Genetic Diversity and Mating System of Two Mangrove Species (Rhizophora apiculata and Avicennia marina) in a Heavily Disturbed Area of China
Abstract
:1. Introduction
2. Materials and Methods
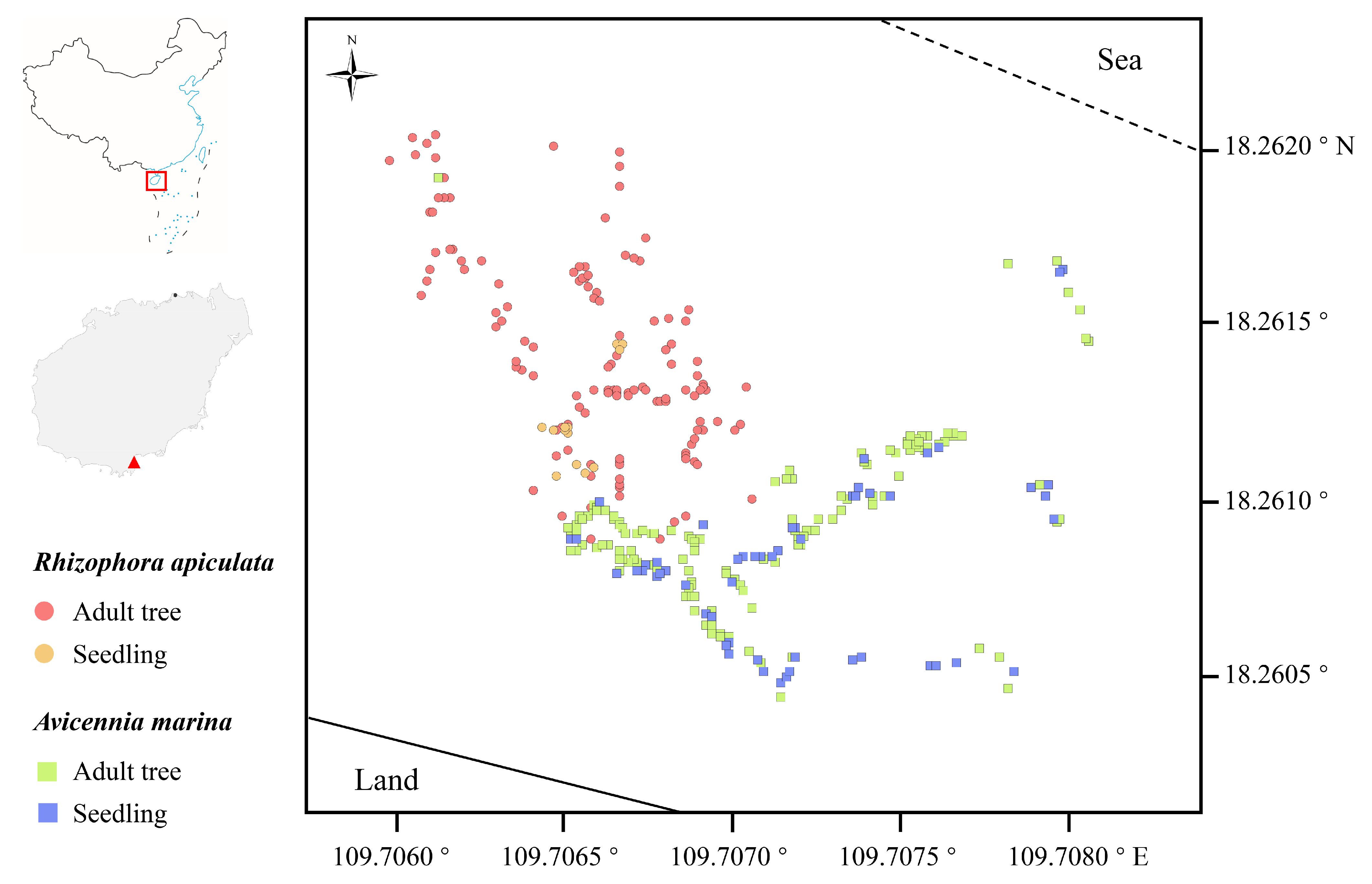
2.1. Study Area and Plant Sampling
2.2. DNA Extraction, PCR Amplification, and Microsatellite Screening
2.3. Statistical Analysis of Genetic Parameters
2.4. Analysis of Bottlenecks and Spatial Genetic Structure
2.5. Analysis of Mating System Parameters and Pollen Dispersal
3. Results
3.1. Genetic Diversity of R. apiculata and A. marina
3.2. Bottlenecks and Spatial Genetic Structure
3.3. Mating System Parameters
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alongi, D.M. Present state and future of the world's mangrove forests. Environ. Conserv. 2002, 29, 331–349. [Google Scholar] [CrossRef] [Green Version]
- Kathiresan, K.; Bingham, B.L. Biology of mangroves and mangrove ecosystems. Adv. Mar. Biol. 2001, 40, 81–251. [Google Scholar]
- Tomlinson, P.B. The Botany of Mangroves, 2nd ed.; Cambridge University Press: Cambridge, UK, 2016; pp. 11–28. [Google Scholar]
- Costanza, R.; d’Arge, R.; de Groot, R.; Farber, S.; Grasso, M.; Hannon, B.; Limburg, K.; Naeem, S.; O’Neill, R.V.; Paruelo, J.; et al. The value of ecosystem service and nature capital in the world. Nature 1997, 387, 235–260. [Google Scholar] [CrossRef]
- Alongi, D.M. Carbon sequestration in mangrove forests. Carbon. Manag. 2012, 3, 313–322. [Google Scholar] [CrossRef]
- Polidoro, B.A.; Carpenter, K.E.; Collins, L.; Duke, N.C.; Ellison, A.M.; Ellison, J.C.; Farnsworth, E.J.; Fernando, E.S.; Kathiresan, K.; Koedam, N.E.; et al. The loss of species: Mangrove extinction risk and geographic areas of global concern. PLoS ONE 2010, 5, e10095. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.; Milantara, N.; Sasmito, S.D.; Kajita, T.; Basyuni, M. Anthropogenic drivers of mangrove loss and associated carbon emissions in South Sumatra, Indonesia. Forests 2021, 12, 187. [Google Scholar] [CrossRef]
- Field, C.D. Impact of expected climate change on mangroves. Hydrobiologia 1995, 295, 75–81. [Google Scholar] [CrossRef]
- Bologna, P.; Campanella, J.J.; Restaino, D.J.; Fetske, Z.A.; Lourenco, M.; Smalley, J.V. Lingering impacts of hurricane Hugo on Rhizophora mangle (red mangrove) population genetics on St. John, USVI. Diversity 2019, 11, 65. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.-Y.; Primavera, J.H.; Dahdouh-Guebas, F.; McKee, K.; Bosire, J.O.; Cannicci, S.; Diele, K.; Fromard, F.; Koedam, N.; Marchand, C.; et al. Ecological role and services of tropical mangrove ecosystems: A reassessment. Glob. Ecol. Biogeogr. 2014, 23, 726–743. [Google Scholar] [CrossRef]
- FAO. The World's Mangroves 1980–2005; Food and Agriculture Organization of the United Nations: Rome, Italy, 2007. [Google Scholar]
- Fan, H.; Wang, W. Some thematic issues for mangrove conservation in China (in Chinese). J. Xiamen Univ. Nat. Sci. 2017, 56, 323–330. [Google Scholar]
- Duke, N.C.; Meynecke, J.-O.; Dittmann, S.; Ellison, A.M.; Anger, K.; Berger, U.; Cannicci, S.; Diele, K.; Ewel, K.C.; Field, C.D.; et al. A world without mangroves? Science 2007, 317, 41–42. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.Q.; Wang, M. The Mangrove of China, 1st ed.; Science Press: Beijing, China, 2007; pp. 1–186. [Google Scholar]
- SFA. National Mangrove Resource Inventory Report; The State Forestry Administration of the People’s Republic of China: Beijing, China, 2002. [Google Scholar]
- Wang, W.; Fu, H.; Lee, S.-Y.; Fan, H.; Wang, M. Can strict protection stop the decline of mangrove ecosystems in China? From rapid destruction to rampant degradation. Forests 2020, 11, 55. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.-C.; Lu, W.-X.; Zou, Z.; Li, S. Mangrove Wetlands: Distribution, Species Composition and Protection in China (in Chinese). Subtrop. Plant Sci. 2017, 46, 301–310. [Google Scholar]
- Jia, M.; Wang, Z.; Zhang, Y.; Mao, D.; Wang, C. Monitoring loss and recovery of mangrove forests during 42 years: The achievements of mangrove conservation in China. Int. J. Appl. Earth Obs. Geoinf. 2018, 73, 535–545. [Google Scholar] [CrossRef]
- Hamilton, S.E.; Casey, D. Creation of a high spatio-temporal resolution global database of continuous mangrove forest cover for the 21st century (CGMFC-21). Glob. Ecol. Biogeogr. 2016, 25, 729–738. [Google Scholar] [CrossRef]
- Wang, M.; Wang, W.; Lin, G.; Ma, W.; Fu, R. The Mangroves of Sanya, 1st ed.; Science Press: Beijing, China, 2019; pp. 1–212. [Google Scholar]
- Ma, Z.; Melville, D.S.; Liu, J.; Chen, Y.; Yang, H.; Ren, W.; Zhang, Z.; Piersma, T.; Li, B. Rethinking China’s new great wall. Science 2014, 346, 912–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Song, L.; Agusti, S.; Duarte, C.; Christakos, G.; Wu, J. Changes of the Macrobenthos Community with Non-native Mangrove Rehabilitation (Kandelia obovata) and Salt Marsh Invasion (Spartina alterniflora) in Ximen Island, Zhejiang, China. Ocean Sci. J. 2021, 56, 395–405. [Google Scholar] [CrossRef]
- Maiti, S.K.; Chowdhury, A. Effects of anthropogenic pollution on mangrove biodiversity: A review. J. Environ. Prot. 2013, 4, 1428–1434. [Google Scholar] [CrossRef] [Green Version]
- Feeley, K.J.; Silman, M.R. Land-use and climate change effects on population size and extinction risk of Andean plants. Glob. Chang. Biol. 2010, 16, 3215–3222. [Google Scholar] [CrossRef]
- Çalişkan, M. Genetic Diversity in Plants, 1st ed.; InTech: Rijeka, Croatia, 2012; pp. 1–493. [Google Scholar]
- Verma, A.K. Genetic diversity as buffer in biodiversity. Indian J. Biol. 2017, 4, 61–63. [Google Scholar]
- Rao, V.R.; Hodgkin, T. Genetic diversity and conservation and utilization of plant genetic resources. Plant Cell Tissue Organ Cult. 2002, 68, 1–19. [Google Scholar]
- Kahilainen, A.; Puurtinen, M.; Kotiaho, J.S. Conservation implications of species–genetic diversity correlations. Glob. Ecol. Conserv. 2014, 2, 315–323. [Google Scholar] [CrossRef] [Green Version]
- Dixo, M.; Metzger, J.P.; Morgante, J.S.; Zamudio, K.R. Habitat fragmentation reduces genetic diversity and connectivity among toad populations in the Brazilian Atlantic Coastal Forest. Biol. Conserv. 2009, 142, 1560–1569. [Google Scholar] [CrossRef]
- Fahrig, L. Effects of habitat fragmentation on biodiversity. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 487–515. [Google Scholar] [CrossRef] [Green Version]
- Tambarussi, E.V.; Boshier, D.H.; Vencovsky, R.; Freitas, M.L.M.; Di-Dio, O.J.; Sebbenn, A.M. Several small: How inbreeding affects conservation of Cariniana legalis Mart. Kuntze (Lecythidaceae) the Brazilian Atlantic Forest's largest tree. Int. For. Rev. 2016, 18, 502–510. [Google Scholar] [CrossRef]
- Coelho, N.H.P.; Tambarussi, E.V.; Aguiar, B.I.; Roque, R.H.; Portela, R.M.; Braga, R.C.; Sanson, D.; Silva, R.A.R.; Ferraz, E.M.; Moreno, M.A.; et al. Understanding genetic diversity, spatial genetic structure, and mating system through microsatellite markers for the conservation and sustainable use of Acrocomia aculeata (Jacq.) Lodd. Ex Mart. Conserv. Genet. 2018, 19, 879–891. [Google Scholar] [CrossRef] [Green Version]
- Eckert, C.G.; Kalisz, S.; Geber, M.A.; Sargent, R.; Elle, E.; Cheptou, P.O.; Goodwillie, C.; Johnston, M.O.; Kelly, J.K.; Moeller, D.A.; et al. Plant mating systems in a changing world. Trends Ecol. Evol. 2010, 25, 35–43. [Google Scholar] [CrossRef]
- Hopper, S.D. OCBIL theory: Towards an integrated understanding of the evolution, ecology and conservation of biodiversity on old, climatically buffered, infertile landscapes. Plant Soil 2009, 322, 49–86. [Google Scholar] [CrossRef]
- Miller, M.P.; Haig, S.M.; Wagner, R.S. Phylogeography and spatial genetic structure of the southern torrent salamander: Implications for conservation and management. J. Hered. 2006, 97, 561–570. [Google Scholar] [CrossRef] [Green Version]
- Caballero, A.; Rodríguez-Ramilo, S.T.; Ávila, V.; Fernández, J. Management of genetic diversity of subdivided populations in conservation programmes. Conserv. Genet. 2010, 11, 409–419. [Google Scholar] [CrossRef]
- Al-Qthanin, R.N.; Alharbi, S.A. Spatial structure and genetic variation of a mangrove species (Avicennia marina (Forssk.) Vierh) in the Farasan Archipelago. Forests 2020, 11, 1287. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.-W.; Yang, Y.; Li, X.-N. Structural and comparative analysis of the complete chloroplast genome of a mangrove plant: Scyphiphora hydrophyllacea Gaertn. f. and related Rubiaceae species. Forests 2019, 10, 1000. [Google Scholar] [CrossRef] [Green Version]
- Yao, Y.-F.; Liao, W.-B.; Song, X.-Y.; Li, C.-S. Current status and conservation of mangrove resources in Tielu Harbor, Sanya, Hainan (in Chinese). Mar. Sci. Bull. 2010, 29, 150–155. [Google Scholar]
- Tao, L.-P.; Huang, S.-M. Research on Mangrove Resources and Communities in Sanya (in Chinese). Nat. Sci. J. Hainan Univ. 2004, 22, 70–74. [Google Scholar]
- Arnaud-Haond, S.; Teixeira, S.; Massa, S.I.; Billot, C.; Saenger, P.; Coupland, G.; Duarte, C.M.; Serrão, E.A. Genetic structure at range edge: Low diversity and high inbreeding in Southeast Asian mangrove (Avicennia marina) populations. Mol. Ecol. 2006, 15, 3515–3525. [Google Scholar] [CrossRef]
- Geng, Q.F.; Lian, C.L.; Tao, J.M.; Li, S.Q.; Hogetsu, T. Isolation and characterization of 10 new compound microsatellite markers for a mangrove tree species, Avicennia marina (Forsk.) Vierh. (Avicenniaceae). Mol. Ecol. Notes 2007, 7, 1208–1210. [Google Scholar] [CrossRef]
- Islam, M.S.; Lian, C.; Kameyama, N.; Wu, B.; Hogetsu, T. Development of microsatellite markers in Rhizophora stylosa using a dual-suppression-polymerase chain reaction technique. Mol. Ecol. Notes 2004, 4, 110–112. [Google Scholar] [CrossRef]
- Shinmura, Y.; Wee, A.K.S.; Takayama, K.; Meenakshisundaram, S.H.; Asakawa, T.; Onrizal, O.; Adjie, B.; Ardli, E.R.; Sungkaew, S.; Malekal, N.B.; et al. Isolation and characterization of 14 microsatellite markers for Rhizophora mucronata (Rhizophoraceae) and their potential use in range-wide population studies. Conserv. Genet. Resour. 2012, 4, 951–954. [Google Scholar] [CrossRef]
- Raymond, M.; Rousset, F. GENEPOP (Version 1.2): Population genetics software for exact tests and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Szpiech, Z.A.; Jakobsson, M.; Rosenberg, N.A. ADZE: A rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 2008, 24, 2498–2504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slate, J.; Marshall, T.; Pemberton, J. A retrospective assessment of the accuracy of the paternity inference program CERVUS. Mol. Ecol. 2000, 9, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Piry, S.; Luikart, G.; Cornuet, J.-M. Computer note. BOTTLENECK: A computer program for detecting recent reductions in the effective size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Geng, Q.; Lian, C.; Goto, S.; Tao, J.; Kimura, M.; Islam, M.S.; Hogetsu, T. Mating system, pollen and propagule dispersal, and spatial genetic structure in a high-density population of the mangrove tree Kandelia candel. Mol. Ecol. 2008, 17, 4724–4739. [Google Scholar] [CrossRef]
- Hardy, O.J.; Vekemans, X. SPAGeDi: A versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2002, 2, 618–620. [Google Scholar] [CrossRef] [Green Version]
- Vekemans, X.; Hardy, O.J. New insights from fine-scale spatial genetic structure analyses in plant populations. Mol. Ecol. 2004, 13, 921–935. [Google Scholar] [CrossRef]
- Ritland, K. Extensions of models for the estimation of mating systems using n independent loci. Heredity 2002, 88, 221–228. [Google Scholar] [CrossRef]
- Thomas, E.; Jalonen, R.; Loo, J.; Boshier, D.; Gallo, L.; Cavers, S.; Bordács, S.; Smith, P.; Bozzano, M. Genetic considerations in ecosystem restoration using native tree species. For. Ecol. Manag. 2014, 333, 66–75. [Google Scholar] [CrossRef] [Green Version]
- Salas-Leiva, D.E.; Mayor-Durán, V.M.; Toro-Perea, N. Genetic diversity of the black mangrove (Avicennia germinans L.) in Colombia. Aquat. Bot. 2009, 91, 187–193. [Google Scholar] [CrossRef]
- Salas-Leiva, D.E.; Mayor-Durán, V.M.; Toro-Perea, N. Genetic diversity of black mangrove (Avicennia germinans) in natural and reforested areas of Salamanca Island Parkway, Colombian Caribbean. Hydrobiologia 2009, 620, 17–24. [Google Scholar] [CrossRef]
- Millán-Aguilar, O.; Manzano-Sarabia, M.; Nettel-Hernanz, A.; Dodd, R.S.; Hurtado-Oliva, M.Á.; Velázquez-Velázquez, E. Genetic diversity of the black mangrove Avicennia germinans (L.) Stearn in Northwestern Mexico. Forests 2016, 7, 197. [Google Scholar] [CrossRef] [Green Version]
- Yahya, A.F.; Hyun, J.O.; Lee, J.H.; Kim, Y.Y.; Lee, K.M.; Hong, K.N.; Kim, S.-C. Genetic variation and population genetic structure of Rhizophora apiculata (Rhizophoraceae) in the greater Sunda Islands, Indonesia using microsatellite markers. J. Plant Res. 2014, 127, 287–297. [Google Scholar] [CrossRef]
- Azman, A.; Ng, K.-K.-S.; Ng, C.-H.; Lee, C.-T.; Tnah, L.-H.; Zakaria, N.-F.; Mahruji, S.; Perdan, K.; Abdul-Kadir, M.-Z.; Cheng, A.; et al. Low genetic diversity indicating the threatened status of Rhizophora apiculata (Rhizophoraceae) in Malaysia: Declined evolution meets habitat destruction. Sci. Rep. 2020, 10, 19112. [Google Scholar] [CrossRef] [PubMed]
- Christensen, B.; Wium-Andersen, S. Seasonal growth of mangrove trees in southern Thailand. I. The phenology of Rhizophora apiculata Bl. Aquat. Bot. 1977, 3, 281–286. [Google Scholar] [CrossRef]
- Maguire, T.L.; Saenger, P.; Baverstock, P.; Henry, R. Microsatellite analysis of genetic structure in the mangrove species Avicennia marina (Forsk.) Vierh.(Avicenniaceae). Mol. Ecol. 2000, 9, 1853–1862. [Google Scholar] [CrossRef] [PubMed]
- Duke, N.C. Mangrove taxonomy, biogeography and evolution-Implications for conservation and management (An Indo West Pacific perspective). Excellence in Research Australia (ERA)-Collection Centre for Marine Studies Publication. In Proceedings of the International Conference and Exhibition on the Mangroves of Indian and Western Pacific Oceans, Kuala Lumpur, Malaysia, 21–24 August 2006; Hooi, T.K., Eong, O.J., Primavera, J.H., Eds.; Maritime Institute of Malaysia (MIMA): Kuala Lumpur, Malaysia, 2008. [Google Scholar]
- Arbeláez-Cortes, E.; Castillo-Cárdenas, M.F.; Toro-Perea, N.; Cárdenas-Henao, H. Genetic structure of the red mangrove (Rhizophora mangle L.) on the Colombian Pacific detected by microsatellite molecular markers. Hydrobiologia 2007, 583, 321–330. [Google Scholar] [CrossRef]
- Chen, X.-Y.; Lin, P.; Lin, Y.-M. Mating systems and spontaneous mutation rates for chlorophyll-deficiency in populations of the mangrove Kandelia candel. Hereditas 1996, 125, 47–52. [Google Scholar]
- Jump, A.S.; Peñuelas, J. Genetic effects of chronic habitat fragmentation in a wind-pollinated tree. Proc. Natl. Acad. Sci. USA 2006, 103, 8096–8100. [Google Scholar] [CrossRef] [Green Version]
- Ge, J.P.; Cai, B.; Ping, W.; Song, G.; Ling, H.; Lin, P. Mating system and population genetic structure of Bruguiera gymnorrhiza (Rhizophoraceae), a viviparous mangrove species in China. J. Exp. Mar. Biol. Ecol. 2005, 326, 48–55. [Google Scholar] [CrossRef]
- Hanson, T.R.; Brunsfeld, S.J.; Finegan, B.; Waits, L.P. Pollen dispersal and genetic structure of the tropical tree Dipteryx panamensis in a fragmented Costa Rican landscape. Mol. Ecol. 2008, 17, 2060–2073. [Google Scholar] [CrossRef]
- Ha, H.T.; Duarte, C.M.; Tri, N.H.; Terrados, J.; Borum, J. Growth and population dynamics during early stages of the mangrove Kandelia candel in Halong Bay, North Viet Nam. Estuar. Coast. Shelf Sci. 2003, 58, 435–444. [Google Scholar]
- Kettenring, K.M.; Mercer, K.L.; Reinhardt Adams, C.; Hines, J. EDITOR’S CHOICE: Application of genetic diversity-ecosystem function research to ecological restoration. J. Appl. Ecol. 2014, 51, 339–348. [Google Scholar] [CrossRef]


| Locus | n | Na | Ne | Ho | He | uHe | I | Pr(Ex1) | Pr(Ex2) |
|---|---|---|---|---|---|---|---|---|---|
| R. apiculata | |||||||||
| Rhst01 | 167 | 2 | 1.519 | 0.018 | 0.342 | 0.343 | 0.525 | 0.962 | 0.880 |
| Rhst02 | 167 | 4 | 1.877 | 0.665 | 0.467 | 0.469 | 0.747 | 0.871 | 0.755 |
| Rhst11 | 167 | 2 | 1.916 | 0.347 | 0.478 | 0.479 | 0.671 | 0.903 | 0.828 |
| Rhst13 | 167 | 7 | 2.065 | 0.413 | 0.516 | 0.517 | 0.920 | 0.861 | 0.743 |
| Rhst20 | 167 | 8 | 4.322 | 0.311 | 0.769 | 0.771 | 1.589 | 0.630 | 0.452 |
| RM113 | 167 | 5 | 2.453 | 0.593 | 0.592 | 0.594 | 1.056 | 0.814 | 0.671 |
| RM114 | 167 | 6 | 2.207 | 0.467 | 0.547 | 0.548 | 0.999 | 0.839 | 0.692 |
| RM116 | 167 | 3 | 1.068 | 0.042 | 0.064 | 0.064 | 0.155 | 0.997 | 0.960 |
| Mean | 167 | 4.6 | 2.178 | 0.357 | 0.472 | 0.473 | 0.833 | / | / |
| A. marina | |||||||||
| Avma01 | 210 | 3 | 1.840 | 0.262 | 0.457 | 0.458 | 0.662 | 0.881 | 0.813 |
| Avma02 | 210 | 5 | 1.479 | 0.248 | 0.324 | 0.325 | 0.596 | 0.930 | 0.823 |
| Avma16 | 210 | 4 | 1.975 | 0.238 | 0.494 | 0.495 | 0.722 | 0.874 | 0.804 |
| Avma17 | 210 | 5 | 2.795 | 0.476 | 0.642 | 0.644 | 1.201 | 0.797 | 0.634 |
| Am3 | 210 | 2 | 1.379 | 0.157 | 0.275 | 0.275 | 0.447 | 0.975 | 0.901 |
| Am32 | 210 | 3 | 1.100 | 0.010 | 0.091 | 0.091 | 0.212 | 0.999 | 0.976 |
| Am40 | 210 | 4 | 1.776 | 0.152 | 0.437 | 0.438 | 0.671 | 0.903 | 0.819 |
| Am81 | 210 | 4 | 1.547 | 0.014 | 0.354 | 0.354 | 0.594 | 0.908 | 0.818 |
| Mean | 210 | 3.5 | 1.750 | 0.197 | 0.387 | 0.389 | 0636 | / | / |
| Species | n | Na | Ne | Ho | He | uHe | I | Fis | AR | PAR |
|---|---|---|---|---|---|---|---|---|---|---|
| R. apiculata | ||||||||||
| Adults | 139 | 4.625 | 2.165 | 0.366 | 0.464 | 0.466 | 0.828 | 0.215 | 3.046 | 0.433 |
| (0.800) | (0.365) | (0.084) | (0.073) | (0.074) | (0.152) | / | (0.441) | (0.130) | ||
| Seedlings | 28 | 3.375 | 2.169 | 0.313 | 0.486 | 0.495 | 0.813 | 0.373 * | 2.941 | 0.327 |
| (0.460) | (0.262) | (0.081) | (0.071) | (0.072) | (0.135) | / | (0.402) | (0.155) | ||
| Total | 167 | 4.625 | 2.178 | 0.357 | 0.472 | 0.473 | 0.833 | 0.246 | / | / |
| (0.800) | (0.341) | (0.083) | (0.072) | (0.073) | (0.149) | / | / | / | ||
| A. marina | ||||||||||
| Adults | 152 | 3.500 | 1.750 | 0.197 | 0.387 | 0.389 | 0.636 | 0.497 * | 2.623 | 0.277 |
| (0.267) | (0.181) | (0.056) | (0.060) | (0.061) | (0.102) | / | (0.233) | (0.111) | ||
| Seedlings | 58 | 2.875 | 1.699 | 0.190 | 0.372 | 0.376 | 0.625 | 0.497 * | 2.704 | 0.358 |
| (0.389) | (0.181) | (0.049) | (0.055) | (0.055) | (0.097) | / | (0.329) | (0.143) | ||
| Total | 210 | 3.750 | 1.736 | 0.195 | 0.384 | 0.385 | 0.638 | 0.497 * | / | / |
| (0.366) | (0.180) | (0.053) | (0.058) | (0.058) | (0.099) | / | / | / | ||
| Model | R. apiculata | A. marina | |||||
|---|---|---|---|---|---|---|---|
| Adults | Seedlings | Total | Adults | Seedlings | Total | ||
| Sign test | TPM | 0.450 | 0.200 | 0.430 | 0.531 | 0.142 | 0.534 |
| IAA | 0.154 | 0.042 | 0.145 | 0.130 | 0.096 | 0.138 | |
| SMM | 0.086 | 0.269 | 0.083 | 0.225 | 0.593 | 0.069 | |
| mean | 0.230 | 0.170 | 0.219 | 0.295 | 0.277 | 0.247 | |
| One-tailed Wilcoxon test | TPM | 0.422 | 0.125 | 0.422 | 0.527 | 0.230 | 0.578 |
| IAA | 0.098 | 0.020 | 0.027 | 0.098 | 0.125 | 0.125 | |
| SMM | 0.963 | 0.473 | 0.902 | 0.973 | 0.527 | 0.980 | |
| mean | 0.494 | 0.206 | 0.451 | 0.533 | 0.294 | 0.561 | |
| Mother Tree | Propagules | tm (SE) | ts (SE) | tm−ts |
|---|---|---|---|---|
| R. apiculata | ||||
| Z12 | 6 | 1.001 (0.001) | 1.138 (0.053) | −0.137 |
| Z85 | 11 | 1.116 (0.041) | 1.551 (0.165) | −0.435 |
| Z110 | 6 | 1.004 (0.001) | 1.082 (0.112) | −0.078 |
| A. marina | ||||
| A1 | 17 | 0.822 (0.209) | 1.027 (0.203) | −0.205 |
| A2 | 10 | 0.186 (0.151) | 0.094 (0.073) | 0.092 |
| A21 | 9 | 0.755 (0.170) | 0.691 (0.199) | 0.064 |
| A48 | 9 | 1.024 (0.001) | 0.589 (0.079) | 0.435 |
| A126 | 6 | 0.849 (0.162) | 0.818 (0.228) | 0.031 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, W.; Zou, Z.; Hu, X.; Yang, S. Genetic Diversity and Mating System of Two Mangrove Species (Rhizophora apiculata and Avicennia marina) in a Heavily Disturbed Area of China. Diversity 2022, 14, 115. https://doi.org/10.3390/d14020115
Lu W, Zou Z, Hu X, Yang S. Genetic Diversity and Mating System of Two Mangrove Species (Rhizophora apiculata and Avicennia marina) in a Heavily Disturbed Area of China. Diversity. 2022; 14(2):115. https://doi.org/10.3390/d14020115
Chicago/Turabian StyleLu, Wenxun, Zhen Zou, Xueying Hu, and Shengchang Yang. 2022. "Genetic Diversity and Mating System of Two Mangrove Species (Rhizophora apiculata and Avicennia marina) in a Heavily Disturbed Area of China" Diversity 14, no. 2: 115. https://doi.org/10.3390/d14020115
APA StyleLu, W., Zou, Z., Hu, X., & Yang, S. (2022). Genetic Diversity and Mating System of Two Mangrove Species (Rhizophora apiculata and Avicennia marina) in a Heavily Disturbed Area of China. Diversity, 14(2), 115. https://doi.org/10.3390/d14020115






