The Efficiency of DNA Barcoding in the Identification of Afromontane Forest Tree Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and Sampling
2.2. DNA Extraction and Sequencing
2.3. Testing the DNA Barcode Accuracy
3. Results
3.1. Sequencing Success
3.2. Taxonomic Update Using BLAST
3.3. Barcode Accuracy
4. Discussion
4.1. Recoverability of DNA Barcode Used
4.2. Tree Species Identification Using DNA Barcode in Ngel Nyaki Montane Forest
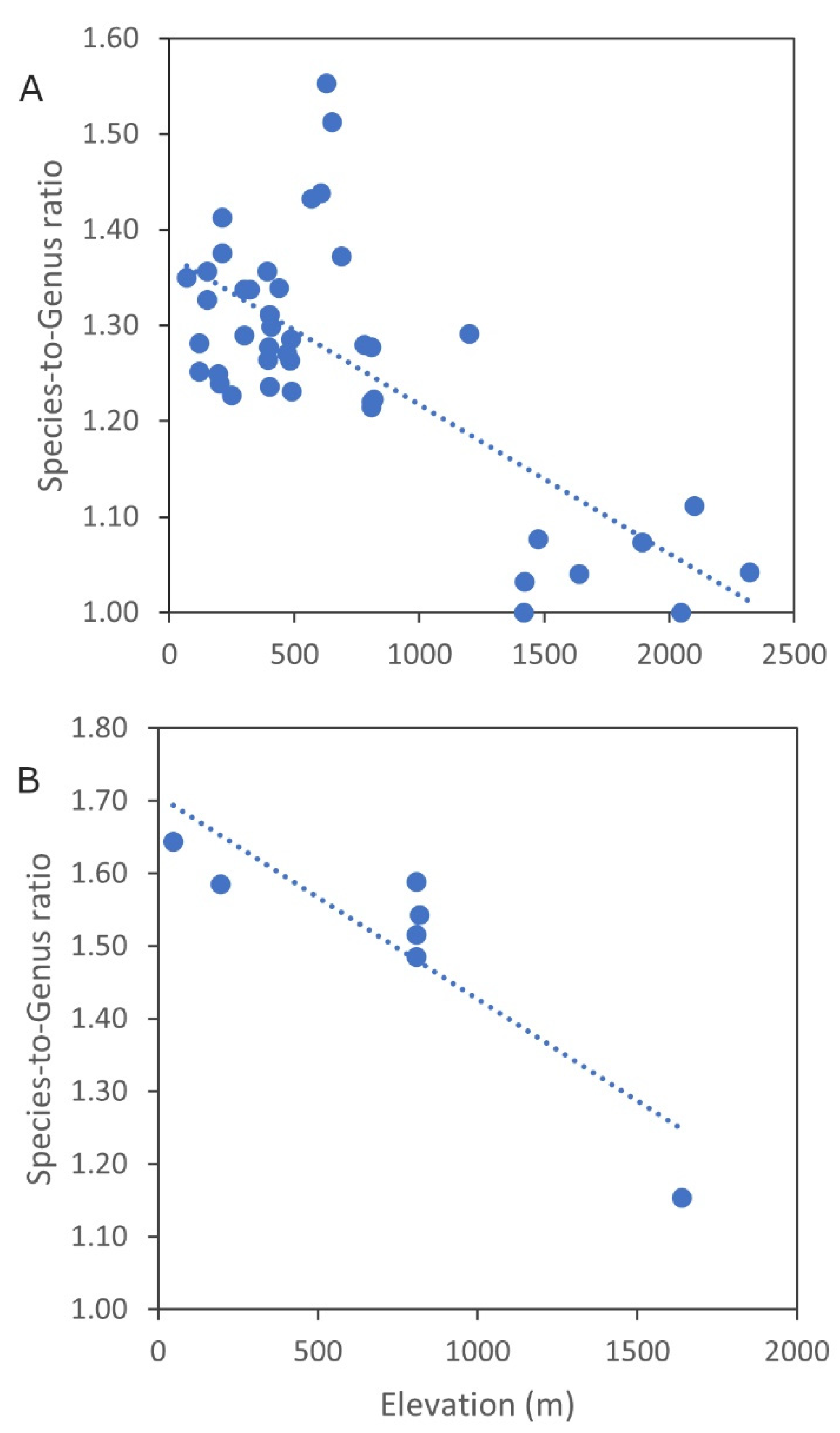
4.3. The Efficiency of DNA Barcoding in the Context of the Afromontane Flora
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Site | Country | Elevation (m) | S | G | S/G | Source |
|---|---|---|---|---|---|---|
| Bwindi-1 | Burundi | 1474 | 42 | 39 | 1.08 | TEAM Network |
| Bwindi-2 | Burundi | 1419 | 28 | 28 | 1.00 | TEAM Network |
| Bwindi-3 | Burundi | 1893 | 44 | 41 | 1.07 | TEAM Network |
| Bwindi-4 | Burundi | 2049 | 27 | 27 | 1.00 | TEAM Network |
| Bwindi-5 | Burundi | 2101 | 30 | 27 | 1.11 | TEAM Network |
| Bwindi-6 | Burundi | 2321 | 25 | 24 | 1.04 | TEAM Network |
| Bidjouka-1 | Cameroon | 392 | 99 | 73 | 1.36 | [42] |
| Bidjouka-2 | Cameroon | 605 | 105 | 73 | 1.44 | [42] |
| Korup 50-ha * | Cameroon | 195 | 87.2 | 48.82 | 1.79 | [33] |
| Lambi-1 | Cameroon | 396 | 106 | 83 | 1.28 | [42] |
| Lambi-2 | Cameroon | 627 | 118 | 76 | 1.55 | [42] |
| Ngovayang-1 | Cameroon | 650 | 121 | 80 | 1.51 | [42] |
| Rumpi-hills-11 | Cameroon | 1450 | 32 | 31 | 1.03 | [43] |
| Takamanda-10 | Cameroon | 210 | 108 | 78.5 | 1.38 | [44] |
| Takamanda-11 | Cameroon | 210 | 113 | 80 | 1.41 | [44] |
| Takamanda-12 | Cameroon | 150 | 105.5 | 79.5 | 1.33 | [44] |
| Takamanda-13 | Cameroon | 150 | 118 | 87 | 1.36 | [44] |
| Takamanda-14 | Cameroon | 120 | 87 | 69.5 | 1.25 | [44] |
| Takamanda-15 | Cameroon | 120 | 91 | 71 | 1.28 | [44] |
| Takamanda-6 | Cameroon | 320 | 103 | 77 | 1.34 | [44] |
| Takamanda-7 | Cameroon | 400 | 97 | 74 | 1.31 | [44] |
| Takamanda-8 | Cameroon | 780 | 64 | 50 | 1.28 | [44] |
| Takamanda-9 | Cameroon | 1200 | 71 | 55 | 1.29 | [44] |
| Dzanga-Sanga-1 | Central African Republic | 471 | 108 | 85 | 1.27 | [45] |
| Dzanga-Sanga-2 | Central African Republic | 482 | 120 | 95 | 1.26 | [45] |
| Dzanga-Sanga-3 | Central African Republic | 393 | 67 | 53 | 1.26 | [45] |
| Dzanga-Sanga-4 | Central African Republic | 489 | 96 | 78 | 1.23 | [45] |
| Dzanga-Sanga-5 | Central African Republic | 485 | 108 | 84 | 1.29 | [45] |
| Edoro-1 (10-ha) * | DR Congo | 808 | 65.4 | 53.6 | 1.22 | [46] |
| Edoro-2 (10-ha) * | DR Congo | 809 | 67.4 | 55.5 | 1.21 | [46] |
| Lenda-1 (10-ha) * | DR Congo | 808 | 60.4 | 47.3 | 1.28 | [46] |
| Lenda-2 (10-ha) * | DR Congo | 819 | 49.9 | 40.8 | 1.22 | [46] |
| Monts de Cristal-1 | Gabon | 400 | 89 | 72 | 1.24 | [47] |
| Monts de Cristal-2 | Gabon | 300 | 89 | 69 | 1.29 | [47] |
| Monts de Cristal-3 | Gabon | 300 | 99 | 74 | 1.34 | [47] |
| Monts de Cristal-4 | Gabon | 200 | 88 | 71 | 1.24 | [47] |
| Monts de Cristal-5 | Gabon | 250 | 108 | 88 | 1.23 | [47] |
| Rabi 25-ha * | Gabon | 47 | 84.6 | 62.68 | 1.35 | [38] |
| Waka-10 | Gabon | 569 | 106 | 74 | 1.43 | [48] |
| Waka-6 | Gabon | 438 | 83 | 62 | 1.34 | [48] |
| Waka-7 | Gabon | 407 | 100 | 77 | 1.30 | [48] |
| Waka-8 | Gabon | 687 | 107 | 78 | 1.37 | [48] |
| Ngel Nyaki (20.28 ha) * | Nigeria | 1639 | 41.1 | 39.5 | 1.04 | [19] |
References
- Mittermeier, R.A.; Mittermeier, C.G.; Brooks, T.M.; Pilgrim, J.D.; Konstant, W.R.; da Fonseca, G.A.B.; Kormos, C. Wilderness and Biodiversity Conservation. Proc. Natl. Acad. Sci. USA 2003, 100, 10309–10313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shapiro, A.C.; Grantham, H.S.; Aguilar-Amuchastegui, N.; Murray, N.J.; Gond, V.; Bonfils, D.; Rickenbach, O. Forest Condition in the Congo Basin for the Assessment of Ecosystem Conservation Status. Ecol. Indic. 2021, 122, 107268. [Google Scholar] [CrossRef]
- Rudel, T.K. The National Determinants of Deforestation in Sub-Saharan Africa. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20120405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, S.L.; Sonké, B.; Sunderland, T.; Begne, S.K.; Lopez-Gonzalez, G.; van der Heijden, G.M.F.; Phillips, O.L.; Affum-Baffoe, K.; Baker, T.R.; Banin, L.; et al. Above-Ground Biomass and Structure of 260 African Tropical Forests. Phil. Trans. R. Soc. B 2013, 368, 20120295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malhi, Y.; Adu-Bredu, S.; Asare, R.A.; Lewis, S.L.; Mayaux, P. African Rainforests: Past, Present and Future. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20120312. [Google Scholar] [CrossRef] [Green Version]
- Brooks, T.M.; Mittermeier, R.A.; da Fonseca, G.A.B.; Gerlach, J.; Hoffmann, M.; Lamoreux, J.F.; Mittermeier, C.G.; Pilgrim, J.D.; Rodrigues, A.S.L. Global Biodiversity Conservation Priorities. Science 2006, 313, 58–61. [Google Scholar] [CrossRef] [Green Version]
- Condit, R. Tropical Forest Census Plots: Methods and Results from Barro Colorado Island, Panama and a Comparison with Other Plots; Springer: Berlin/Heidelberg, Germany, 1998; ISBN 978-3-540-64144-5. [Google Scholar]
- Gonzalez, M.A.; Baraloto, C.; Engel, J.; Mori, S.A.; Pétronelli, P.; Riéra, B.; Roger, A.; Thébaud, C.; Chave, J. Identification of Amazonian Trees with DNA Barcodes. PLoS ONE 2009, 4, e7483. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological Identifications through DNA Barcodes. Proc. R. Soc. Lond. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [Green Version]
- Taylor, H.R.; Harris, W.E. An Emergent Science on the Brink of Irrelevance: A Review of the Past 8 Years of DNA Barcoding. Mol. Ecol. Resour. 2012, 12, 377–388. [Google Scholar] [CrossRef]
- CBOL Plant Working Group; Hollingsworth, P.M.; Forrest, L.L.; Spouge, J.L.; Hajibabaei, M.; Ratnasingham, S.; van der Bank, M.; Chase, M.W.; Cowan, R.S.; Erickson, D.L.; et al. A DNA Barcode for Land Plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef] [Green Version]
- Kress, W.J.; Erickson, D.L. A Two-Locus Global DNA Barcode for Land Plants: The Coding RbcL Gene Complements the Non-Coding TrnH-PsbA Spacer Region. PLoS ONE 2007, 2, e508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gill, B.A.; Musili, P.M.; Kurukura, S.; Hassan, A.A.; Goheen, J.R.; Kress, W.J.; Kuzmina, M.; Pringle, R.M.; Kartzinel, T.R. Plant DNA-Barcode Library and Community Phylogeny for a Semi-Arid East African Savanna. Mol. Ecol. Resour. 2019, 19, 838–846. [Google Scholar] [CrossRef] [PubMed]
- Bezeng, B.S.; Davies, T.J.; Daru, B.H.; Kabongo, R.M.; Maurin, O.; Yessoufou, K.; van der Bank, H.; van der Bank, M. Ten Years of Barcoding at the African Centre for DNA Barcoding. Genome 2017, 60, 629–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boatwright, J.S.; Maurin, O.; van der Bank, M. Phylogenetic Position of Madagascan Species of Acacia s.l. and New Combinations in Senegalia and Vachellia (Fabaceae, Mimosoideae, Acacieae). Bot. J. Linn. Soc. 2015, 179, 288–294. [Google Scholar] [CrossRef] [Green Version]
- Gere, J.; Yessoufou, K.; Daru, B.H.; Mankga, L.T.; Maurin, O.; van der Bank, M. Incorporating TrnH-PsbA to the Core DNA Barcodes Improves Significantly Species Discrimination within Southern African Combretaceae. Zookeys 2013, 365, 129–147. [Google Scholar] [CrossRef]
- Daru, B.H.; Manning, J.C.; Boatwright, J.S.; Maurin, O.; Maclean, N.; Schaefer, H.; Kuzmina, M.; van der Bank, M. Molecular and Morphological Analysis of Subfamily Alooideae (Asphodelaceae) and the Inclusion of Chortolirion in Aloe. Taxon 2013, 62, 62–76. [Google Scholar] [CrossRef] [Green Version]
- Parmentier, I.; Duminil, J.; Kuzmina, M.; Philippe, M.; Thomas, D.W.; Kenfack, D.; Chuyong, G.B.; Cruaud, C.; Hardy, O.J. How Effective Are DNA Barcodes in the Identification of African Rainforest Trees? PLoS ONE 2013, 8, e54921. [Google Scholar] [CrossRef]
- Abiem, I.; Arellano, G.; Kenfack, D.; Chapman, H. Afromontane Forest Diversity and the Role of Grassland-Forest Transition in Tree Species Distribution. Diversity 2020, 12, 30. [Google Scholar] [CrossRef] [Green Version]
- Davies, S.J.; Abiem, I.; Abu Salim, K.; Aguilar, S.; Allen, D.; Alonso, A.; Anderson-Teixeira, K.; Andrade, A.; Arellano, G.; Ashton, P.S.; et al. ForestGEO: Understanding Forest Diversity and Dynamics through a Global Observatory Network. Biol. Conserv. 2021, 253, 108907. [Google Scholar] [CrossRef]
- Chapman, J.D.; Chapman, H.M. The Forests of Taraba and Adamawa States, Nigeria: An Ecological Account and Plant Species Checklist; University of Canterbury: Christchurch, New Zealand, 2001. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Newmaster, S.G.; Fazekas, A.J.; Ragupathy, S. DNA Barcoding in Land Plants: Evaluation of RbcL in a Multigene Tiered Approach. Can. J. Bot. 2006, 84, 335–341. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Xue, J.-H.; Zhou, S.-L. New Universal MatK Primers for DNA Barcoding Angiosperms. J. Syst. Evol. 2011, 49, 176–181. [Google Scholar] [CrossRef]
- Kress, W.J.; Erickson, D.L.; Jones, F.A.; Swenson, N.G.; Perez, R.; Sanjur, O.; Bermingham, E. Plant DNA Barcodes and a Community Phylogeny of a Tropical Forest Dynamics Plot in Panama. Proc. Natl. Acad. Sci. USA 2009, 106, 18621–18626. [Google Scholar] [CrossRef] [Green Version]
- Amandita, F.Y.; Rembold, K.; Vornam, B.; Rahayu, S.; Siregar, I.Z.; Kreft, H.; Finkeldey, R. DNA Barcoding of Flowering Plants in Sumatra, Indonesia. Ecol. Evol. 2019, 9, 1858–1868. [Google Scholar] [CrossRef] [Green Version]
- Hollingsworth, P.M.; Graham, S.W.; Little, D.P. Choosing and Using a Plant DNA Barcode. PLoS ONE 2011, 6, e19254. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Ci, X.; Conran, J.G.; Li, J. Application of DNA Barcodes in Asian Tropical Trees—A Case Study from Xishuangbanna Nature Reserve, Southwest China. PLoS ONE 2015, 10, e0129295. [Google Scholar] [CrossRef] [Green Version]
- Newmaster, S.G.; Ragupathy, S.; Janovec, J. A Botanical Renaissance: State-of-the-Art DNA Bar Coding Facilitates an Automated Identification Technology System for Plants. Int. J. Comput. Appl. Technol. 2009, 35, 50–60. [Google Scholar] [CrossRef] [Green Version]
- Kress, W.J.; Wurdack, K.J.; Zimmer, E.A.; Weigt, L.A.; Janzen, D.H. Use of DNA Barcodes to Identify Flowering Plants. Proc. Natl. Acad. Sci. USA 2005, 102, 8369–8374. [Google Scholar] [CrossRef] [Green Version]
- Kenfack, D.; Thomas, D.W.; Chuyong, G.; Condit, R. Rarity and Abundance in a Diverse African Forest. Biodivers. Conserv. 2007, 16, 2045–2074. [Google Scholar] [CrossRef]
- Mensah, S.; Egeru, A.; Assogbadjo, A.E.; Glèlè Kakaï, R. Vegetation Structure, Dominance Patterns and Height Growth in an Afromontane Forest, Southern Africa. J. For. Res. 2020, 31, 453–462. [Google Scholar] [CrossRef]
- Gebeyehu, G.; Soromessa, T.; Bekele, T.; Teketay, D. Species Composition, Stand Structure, and Regeneration Status of Tree Species in Dry Afromontane Forests of Awi Zone, Northwestern Ethiopia. Ecosyst. Health Sustain. 2019, 5, 199–215. [Google Scholar] [CrossRef] [Green Version]
- Pitman, N.C.A.; Terborgh, J.W.; Silman, M.R.; Núñez, V.P.; Neill, D.A.; Cerón, C.E.; Palacios, W.A.; Aulestia, M. A Comparison of Tree Species Diversity in Two Upper Amazonian Forests. Ecology 2002, 83, 3210–3224. [Google Scholar] [CrossRef]
- LaFrankie, J.V.; Ashton, P.S.; Chuyong, G.B.; Co, L.; Condit, R.; Davies, S.J.; Foster, R.; Hubbell, S.P.; Kenfack, D.; Lagunzad, D.; et al. Contrasting Structure and Composition of the Understory in Species-Rich Tropical Rain Forests. Ecology 2006, 87, 2298–2305. [Google Scholar] [CrossRef] [Green Version]
- Memiaghe, H.R.; Lutz, J.A.; Korte, L.; Alonso, A.; Kenfack, D. Ecological Importance of Small-Diameter Trees to the Structure, Diversity and Biomass of a Tropical Evergreen Forest at Rabi, Gabon. PLoS ONE 2016, 11, e0154988. [Google Scholar] [CrossRef]
- Bhat, J.A.; Kumar, M.; Negi, A.K.; Todaria, N.P.; Malik, Z.A.; Pala, N.A.; Kumar, A.; Shukla, G. Species Diversity of Woody Vegetation along Altitudinal Gradient of the Western Himalayas. Glob. Ecol. Conserv. 2020, 24, e01302. [Google Scholar] [CrossRef]
- Song, X.; Cao, M.; Li, J.; Kitching, R.L.; Nakamura, A.; Laidlaw, M.J.; Tang, Y.; Sun, Z.; Zhang, W.; Yang, J. Different Environmental Factors Drive Tree Species Diversity along Elevation Gradients in Three Climatic Zones in Yunnan, Southern China. Plant Divers. 2021, 43, 433–443. [Google Scholar] [CrossRef]
- Jaccard, P. The Distribution of the Flora in the Alpine Zone. New Phytol. 1912, 11, 37–50. [Google Scholar] [CrossRef]
- Gonmadje, C.F.; Doumenge, C.; McKey, D.; Tchouto, G.P.M.; Sunderland, T.C.H.; Balinga, M.P.B.; Sonké, B. Tree Diversity and Conservation Value of Ngovayang’s Lowland Forests, Cameroon. Biodivers. Conserv. 2011, 20, 2627–2648. [Google Scholar] [CrossRef]
- Sainge, M.N.; Lyonga, N.M.; Mbatchou, G.P.T.; Kenfack, D.; Nchu, F.; Peterson, A.T. Vegetation, Floristic Composition and Structure of a Tropical Montane Forest in Cameroon. Bothalia 2019, 49, 12. [Google Scholar] [CrossRef]
- Comiskey, J.A.; Sunderland, T.C.; Sunderland-Groves, J.L. Takamanda: The Biodiversity of an African Rainforest; Smithsonian Institution, SI/MAB Biodiversity Program: Washington, DC, USA, 2003; ISBN 1-893912-12-4. [Google Scholar]
- Balinga, M.; Moses, S.; Fombod, E.; Sunderland, T.C.; Chantal, S.; Asaha, S. A Preliminary Assessment of the Vegetation of the Dzanga Sangha Protected Area Complex, Central African Republic; WWF: Gland, Switzerland; Smithsonian Institution: Washington, DC, USA; Forests, Resources and People: Limbe, Cameroon; CARPE: Kinshasa, Republic of the Congo, 2006; 124p. [Google Scholar]
- Makana, J.R.; Terese, B.H.; Hibbs, D.E.; Condit, R.; Losos, E.; Leigh, E. Stand Structure and Species Diversity in the Ituri Forest Dynamics Plots: A Comparison of Monodominant and Mixed Forest Stands. Trop. For. Divers. Dynamism 2004, 159–174. [Google Scholar]
- Sunderland, T.; Walters, G.; Issembe, Y. A Preliminary Vegetation Assessment of the Mbé National Park, Monts de Cristal, Gabon. 2004. 47p. Available online: https://carpe.umd.edu/sites/default/files/documentsarchive/Monts_de_Cristal_final_report.pdf (accessed on 10 February 2022).
- Balinga, M.P.B.; Sunderland, T.; Walters, G.; Issembe, Y.; Asaha, S.; Fombod, E. A Vegetation Assessment of the Waka National Park, Gabon. CARPE Report; Wildlife Conservation Society: Libreville, Gabon, 2006; 155p, Available online: https://carpe.umd.edu/sites/default/files/documentsarchive/SI_WakaNPGabon_Rpt_Nov2006.pdf (accessed on 10 February 2022).
| matK | rbcLa | trnH-psbA | |
|---|---|---|---|
| Number of individuals tested | 274 | 274 | 274 |
| Sequencing success: N ind. (% ind.) | 216 (78.9) | 267 (97.5) | 261 (95.3) |
| Sequencing success: N sp. (% sp.) | 94 (93.1) | 101 (100) | 101 (100) |
| N sp. with sequences ≥ 2 samples | 71 (70.3) | 88 (87.2) | 85 (84.2) |
| Barcoding Accuracy | Query Samples | |||||
|---|---|---|---|---|---|---|
| Correct ID | Multiple ID | Wrong ID | N. ind. | N. sp. | N. Gen. | |
| Species identification | ||||||
| rbcLa | 93.8 | 6.15 | 0 | 244 | 92 | 92 |
| matK | 98.3 | 1.1 | 0.55 | 181 | 67 | 59 |
| matK + rbcLa | 98.9 | 0.5 | 0.54 | 186 | 72 | 63 |
| Genus identification | ||||||
| rbcLa | 98.4 | 1.6 | 0 | 244 | 92 | 92 |
| matK | 100 | 0 | 0 | 181 | 67 | 59 |
| matK + rbcLa | 100 | 0 | 0 | 186 | 72 | 63 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kenfack, D.; Abiem, I.; Chapman, H. The Efficiency of DNA Barcoding in the Identification of Afromontane Forest Tree Species. Diversity 2022, 14, 233. https://doi.org/10.3390/d14040233
Kenfack D, Abiem I, Chapman H. The Efficiency of DNA Barcoding in the Identification of Afromontane Forest Tree Species. Diversity. 2022; 14(4):233. https://doi.org/10.3390/d14040233
Chicago/Turabian StyleKenfack, David, Iveren Abiem, and Hazel Chapman. 2022. "The Efficiency of DNA Barcoding in the Identification of Afromontane Forest Tree Species" Diversity 14, no. 4: 233. https://doi.org/10.3390/d14040233
APA StyleKenfack, D., Abiem, I., & Chapman, H. (2022). The Efficiency of DNA Barcoding in the Identification of Afromontane Forest Tree Species. Diversity, 14(4), 233. https://doi.org/10.3390/d14040233







