Gradients in the Diversity of Plants and Large Herbivores Revealed with DNA Barcoding in a Semi-Arid African Savanna
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Location
2.2. Woody Plant Distribution and DNA Barcoding
2.3. Large Mammalian Herbivore Community Data
2.4. Community Diversity Indices
2.5. Hypothesis Testing
3. Results
3.1. Ecological Characteristics of the Plot
3.2. Hypothesis Testing
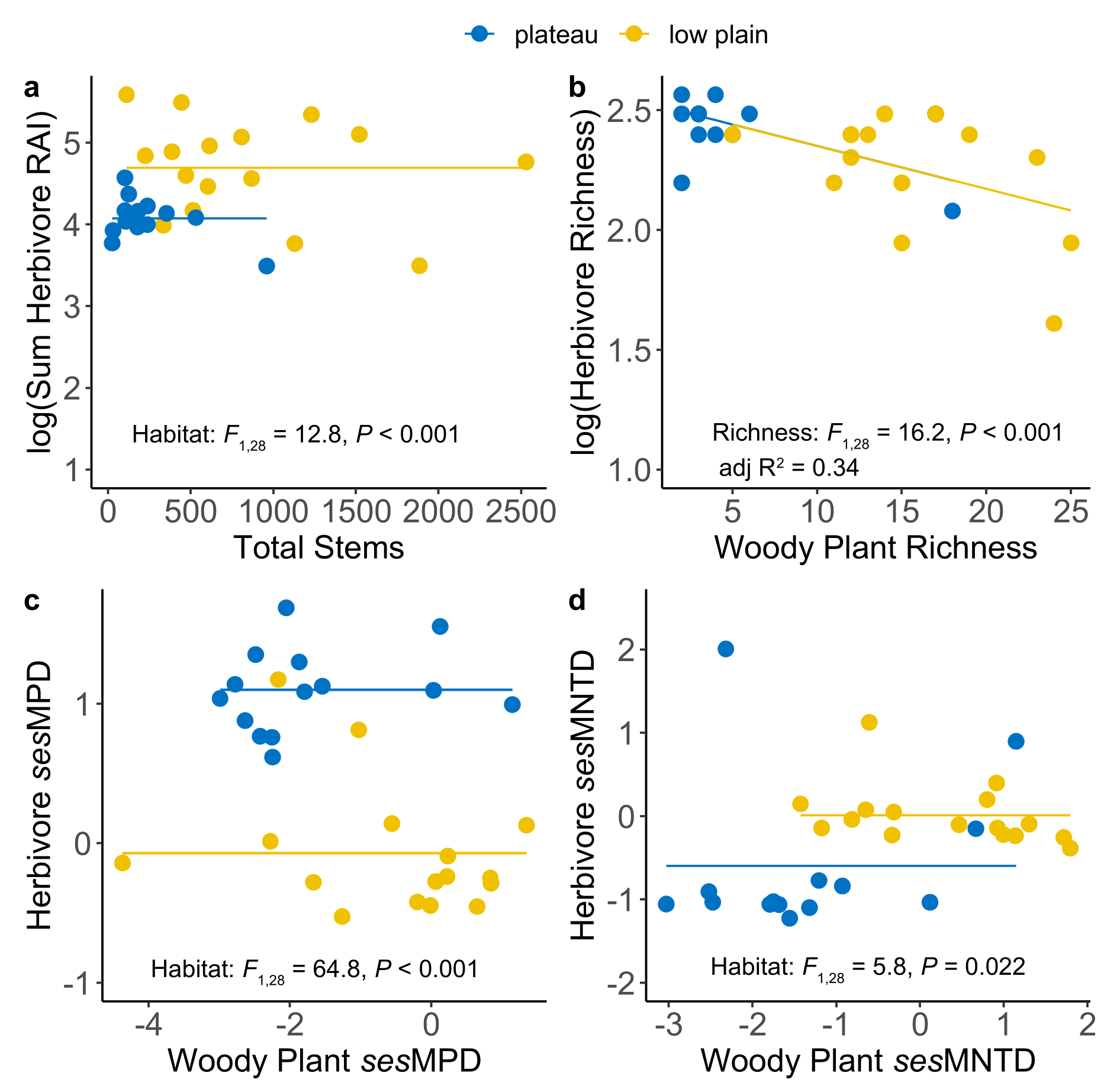
3.2.1. Hypothesis 1: Dense Habitats Should Have High Plant and Animal Diversity
3.2.2. Hypothesis 2: Herbivore and Plant Diversity Should Be Positively Correlated
3.2.3. Hypothesis 3: Spatial Links in the Composition of Plant and Animal Communities
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Galetti, M.; Moleón, M.; Jordano, P.; Pires, M.M.; Guimarães, P.R.; Pape, T.; Nichols, E.; Hansen, D.; Olesen, J.M.; Munk, M.; et al. Ecological and evolutionary legacy of megafauna extinctions. Biol. Rev. Camb. Philos. Soc. 2018, 93, 845–862. [Google Scholar] [CrossRef] [PubMed]
- Ripple, W.J.; Newsome, T.M.; Wolf, C.; Dirzo, R.; Everatt, K.T.; Galetti, M.; Hayward, M.W.; Kerley, G.I.H.; Levi, T.; Lindsey, P.A.; et al. Collapse of the world’s largest herbivores. Sci. Adv. 2015, 1, e1400103. [Google Scholar] [CrossRef] [Green Version]
- Estes, J.A.; Terborgh, J.; Brashares, J.S.; Power, M.E.; Berger, J.; Bond, W.J.; Carpenter, S.R.; Essington, T.E.; Holt, R.D.; Jackson, J.B.C.; et al. Trophic downgrading of planet earth. Science 2011, 333, 301–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ford, A.T.; Goheen, J.R.; Otieno, T.O.; Bidner, L.; Isbell, L.A.; Palmer, T.M.; Ward, D.; Woodroffe, R.; Pringle, R.M. Large carnivores make savanna tree communities less thorny. Science 2014, 346, 346–349. [Google Scholar] [CrossRef] [PubMed]
- Owen-Smith, N.; Roux, E.L.; Macandza, V. Are relatively rare antelope narrowly selective feeders? A sable antelope and zebra comparison. J. Zool. 2013, 291, 163–170. [Google Scholar] [CrossRef]
- Riginos, C. Climate and the landscape of fear in an African savanna. J. Anim. Ecol. 2015, 84, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Thouless, C.R. Long distance movements of elephants in northern Kenya. Afr. J. Ecol. 1995, 33, 321–334. [Google Scholar] [CrossRef]
- Loreau, M.; Naeem, S.; Inchausti, P.; Bengtsson, J.; Grime, J.P.; Hector, A.; Hooper, D.U.; Huston, M.A.; Raffaelli, D.; Schmid, B.; et al. Biodiversity and ecosystem functioning: Current knowledge and future challenges. Science 2001, 294, 804–808. [Google Scholar] [CrossRef] [Green Version]
- Mittelbach, G.G.; Schemske, D.W.; Cornell, H.V.; Allen, A.P.; Brown, J.M.; Bush, M.B.; Harrison, S.P.; Hurlbert, A.H.; Knowlton, N.; Lessios, H.A.; et al. Evolution and the latitudinal diversity gradient: Speciation, extinction and biogeography. Ecol. Lett. 2007, 10, 315–331. [Google Scholar] [CrossRef]
- Thompson, J.N. Evolutionary ecology and the conservation of biodiversity. Trends Ecol. Evol. 1996, 11, 300–303. [Google Scholar] [CrossRef]
- Wootton, J.T. Effects of disturbance on species diversity: A multitrophic perspective. Am. Nat. 1998, 152, 803–825. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Qian, H.; Girardello, M.; Pellissier, V.; Nielsen, S.E.; Svenning, J.-C. Trophic interactions among vertebrate guilds and plants shape global patterns in species diversity. Proc. R. Soc. B 2018, 285, 20180949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cromsigt, J.P.G.M.; Prins, H.H.T.; Olff, H. Habitat heterogeneity as a driver of ungulate diversity and distribution patterns: Interaction of body mass and digestive strategy. Divers. Distrib. 2009, 15, 513–522. [Google Scholar] [CrossRef] [Green Version]
- Jetz, W.; Kreft, H.; Ceballos, G.; Mutke, J. Global associations between terrestrial producer and vertebrate consumer diversity. Proc. R. Soc. B 2009, 276, 269–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kartzinel, T.R.; Chen, P.A.; Coverdale, T.C.; Erickson, D.L.; Kress, W.J.; Kuzmina, M.L.; Rubenstein, D.I.; Wang, W.; Pringle, R.M. DNA metabarcoding illuminates dietary niche partitioning by African large herbivores. Proc. Nat. Acad. Sci. USA 2015, 112, 8019–8024. [Google Scholar] [CrossRef] [Green Version]
- Haddad, N.M.; Crutsinger, G.M.; Gross, K.; Haarstad, J.; Knops, J.M.H.; Tilman, D. Plant species loss decreases arthropod diversity and shifts trophic structure. Ecol. Lett. 2009, 12, 1029–1039. [Google Scholar] [CrossRef]
- Hutchinson, G.E. Homage to Santa Rosalia or why are there so many kinds of animals? Am. Nat. 1959, 93, 145–159. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, D.; Lawton, J. Why more productive sites have more species: An experimental test of theory using tree-hold communities. Am. Nat. 1998, 152, 510–529. [Google Scholar] [CrossRef]
- Kartzinel, T.R.; Pringle, R.M. Multiple dimensions of dietary diversity in large mammalian herbivores. J. Anim. Ecol. 2020, 89, 1482–1496. [Google Scholar] [CrossRef]
- Swenson, N.G.; Stegen, J.C.; Davies, S.J.; Erickson, D.L.; Forero-Montaña, J.; Hurlbert, A.H.; Kress, W.J.; Thompson, J.; Uriarte, M.; Wright, S.J.; et al. Temporal turnover in the composition of tropical tree communities: Functional determinism and phylogenetic stochasticity. Ecology 2012, 93, 490–499. [Google Scholar] [CrossRef]
- Cavender-Bares, J.; Kozak, K.H.; Fine, P.V.; Kembel, S.W. The merging of community ecology and phylogenetic biology. Ecology Lett. 2009, 12, 693–715. [Google Scholar] [CrossRef] [PubMed]
- Mutuku, P.M.; Kenfack, D. Effect of local topographic heterogeneity on tree species assembly in an Acacia-dominated African savanna. J. Trop. Ecol. 2019, 35, 46–56. [Google Scholar] [CrossRef]
- Gill, B.A.; Musili, P.M.; Kurukura, S.; Hassan, A.A.; Goheen, J.R.; Kress, W.J.; Kuzmina, M.; Pringle, R.M.; Kartzinel, T.R. Plant DNA-barcode library and community phylogeny for a semi-arid East African savanna. Mol. Ecol. Resour. 2019, 19, 838–846. [Google Scholar] [CrossRef] [PubMed]
- Lepore, M.; Arellano, G.; Condit, R.; Detto, M.; Harms, K.; Lao, S.; Russo, S.; Zuleta, D. fgeo.analyze: Analyze ForestGEO Data, R Package Version 1.1.14. 2020. Available online: https://CRAN.R-project.org/package=fgeo.analyze (accessed on 12 December 2021).
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Hopcraft, J.G.C.; Anderson, T.M.; Pérez-Vila, S.; Mayeba, E.; Olff, H. Body size and the division of niche space: Food and predation differentially shape the distribution of Serengeti grazers. J. Anim. Ecol. 2012, 81, 201–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metcalfe, P.; Beven, K.; Freer, J. Dynamic TOPMODEL: A new implementation in R and its sensitivity to time and space steps. Environ. Model. Softw. 2015, 72, 155–172. [Google Scholar] [CrossRef] [Green Version]
- Kress, W.J.; Erickson, D.L.; Swenson, N.G.; Thompson, J.; Uriarte, M.; Zimmerman, J.K. Advances in the use of DNA barcodes to build a community phylogeny for tropical trees in a Puerto Rican forest dynamics plot. PLoS ONE 2010, 5, e15409. [Google Scholar] [CrossRef] [Green Version]
- Bolger, D.T.; Morrison, T.A.; Vance, B.; Lee, D.; Farid, H. Development and application of a computer-assisted system for photographic mark-recapture analysis. Methods Ecol. Evolut. 2012, 3, 813–822. [Google Scholar] [CrossRef]
- Palmer, M.S.; Swanson, A.; Kosmala, M.; Arnold, T.; Packer, C. Evaluating relative abundance indices for terrestrial herbivores from large-scale camera trap surveys. Afr. J. Ecol. 2018, 56, 791–803. [Google Scholar] [CrossRef] [Green Version]
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef] [Green Version]
- Mazel, F.; Davies, T.J.; Gallien, L.; Renaud, J.; Groussin, M.; Münkemüller, T.; Thuiller, W. Influence of tree shape and evolutionary time-scale on phylogenetic diversity metrics. Ecography 2016, 39, 913–920. [Google Scholar] [CrossRef] [Green Version]
- Webb, C.O.; Ackerly, D.D.; McPeek, M.A.; Donoghue, M.J. Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 2002, 31, 475–505. [Google Scholar] [CrossRef] [Green Version]
- Ives, A.R.; Helmus, M.R. Phylogenetic metrics of community similarity. Am. Nat. 2010, 176, E128–E142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riginos, C.; Grace, J.B. Savanna tree density, herbivores, and the herbaceous community: Bottom-up vs. top-down effects. Ecology 2008, 89, 2228–2238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimuyu, D.M.; Kenfack, D.; Musili, P.M.; Ang’ila, R.O. Fine-scale habitat heterogeneity influences browsing damage by elephant and giraffe. Biotropica 2021, 53, 86–96. [Google Scholar] [CrossRef]
- Colwell, R.K.; Chao, A.; Gotelli, N.J.; Lin, S.-Y.; Mao, C.X.; Chazdon, R.L.; Longino, J.T. Models and estimators linking individual-based and sample-based rarefaction, extrapolation and comparison of assemblages. J. Plant Ecol. 2012, 5, 3–21. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package; R Package Version 2.5-7:2020; R Core Team: Vienna, Austria, 2020. [Google Scholar]
- De Caceres, M.; Legendre, P. Associations between species and groups of sites: Indices and statistical inference. Ecology 2009, 90, 3566–3574. [Google Scholar] [CrossRef]
- Pellissier, L.; Ndiribe, C.; Dubuis, A.; Pradervand, J.-N.; Salamin, N.; Guisan, A.; Rasmann, S. Turnover of plant lineages shapes herbivore phylogenetic beta diversity along ecological gradients. Ecol. Lett. 2013, 16, 600–608. [Google Scholar] [CrossRef]
- Kemp, J.E.; Linder, H.P.; Ellis, A.G. Beta diversity of herbivorous insects is coupled to high species and phylogenetic turnover of plant communities across short spatial scales in the Cape Floristic Region. J. Biogeogr. 2017, 44, 1813–1823. [Google Scholar] [CrossRef]
- Dinnage, R.; Cadotte, M.W.; Haddad, N.M.; Crutsinger, G.M.; Tilman, D. Diversity of plant evolutionary lineages promotes arthropod diversity. Ecol. Lett. 2012, 15, 1308–1317. [Google Scholar] [CrossRef]
- Schuldt, A.; Ebeling, A.; Kunz, M.; Staab, M.; Guimarães-Steinicke, C.; Bachmann, D.; Buchmann, N.; Durka, W.; Fichtner, A.; Fornoff, F.; et al. Multiple plant diversity components drive consumer communities across ecosystems. Nat. Commun. 2019, 10, 1460. [Google Scholar] [CrossRef] [Green Version]
- Wells, H.B.M.; Crego, R.D.; Odepal, Ø.H.; Khasoha, L.M.; Alston, J.M.; Reed, C.G.; Weiner, S.; Kurukura, S.; Hassan, A.A.; Namoni, M.; et al. Experimental evidence that effects of megaherbivores on mesoherbivore space use are influenced by species’ traits. J. Anim. Ecol. 2021, 90, 2510–2522. [Google Scholar] [CrossRef] [PubMed]
- Hairston, N.G.; Smith, F.E.; Slobodkin, L. Community structure, population control, and competition. Am. Nat. 1960, 879, 421–425. [Google Scholar] [CrossRef]
- Polis, G.A. Why are parts of the world green? Multiple factors control productivity and the distribution of biomes. Oikos 1999, 86, 3–15. [Google Scholar] [CrossRef]
- Beschta, R.L.; Ripple, W.J. Large predators and trophic cascades in ecosystems of the western United States. Biol. Conserv. 2009, 142, 2401–2414. [Google Scholar] [CrossRef]
- Coverdale, T.C.; Kartzinel, T.R.; Grabowski, K.L.; Shriver, R.K.; Hassan, A.A.; Goheen, J.R.; Palmer, T.M.; Pringle, R.M. Elephants in the understory: Opposing direct and indirect effects of consumption and ecosystem engineering by megaherbivores. Ecology 2016, 97, 3219–3230. [Google Scholar] [CrossRef] [PubMed]
- Coverdale, T.C.; O’Connell, R.D.; Hutchinson, M.C.; Savagian, A.; Kartzinel, T.R.; Palmer, T.M.; Goheen, J.R.; Augustine, D.J.; Sankaran, M.; Tarnita, C.E.; et al. Large herbivores suppress liana infestation in an African savanna. Proc. Nat. Acad. Sci. USA 2021, 118, e2101676118. [Google Scholar] [CrossRef]
- Goheen, J.R.; Augustine, D.J.; Veblen, K.E.; Kimuyu, D.M.; Palmer, T.M.; Porensky, L.M.; Pringle, R.M.; Ratnam, J.; Riginos, C.; Sankaran, M.; et al. Conservation lessons from large-mammal manipulations in East African savannas: The KLEE, UHURU, and GLADE experiments. Ann. N. Y. Acad. Sci. 2018, 1429, 31–49. [Google Scholar] [CrossRef]
- Hattas, D.; Hjältén, J.; Julkunen-Tiitto, R.; Scogings, P.F.; Rooke, T. Differential phenolic profiles in six African savanna woody species in relation to antiherbivore defense. Phytochemistry 2011, 72, 1796–1803. [Google Scholar] [CrossRef]
- Kartzinel, T.R.; Hsing, J.C.; Musili, P.M.; Brown, B.R.P.; Pringle, R.M. Covariation of diet and gut microbiome in African megafauna. Proc. Nat. Acad. Sci. USA 2019, 116, 23588–23593. [Google Scholar] [CrossRef] [Green Version]
- Kimuyu, D.M.; Veblen, K.E.; Riginos, C.; Chira, R.M.; Githaiga, J.M.; Young, T.P. Influence of cattle on browsing and grazing wildlife varies with rainfall and presence of megaherbivores. Ecol. Appl. 2017, 27, 786–798. [Google Scholar] [CrossRef]
- MacKenzie, D.I.; Nichols, J.D.; Hines, J.E.; Knutson, M.G.; Franklin, A.B. Estimating site occupancy, colonization, and local extinction when a species is detected imperfectly. Ecology 2003, 84, 2200–2207. [Google Scholar] [CrossRef] [Green Version]
- Becker, J.A.; Hutchinson, M.C.; Potter, A.B.; Park, S.; Guyton, J.A.; Abernathy, K.; Americo, V.F.; da Conceiçāo, A.; Kartzinel, T.R.; Kuziel, L.; et al. Ecological and behavioral mechanisms of density-dependent habitat expansion in a recovering African ungulate population. Ecol. Monogr. 2021, 91, e01476. [Google Scholar] [CrossRef]
- Milchunas, D.G.; Lauenroth, W.K. Quantitative effects of grazing on vegetation and soils over a global range of environments. Ecol. Monogr. 1993, 63, 327–366. [Google Scholar] [CrossRef]
- Wood, K.A.; O’Hare, M.T.; McDonald, C.; Searle, K.R.; Daunt, F.; Stillman, R.A. Herbivore regulation of plant abundance in aquatic ecosystems. Biol. Rev. 2017, 92, 1128–1141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brooks, A.W.; Kohl, K.D.; Brucker, R.M.; van Opstal, E.J.; Bordenstein, S.R. Phylosymbiosis: Relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol. 2017, 15, e1002587. [Google Scholar] [CrossRef] [Green Version]
- Park, A.W.; Farrell, M.J.; Schmidt, J.P.; Huang, S.; Dallas, T.A.; Pappalardo, P.; Drake, J.M.; Stephens, P.R.; Poulin, R.; Nunn, C.L.; et al. Characterizing the phylogenetic specialism–generalism spectrum of mammal parasites. Proc. R. Soc. B 2018, 285, 20172613. [Google Scholar] [CrossRef] [Green Version]
- Shefferson, R.P.; Bunch, W.; Cowden, C.C.; Lee, Y.I.; Kartzinel, T.R.; Yukawa, T.; Downing, J.; Jiang, H. Does evolutionary history determine specificity in broad ecological interactions? J. Ecol. 2019, 107, 1582–1593. [Google Scholar] [CrossRef]
- Corro, E.J.; Villalobos, F.; Lira-Noriega, A.; Guevara, R.; Guimarães, P.R.; Dáttilo, W. Annual precipitation predicts the phylogenetic signal in bat-fruit interaction networks across the Neotropics. Biol. Lett. 2021, 17, 20210478. [Google Scholar] [CrossRef]
- Staab, M.; Liu, X.; Assmann, T.; Bruelheide, H.; Buscot, F.; Durka, W.; Erfmeier, A.; Klein, A.-M.; Ma, K.; Michalski, S.; et al. Tree phylogenetic diversity structures multitrophic communities. Funct. Ecol. 2020, 35, 521–534. [Google Scholar] [CrossRef]
- Simth, G.F.; Figueiredo, E. Conserving Acacia Mill. with a conserved type: What happenedin Melbourne? Taxonomy 2018, 60, 1504–1506. [Google Scholar] [CrossRef] [Green Version]
- Antonellia, A.; Nylanderb, J.A.A.; Perssona, C.; Sanmartin, I. Tracing the impact of the Andean uplift on Neotropical plant evolution. Proc. Nat. Acad. Sci. USA 2009, 106, 9749–9754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janssens, S.; Couvreur, T.L.; Mertens, A.; Dauby, G.; Dagallier, L.-P.; Abeele, S.V.; Vandelook, F.; Mascarello, M.; Beeckman, H.; Sosef, M.; et al. A large-scale species level dated angiosperm phylogeny for evolutionary and ecological analyses. Biodivers. Data J. 2020, 8, e39677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
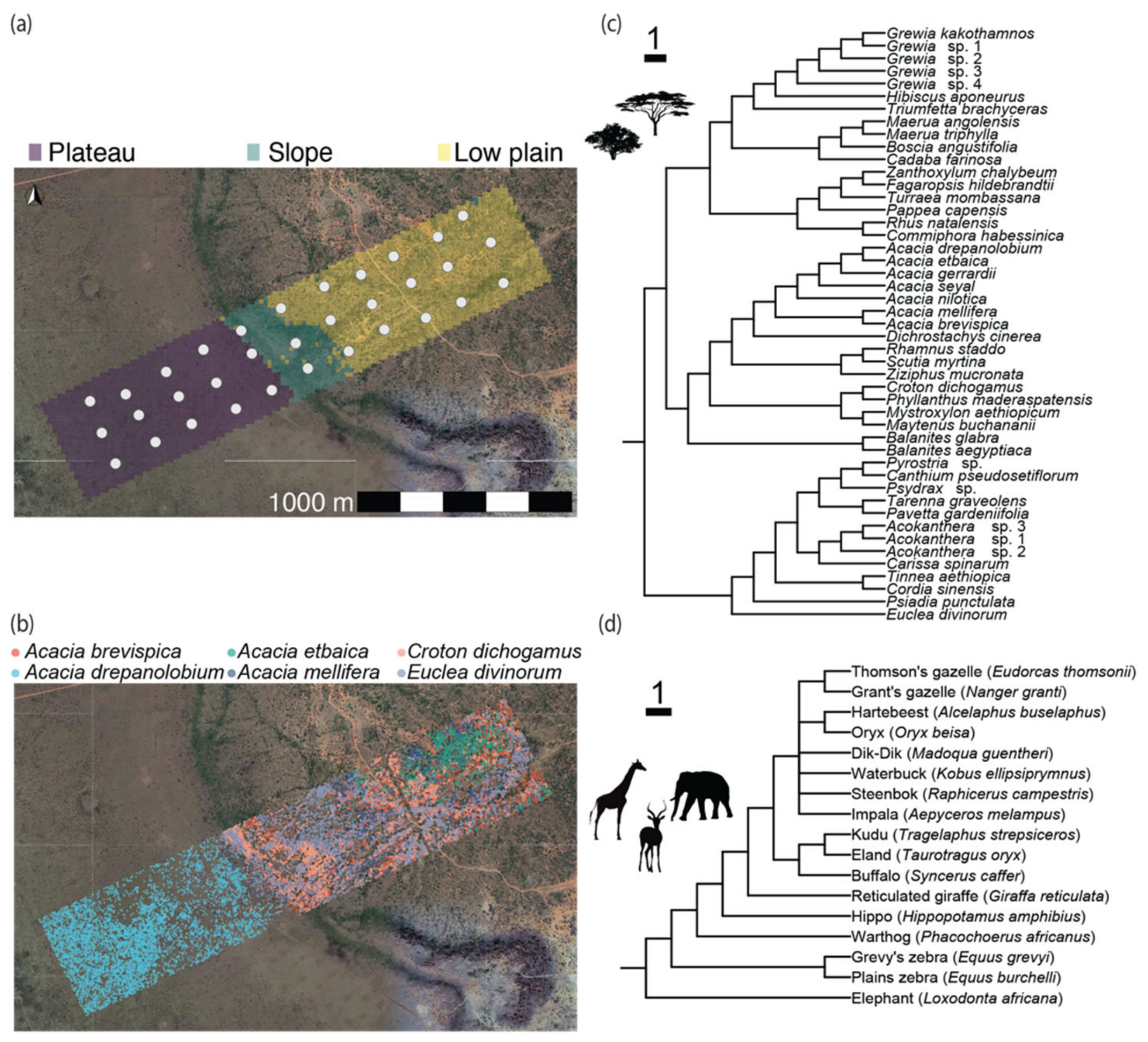


| Variable | Measure | Plateau | Slope | Low Plain |
|---|---|---|---|---|
| Abiotic characteristics | Number of sites | 14 | 3 | 16 |
| Mean elevation (m) | 1787 | 1754 | 1692 | |
| Mean convexity | 0.019 | 0.556 | −0.022 | |
| Mean slope (rad) | 1.04 | 12.30 | 4.10 | |
| Mean topographic water index (TWI) | 10.6 | 7.9 | 9.0 | |
| Woody plants | Mean total stems | 238 | 1602 | 855 |
| Mean richness | 5.4 | 25.0 | 15.4 | |
| Mean sesMPD | −1.7 | 0.9 | −0.6 | |
| Mean sesMNTD | −1.3 | 1.8 | 0.3 | |
| Large herbivores | Mean cumulative RAI | 60.4 | 49.0 | 127.0 |
| Mean richness | 11.4 | 6.3 | 9.8 | |
| Mean sesMPD | 1.1 | 0.2 | −0.1 | |
| Mean sesMPD | −0.6 | −0.3 | 0.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Freeman, P.T.; Ang’ila, R.O.; Kimuyu, D.; Musili, P.M.; Kenfack, D.; Lokeny Etelej, P.; Magid, M.; Gill, B.A.; Kartzinel, T.R. Gradients in the Diversity of Plants and Large Herbivores Revealed with DNA Barcoding in a Semi-Arid African Savanna. Diversity 2022, 14, 219. https://doi.org/10.3390/d14030219
Freeman PT, Ang’ila RO, Kimuyu D, Musili PM, Kenfack D, Lokeny Etelej P, Magid M, Gill BA, Kartzinel TR. Gradients in the Diversity of Plants and Large Herbivores Revealed with DNA Barcoding in a Semi-Arid African Savanna. Diversity. 2022; 14(3):219. https://doi.org/10.3390/d14030219
Chicago/Turabian StyleFreeman, Patrick T., Robert O. Ang’ila, Duncan Kimuyu, Paul M. Musili, David Kenfack, Peter Lokeny Etelej, Molly Magid, Brian A. Gill, and Tyler R. Kartzinel. 2022. "Gradients in the Diversity of Plants and Large Herbivores Revealed with DNA Barcoding in a Semi-Arid African Savanna" Diversity 14, no. 3: 219. https://doi.org/10.3390/d14030219
APA StyleFreeman, P. T., Ang’ila, R. O., Kimuyu, D., Musili, P. M., Kenfack, D., Lokeny Etelej, P., Magid, M., Gill, B. A., & Kartzinel, T. R. (2022). Gradients in the Diversity of Plants and Large Herbivores Revealed with DNA Barcoding in a Semi-Arid African Savanna. Diversity, 14(3), 219. https://doi.org/10.3390/d14030219







