Phylogenetics and Biogeography of Lilium ledebourii from the Hyrcanian Forest
Abstract
1. Introduction
2. Materials and Methods
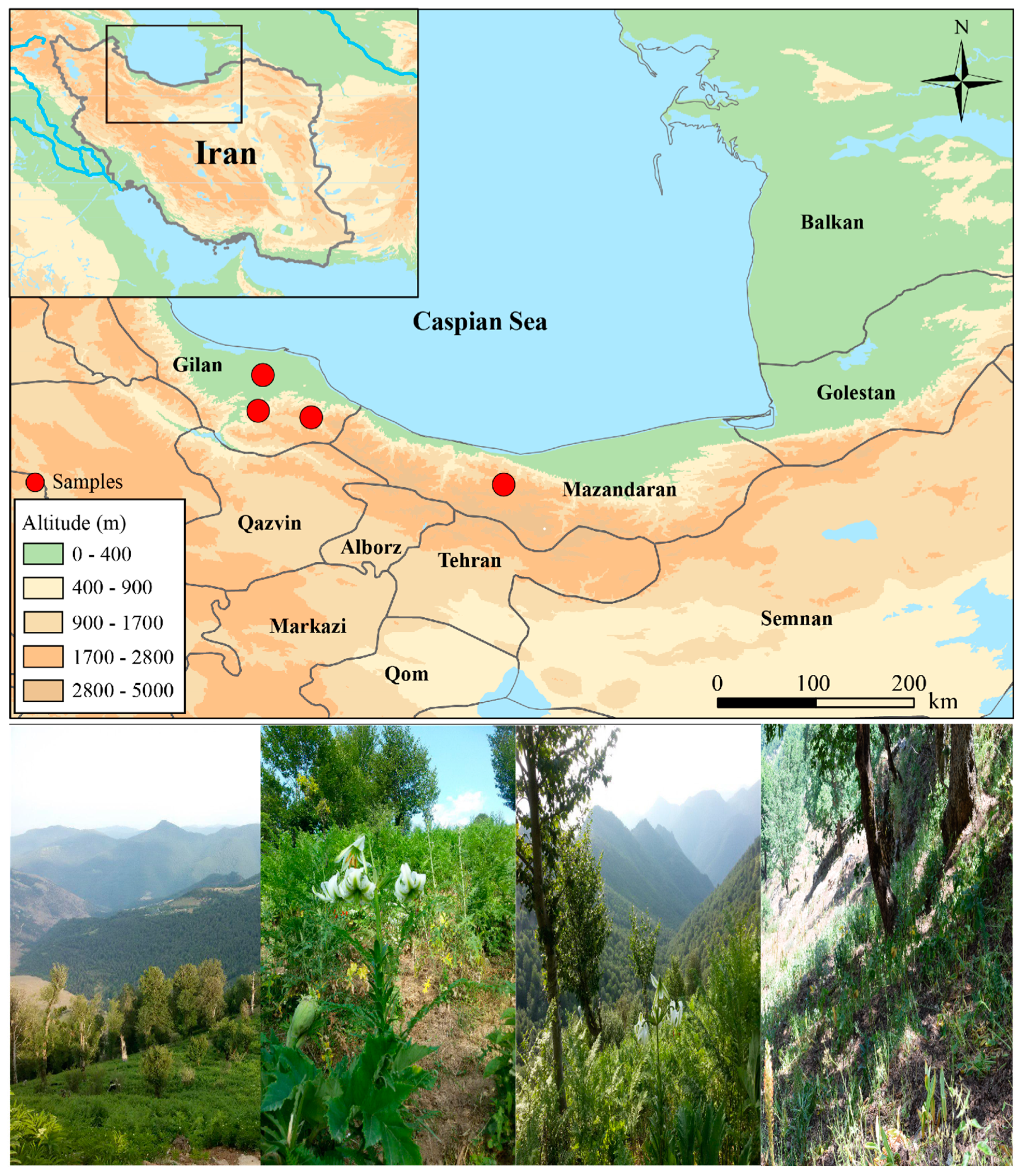
2.1. Sampling and DNA Extraction
2.2. PCR Amplification and Sequencing
2.3. Phylogenetic Analysis and ITS2 Secondary Structure
2.4. Divergence Time Estimate and Biogeographic Analysis
3. Results
3.1. ITS and Plastid Regions (MatK, TrnL-F and TrnH-PsbA)
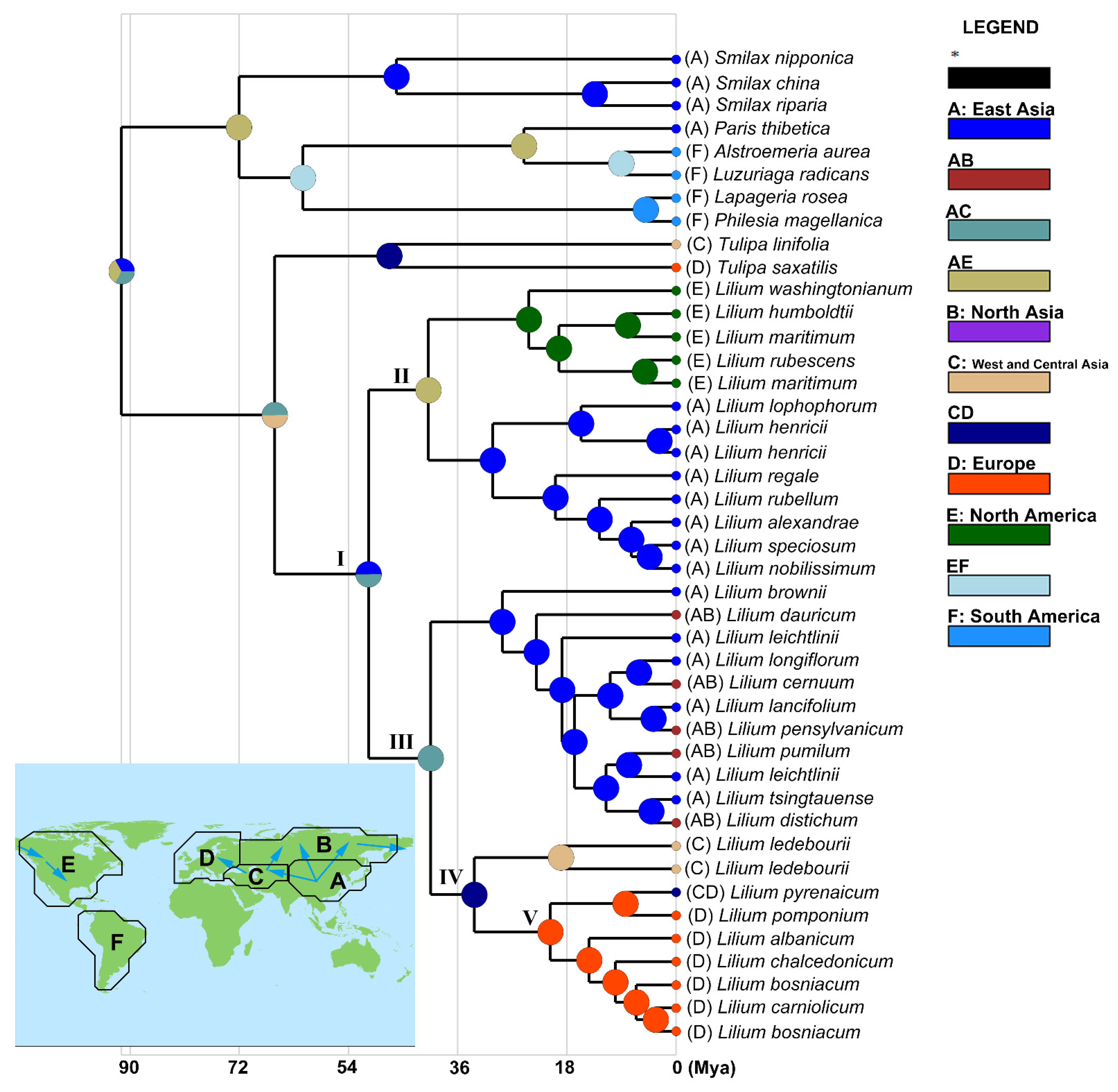
3.2. Phylogenetic Position of L. ledebourii
3.3. Biogeography of the Genus Lilium
4. Discussion
4.1. Phylogeny and Biogeography of Lilies
4.2. Biogeography of Lilium Emphasis on West Asia
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Woodcock, H.; Stearn, W.T. Lilies of the World, Their Cultivation and Classification; Country Life Limited: London, UK, 1950. [Google Scholar]
- Gong, X.; Huang, H.H.; Ting, Y.W.; Hsu, T.W.; Malikova, L.; Tran, H.T.; Huang, C.L.; Liu, S.H.; Chiang, T.Y. Frequent gene flow blurred taxonomic boundaries of sections in Lilium L. (Liliaceae). PLoS ONE 2017, 12, e0183209. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.T.; Lim, K.B.; Kim, J.S. New insights on Lilium phylogeny based on a comparative phylogenomic study using complete plastome sequences. Plants 2019, 8, 547. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Kim, S.C.; Yeau, S.H. Major lineages of the genus Lilium (Liliaceae) based on nrDNA ITS sequences, with special emphasis on the Korean species. J. Plant Biol. 2011, 54, 159–171. [Google Scholar] [CrossRef]
- Gao, Y.-D.; Harris, A.; Zhou, S.-D.; He, X.-J. Evolutionary events in Lilium (including Nomocharis, Liliaceae) are temporally correlated with orogenies of the Q–T plateau and the Hengduan Mountains. Mol. Phylogenet. Evol. 2013, 68, 443–460. [Google Scholar] [CrossRef]
- Patterson, T.B.; Givnish, T.J. Phylogeny, concerted convergence, and phylogenetic niche conservatism in the core Liliales: Insights from rbcL and ndhF sequence data. Evolution 2002, 56, 233–252. [Google Scholar] [CrossRef]
- Baranova, M. The lily species in the flora of the former Soviet Union and their classification within the genus Lilium. In International Symposium on the Genus Lilium 414; ISHS: Leuven, Belgium, 1994; pp. 133–136. [Google Scholar]
- Comber, H.F. A new classification of the genus Lilium. Lily Year Book RHS 1949, 86–105. [Google Scholar]
- Endlicher, S.L. Genera Plantarum Secundum Ordines Naturales Disposita: Genera Plantarum… 1836–1840; Beck: Munich, Germany, 1840; Volume 2. [Google Scholar]
- Ikinci, N. Molecular phylogeny and divergence times estimates of Lilium section Liriotypus (Liliaceae) based on plastid and nuclear ribosomal ITS DNA sequence data. Turk. J. Bot. 2011, 35, 319–330. [Google Scholar]
- Naqinezhad, A.; Hamzeh’ee, B.; Attar, F. Vegetation–environment relationships in the alderwood communities of Caspian lowlands, N. Iran (toward an ecological classification). Flora-Morphol. Distrib. Funct. Ecol. Plants 2008, 203, 567–577. [Google Scholar] [CrossRef]
- Miller, A.G. Hyrcanian forests, Iran and Azerbaijan. Centres of Plant Diversity, A Guide and Strategy for their Conservation Oxford. IUCN 1994, 1, 343–344. [Google Scholar]
- Olteanu, R.; Jipa, D. Dacian Basin environmental evolution during Upper Neogene within the Paratethys domain. Geo-Eco-Marina 2006, 12, 91–105. [Google Scholar]
- Tralau, H. Asiatic Dicotyledonous Affinities in the Cainozoic Flora of Europe; Almqvist & Wiksell: Tofnes, UK, 1963. [Google Scholar]
- Zohary, M. Geobotanical Foundation of the Middle East; Fischer-Verlag: Stuttgart, Germany, 1983. [Google Scholar]
- Mimaki, Y.; Satou, T.; Kuroda, M.; Sashida, Y.; Hatakeyama, Y. Steroidal saponins from the bulbs of Lilium candidum. Phytochemistry 1999, 51, 567–573. [Google Scholar] [CrossRef]
- Fataei, E. Recognition of new habitats of Lilium ledebourii Baker in Northwest Ardabil Province of Iran using GIS technology. In II International Symposium on the Genus Lilium 900; ISHS: Leuven, Belgium, 2010; pp. 65–70. [Google Scholar]
- Azadi, P.; Khosh-Khui, M. Micropropagation of Lilium ledebourii (Baker) Boiss as affected by plant growth regulator, sucrose concentration, harvesting season and cold treatments. Electron. J. Biotechnol. 2007, 10, 582–591. [Google Scholar] [CrossRef]
- Salehi, M.; Hatamzadeh, A.; Jafarian, V.; Zarre, S. New molecular record and some biochemical features of the rare plant species of Iranian lily (Lilium ledebourii Boiss.). Hortic. Environ. Biotechnol. 2019, 60, 585–593. [Google Scholar] [CrossRef]
- Ghanbari, S.; Fakheri, B.A.; Naghavi, M.R.; Mahdinezhad, N. Evaluating phylogenetic relationships in the Lilium family using the ITS marker. J. Plant Biotechnol. 2018, 45, 236–241. [Google Scholar] [CrossRef]
- Rockwell, F.F. Complete Book of Lilies; Doubleday: New York, NY, USA, 1961. [Google Scholar]
- Woodcock, H.; Stearn, W.T. Lilies of the World; Country Life Limited: London, UK, 1950. [Google Scholar]
- Vassou, S.L.; Kusuma, G.; Parani, M. DNA barcoding for species identification from dried and powdered plant parts: A case study with authentication of the raw drug market samples of Sida cordifolia. Gene 2015, 559, 86–93. [Google Scholar] [CrossRef]
- Tripathi, A.M.; Tyagi, A.; Kumar, A.; Singh, A.; Singh, S.; Chaudhary, L.B.; Roy, S. The internal transcribed spacer (ITS) region and trnhH-psbA are suitable candidate loci for DNA barcoding of tropical tree species of India. PLoS ONE 2013, 8, e57934. [Google Scholar] [CrossRef]
- Chen, C.W.; Huang, Y.M.; Kuo, L.Y.; Nguyen, Q.D.; Luu, H.T.; Callado, J.R.; Farrar, D.R.; Chiou, W.L. trnL-F is a powerful marker for DNA identification of field vittarioid gametophytes (Pteridaceae). Ann. Bot. 2013, 111, 663–673. [Google Scholar] [CrossRef]
- Janfaza, S.; Yousefzadeh, H.; Hosseini Nasr, S.M.; Botta, R.; Asadi Abkenar, A.; Torello Marinoni, D. Genetic diversity of Castanea sativa an endangered species in the Hyrcanian forest. Silva Fenn. 2017, 51, 1–15. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Johnson, L.A.; Soltis, D.E. matK DNA sequences and phylogenetic reconstruction in Saxifragaceae s. str. Syst. Bot. 1994, 143–156. [Google Scholar] [CrossRef]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Sang, T.; Crawford, D.J.; Stuessy, T.F. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 1997, 84, 1120–1136. [Google Scholar] [CrossRef] [PubMed]
- Hayashi1, K.; Kawano2, S. Molecular systematics of Lilium and allied genera (Liliaceae): Phylogenetic relationships among Lilium and related genera based on the rbcL and matK gene sequence data. Plant Species Biol. 2000, 15, 73–93. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Bandelt, H.-J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Farris, J.S.; Kallersjo, M.; Kluge, A.G.; Bult, C. Testing Significance of Incongruence; Elsevier: Amsterdam, The Netherlands, 1994. [Google Scholar]
- Swofford, D.L. PAUP: Phylogenetic analysis using parsimony version 3.11. In Computer Program Distributed by the Illinois Natural History Survey; University of Illinois at Urbana-Champaign: Champaign, IL, USA, 1993. [Google Scholar]
- Kim, J.S.; Kim, J.-H. Updated molecular phylogenetic analysis, dating and biogeographical history of the lily family (Liliaceae: Liliales). Bot. J. Linn. Soc. 2018, 187, 579–593. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Rambaut, A. Available online: FigTree.tree.bio.ed.ac.uk/software/figtree (accessed on 12 December 2014).
- Yu, Y.; Harris, A.J.; Blair, C.; He, X. RASP (Reconstruct Ancestral State in Phylogenies): A tool for historical biogeography. Mol. Phylogenet. Evol. 2015, 87, 46–49. [Google Scholar] [CrossRef]
- Baker, J.G. A new synopsis of all the known lilies. Gard. Chron. 1871, 104, 1650. [Google Scholar]
- Nishikawa, T.; Okazaki, K.; Uchino, T.; Arakawa, K.; Nagamine, T. A molecular phylogeny of Lilium in the internal transcribed spacer region of nuclear ribosomal DNA. J. Mol. Evol. 1999, 49, 238–249. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.T. The Acceleration of β-Particles in Strong Electric Fields Such as Those of Thunderclouds. Math. Proc. Camb. Philos. Soc. 1925, 22, 534–538. [Google Scholar] [CrossRef]
- McRae, K.; Boisvert, S. Automatic semantic similarity priming. J. Exp. Psychol. Learn. Mem. Cogn. 1998, 24, 558. [Google Scholar] [CrossRef]
- Leslie, A.C. The International Lily Register, Including 17 Additions (1984–1998); Royal Horticultural Society: London, UK, 1982. [Google Scholar]
- Tiffney, B.H.; Manchester, S.R. The use of geological and paleontological evidence in evaluating plant phylogeographic hypotheses in the Northern Hemisphere Tertiary. Int. J. Plant Sci. 2001, 162 (Suppl. 6), S3–S17. [Google Scholar] [CrossRef]
- İkinci, N.; Oberprieler, C.; Güner, A. On the origin of European lilies: Phylogenetic analysis of Lilium section Liriotypus (Liliaceae) using sequences of the nuclear ribosomal transcribed spacers. Willdenowia 2006, 36, 647–656. [Google Scholar] [CrossRef][Green Version]
- Joger, U.; Fritz, U.; Guicking, D.; Kalyabina-Hauf, S.; Nagy, Z.T.; Wink, M. Relict populations and endemic clades in palearctic reptiles: Evolutionary history and implications for conservation. In Relict Species; Springer: Cham, Switzerland, 2010; pp. 119–143. [Google Scholar]
- Khalilzadeh, P.; Rezaei, H.R.; Fadakar, D.; Serati, M.; Aliabadian, M.; Haile, J.; Goshtasb, H. Contact zone of Asian and European wild boar at North West of Iran. PLoS ONE 2016, 11, e0159499. [Google Scholar]
- Colagar, A.H.; Yousefzadeh, H.; Shayanmehr, F.; Jalali, S.G.; Zare, H.; Tippery, N.P. Molecular taxonomy of Hyrcanian Alnus using nuclear ribosomal ITS and chloroplast trnH-psbA DNA barcode markers. Syst. Biodivers. 2016, 14, 88–101. [Google Scholar] [CrossRef]
- Mostajeran, F.; Yousefzadeh, H.; Davitashvili, N.; Kozlowski, G.; Akbarinia, M. Phylogenetic relationships of Pterocarya (Juglandaceae) with an emphasis on the taxonomic status of Iranian populations using ITS and trnH-psb A sequence data. Plant Biosyst. 2017, 151, 1012–1021. [Google Scholar] [CrossRef]
- Manafzadeh, S.; Salvo, G.; Conti, E. A tale of migrations from east to west: The Irano-Turanian floristic region as a source of Mediterranean xerophytes. J. Biogeogr. 2014, 41, 366–379. [Google Scholar] [CrossRef]

| Region | Conserved Sites | Variable Sites | Parsim Informative Sites | Singleton Sites | Length (bp) | A% | T% | C% | G% | Model | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Iranian Sample | ITS | 549 | 80 | 27 | 52 | 644–654 | 18.8 | 21.1 | 27.2 | 32.9 | T93 + G |
| trnH-psbA | 190 | 244 | 10 | 234 | 465–485 | 36.8 | 38.7 | 16.5 | 18 | T92 | |
| matk | 849 | 19 | - | 9 | 853–862 | 31.1 | 38.5 | 15.8 | 14.6 | T92 | |
| trnL-F | 246 | 13 | - | 13 | 270 | 30.5 | 34.5 | 18.5 | 16.6 | T92 | |
| Other species | ITS | 436 | 171 | 108 | 63 | 644–660 | 18.4 | 20.7 | 27.8 | 33 | T93 + G |
| trnH-psbA | 251 | 170 | 12 | 153 | 460–480 | 31 | 36.1 | 15.9 | 17 | T92 | |
| matk | 882 | 36 | 19 | 17 | 872–881 | 31 | 38.6 | 15.8 | 14.6 | T92 | |
| trnL-F | 255 | 4 | 2 | 2 | 260–270 | 30.4 | 35.5 | 13.8 | 15.9 | T92 |
| Age Estimation (MYA) | S-DIVA | BBM | ||||||
|---|---|---|---|---|---|---|---|---|
| Nodes | Mean | 95% HPDlower | 95% HPDupper | AR | MP(%) | AR | MP | Support (PP) |
| I | 50.71 | 36.8 | 68.8 | A/AF | 49/36 | A/AD | 60/16 | 0.90 |
| II | 40.84 | 23.0 | 63.2 | AF | 99.71 | AF/A | 34/26 | 0.80 |
| III | 40.45 | 28.2 | 56.5 | AC | 93.40 | A/AD | 42/23 | 0.95 |
| IV | 33.25 | 20.5 | 49.8 | C | 93.30 | C/CD | 46/38 | 1.00 |
| V | 27.08 | 15.7 | 44.0 | CD | 78.00 | C/CD | 49/42 | 0.90 |
| S-DIVA | BBM | ||||||
| Distribution Range | Dispersal from | Dispersal to | Within | Distribution Range | Dispersal from | Dispersal to | Within |
| A | 44.00 | 0.00 | 28.0 | A | 64.0 | 0.00 | 28.0 |
| B | 0.00 | 5.00 | 1.00 | B | 0.00 | 9.00 | 1.00 |
| C | 2.00 | 5.00 | 8.00 | C | 2.00 | 7.00 | 9.00 |
| D | 1.00 | 19.0 | 15.0 | D | 2.00 | 28.00 | 26.0 |
| E | 0.00 | 3.00 | 0.00 | E | 0.00 | 4.00 | 0.00 |
| F | 1.00 | 16.0 | 9.00 | F | 4.00 | 24.00 | 18.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shokrollahi, S.; Yousefzadeh, H.; Parisod, C.; Heshmati, G.; Bina, H.; Ali, S.; Amirchakhmaghi, N.; Song, Y. Phylogenetics and Biogeography of Lilium ledebourii from the Hyrcanian Forest. Diversity 2022, 14, 137. https://doi.org/10.3390/d14020137
Shokrollahi S, Yousefzadeh H, Parisod C, Heshmati G, Bina H, Ali S, Amirchakhmaghi N, Song Y. Phylogenetics and Biogeography of Lilium ledebourii from the Hyrcanian Forest. Diversity. 2022; 14(2):137. https://doi.org/10.3390/d14020137
Chicago/Turabian StyleShokrollahi, Shekoofeh, Hamed Yousefzadeh, Christian Parisod, Gholamali Heshmati, Hamid Bina, Shujait Ali, Narjes Amirchakhmaghi, and Yigang Song. 2022. "Phylogenetics and Biogeography of Lilium ledebourii from the Hyrcanian Forest" Diversity 14, no. 2: 137. https://doi.org/10.3390/d14020137
APA StyleShokrollahi, S., Yousefzadeh, H., Parisod, C., Heshmati, G., Bina, H., Ali, S., Amirchakhmaghi, N., & Song, Y. (2022). Phylogenetics and Biogeography of Lilium ledebourii from the Hyrcanian Forest. Diversity, 14(2), 137. https://doi.org/10.3390/d14020137







