. [Google Scholar] [CrossRef] [Green Version]
- Xie, R.; Shi, L.; Liu, J.; Deng, T.; Wang, L.; Liu, Y.; Zhao, F. Genome-Wide Scan for Runs of Homozygosity Identifies Candidate Genes in Three Pig Breeds. Animals 2019, 9, 518. [Google Scholar] [CrossRef] [Green Version]
- Peripolli, E.; Munari, D.P.; Silva, M.V.G.B.; Lima, A.L.F.; Irgang, R.; Baldi, F. Runs of homozygosity: Current knowledge and applications in livestock. Animal Genetics 2016, 48, 255–271. [Google Scholar] [CrossRef]
- Addo, S.; Klingel, S.; Hinrichs, D.; Thaller, G. Runs of Homozygosity and NetView analyses provide new insight into the genome-wide diversity and admixture of three German cattle breeds. PLoS ONE 2019, 14, e0225847. [Google Scholar] [CrossRef]
- Szmatoła, T.; Gurgul, A.; Jasielczuk, I.; Ząbek, T.; Ropka-Molik, K.; Litwińczuk, Z.; Bugno-Poniewierska, M. Comprehensive Analysis of Runs of Homozygosity of Eleven Cattle Breeds Representing Different Production Types. Animals 2019, 9, 1024. [Google Scholar] [CrossRef] [Green Version]
- Peripolli, E.; Metzger, J.; De Lemos, M.V.A.; Stafuzza, N.B.; Kluska, S.; Olivieri, B.F.; Feitosa, F.L.B.; Berton, M.P.; Lopes, F.B.; Munari, D.P.; et al. Autozygosity islands and ROH patterns in Nellore lineages: Evidence of selection for functionally important traits. BMC Genom. 2018, 19, 1–14. [Google Scholar] [CrossRef]
- Peripolli, E.; Stafuzza, N.B.; Amorim, S.T.; De Lemos, M.V.A.; Grigoletto, L.; Kluska, S.; Ferraz, J.B.S.; Eler, J.; Mattos, E.C.; Baldi, F. Genome-wide scan for runs of homozygosity in the composite Montana Tropical ® beef cattle. J. Anim. Breed. Genet. 2019, 137, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Marras, G.; Gaspa, G.; Sorbolini, S.; Dimauro, C.; Marsan, P.A.; Valentini, A.; Williams, J.; Macciotta, N.P.P. Analysis of runs of homozygosity and their relationship with inbreeding in five cattle breeds farmed in Italy. Anim. Genet. 2014, 46, 110–121. [Google Scholar] [CrossRef]
- Figeac, N.; Serralbo, O.; Marcelle, C.; Zammit, P.S. ErbB3 binding protein-1 (Ebp1) controls proliferation and myogenic differentiation of muscle stem cells. Dev. Biol. 2014, 386, 135–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cobanoglu, O.; Gurcan, E.K.; Cankaya, S.; Kul, E.; Abaci, H.S. The Detection of STAT1 Gene Influencing Milk Related Traits in Turkish Holstein and Jersey Cows. J. Agric. Sci. Technol. A 2016, 6, 261–269. [Google Scholar] [CrossRef] [Green Version]
- Nobusawa, S.; Stawski, R.; Kim, Y.-H.; Nakazato, Y.; Ohgaki, H. Amplification of the PDGFRA, KIT and KDR genes in glioblastoma: A population-based study. Neuropathology 2011, 31, 583–588. [Google Scholar] [CrossRef]
- Kadokawa, H.; Pandey, K.; Nahar, A.; Nakamura, U.; Rudolf, F.O. Gonadotropin-releasing hormone (GnRH) receptors of cattle aggregate on the surface of gonadotrophs and are increased by elevated GnRH concentrations. Anim. Reprod. Sci. 2014, 150, 84–95. [Google Scholar] [CrossRef]
- Marras, G.; Gaspa, G.; Sorbolini, S.; Dimauro, C.; Ajmone-Marsan, P.; Valentini, A.; Williams, J.L.; Rudolf, F.O.; MacCiotta NPP. Analysis of runs of homozygosity and their relationship with inbreeding in five cattle breeds farmed in Italy. Anim. Genet. 2015, 46, 110–121. [Google Scholar] [CrossRef]
| Breed | Statistics | 1–2 Mb | 2–4 Mb | 4–8 Mb | 8–16 Mb | >16 Mb | |
|---|---|---|---|---|---|---|---|
| KWh | Number of ROH per animal | Mean | 27.15 | 24.12 | 11.34 | 3.3 | 1.71 |
| SD | 12.48 | 11.61 | 5.9 | 2.1 | 1.1 | ||
| Max | 84 | 84 | 36 | 14 | 8 | ||
| Min | 2 | 2 | 1 | 1 | 1 | ||
| Length (Mb) of ROH per animal | Mean | 211.59 | 171.54 | 104.43 | 46.83 | 39.88 | |
| SD | 92.98 | 79.27 | 56.33 | 37.69 | 32.49 | ||
| Max | 498.91 | 433.03 | 321.21 | 288.74 | 241.67 | ||
| Min | 8.02 | 4.25 | 4.71 | 8.03 | 16.01 | ||
| AK | Number of ROH per animal | Mean | 12.1 | 8.67 | 4.64 | 2.17 | 1.56 |
| SD | 3.5 | 3.12 | 2.11 | 1.36 | 1.43 | ||
| Max | 24 | 21 | 12 | 10 | 13 | ||
| Min | 2 | 1 | 1 | 1 | 1 | ||
| Length (MB) of ROH per animal | Mean | 99.62 | 81.98 | 57.88 | 37.49 | 39.84 | |
| SD | 46.48 | 46.05 | 43.86 | 40.99 | 42.36 | ||
| Max | 510.25 | 495.58 | 463.63 | 446.77 | 362.13 | ||
| Min | 29.92 | 13.69 | 4.61 | 8.01 | 16.02 |
| Breed | Statistics | 1–2 (Mb) | 2–4 (Mb) | 4–8 (Mb) | 8–16 (Mb) | >16 (Mb) |
|---|---|---|---|---|---|---|
| KWh | Mean | 0.084 | 0.068 | 0.041 | 0.018 | 0.016 |
| SD | 0.037 | 0.031 | 0.022 | 0.015 | 0.013 | |
| Max | 0.197 | 0.172 | 0.127 | 0.114 | 0.095 | |
| Min | 0.003 | 0.002 | 0.002 | 0.003 | 0.006 | |
| AK | Mean | 0.039 | 0.032 | 0.023 | 0.015 | 0.016 |
| SD | 0.018 | 0.018 | 0.017 | 0.016 | 0.016 | |
| Max | 0.202 | 0.196 | 0.184 | 0.177 | 0.144 | |
| Min | 0.011 | 0.005 | 0.002 | 0.003 | 0.006 |
| Group | Chr | SNPs | Start (bp) | End (bp) | Length (bp) |
|---|---|---|---|---|---|
| AK | BTA1 | 10 | 1,578,430 | 1,983,902 | 405,472 |
| AK | BTA14 | 16 | 24,716,826 | 24,828,922 | 112,096 |
| AK | BTA5 | 14 | 47,559,492 | 48,019,679 | 460,187 |
| AK | BTA5 | 30 | 48,094,474 | 48,280,390 | 185,916 |
| AK | BTA5 | 209 | 53,600,273 | 60,322,619 | 6,722,346 |
| KWh | BTA13 | 13 | 64,228,423 | 64,660,314 | 431,891 |
| KWh | BTA13 | 5 | 65,140,902 | 65,236,809 | 95,907 |
| KWh | BTA14 | 16 | 81,915,831 | 82,174,926 | 259,095 |
| KWh | BTA2 | 216 | 67,226,923 | 72,583,890 | 5,356,967 |
| KWh | BTA2 | 42 | 73,165,500 | 74,246,725 | 1,081,225 |
| KWh | BTA2 | 9 | 74,322,634 | 74,444,101 | 121,467 |
| KWh | BTA26 | 119 | 48,407,133 | 50,450,382 | 2,043,249 |
| KWh | BTA5 | 69 | 17,125,373 | 19,048,571 | 1,923,198 |
| KWh | BTA6 | 982 | 64,466,274 | 88,186,013 | 23,719,739 |
| Biological Process/Breeds | The Number of Genes Involved in the Process | |
|---|---|---|
| KWh | Ak | |
| biological phase (GO:0044848) | 1 | 0 |
| biological regulation (GO:0065007) | 39 | 39 |
| biomineralization (GO:0110148) | 1 | 0 |
| biological adhesion (GO:0022610) | 0 | 1 |
| cellular process (GO:0009987) | 69 | 59 |
| developmental process (GO:0032502) | 10 | 6 |
| growth (GO:0040007) | 0 | 1 |
| immune system process (GO:0002376) | 3 | 3 |
| interspecies interaction between organisms (GO:0044419) | 1 | 1 |
| localization (GO:0051179) | 20 | 12 |
| locomotion (GO:0040011) | 3 | 1 |
| metabolic process (GO:0008152) | 46 | 44 |
| multi-organism process (GO:0051704) | 1 | 0 |
| multicellular organismal process (GO:0032501) | 14 | 11 |
| pigmentation (GO:0043473) | 1 | 1 |
| reproduction (GO:0000003) | 2 | 0 |
| reproductive process (GO:0022414) | 2 | 0 |
| response to stimulus (GO:0050896) | 22 | 22 |
| rhythmic process (GO:0048511) | 1 | 0 |
| signaling (GO:0023052) | 18 | 18 |
| Breed | Pathways | Genes |
|---|---|---|
| Auliekol | PDGF signaling pathway (P00047) | STAT6, STAT2, ARHGAP9 |
| JAK/STAT signaling pathway (P00038) | STAT6, STAT2 | |
| TGF-beta signaling pathway (P00052) | INHBC, INHBE, GDF11 | |
| EGF receptor signaling pathway (P00018) | ERBB3, STAT6, STAT2 | |
| Interleukin signaling pathway (P00036) | IL10RB, IL23A, STAT6, STAT2 | |
| Alzheimer’s disease-presenilin pathway (P00004) | MMP19, LRP1 | |
| Inflammation mediated by chemokine and cytokine signaling pathway (P00031) | IL10RB, STAT6 | |
| p53 pathway feedback loops 2 (P04398) | CDK2, DGKA | |
| p53 pathway (P00059) | CDK2, DGKA | |
| Kazakh White-headed | Angiogenesis (P00005) | PDGFRA, KDR |
| Heterotrimeric G-protein signaling pathway, Gi alpha and Gs alpha-mediated pathways (P00026) | LOC523484, GNRHR | |
| Ubiquitin proteasome pathway (P00060) | UBA6, ITCH |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beishova, I.; Dossybayev, K.; Shamshidin, A.; Belaya, A.; Bissembayev, A.; Khamzin, K.; Kovalchuk, A.; Nametov, A. Distribution of Homozygosity Regions in the Genome of Kazakh Cattle Breeds. Diversity 2022, 14, 279. https://doi.org/10.3390/d14040279
Beishova I, Dossybayev K, Shamshidin A, Belaya A, Bissembayev A, Khamzin K, Kovalchuk A, Nametov A. Distribution of Homozygosity Regions in the Genome of Kazakh Cattle Breeds. Diversity. 2022; 14(4):279. https://doi.org/10.3390/d14040279
Chicago/Turabian StyleBeishova, Indira, Kairat Dossybayev, Alzhan Shamshidin, Alena Belaya, Anuarbek Bissembayev, Kadyrzhan Khamzin, Alexandr Kovalchuk, and Askar Nametov. 2022. "Distribution of Homozygosity Regions in the Genome of Kazakh Cattle Breeds" Diversity 14, no. 4: 279. https://doi.org/10.3390/d14040279
APA StyleBeishova, I., Dossybayev, K., Shamshidin, A., Belaya, A., Bissembayev, A., Khamzin, K., Kovalchuk, A., & Nametov, A. (2022). Distribution of Homozygosity Regions in the Genome of Kazakh Cattle Breeds. Diversity, 14(4), 279. https://doi.org/10.3390/d14040279






