Noninvasive Genetics Knowledge from the Brown Bear Populations to Assist Biodiversity Conservation
Abstract
1. Introduction
2. Materials and Methods
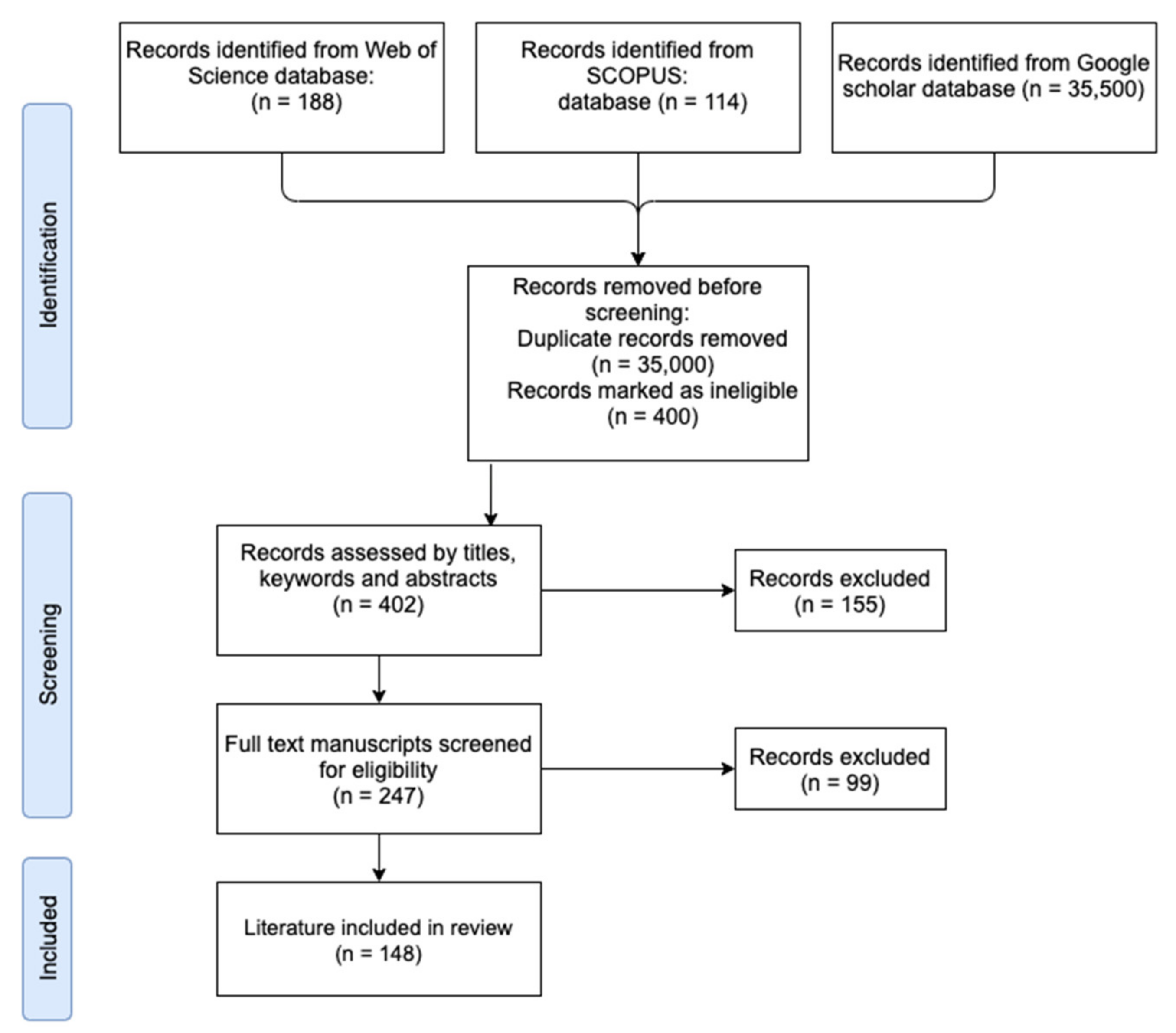
3. Results
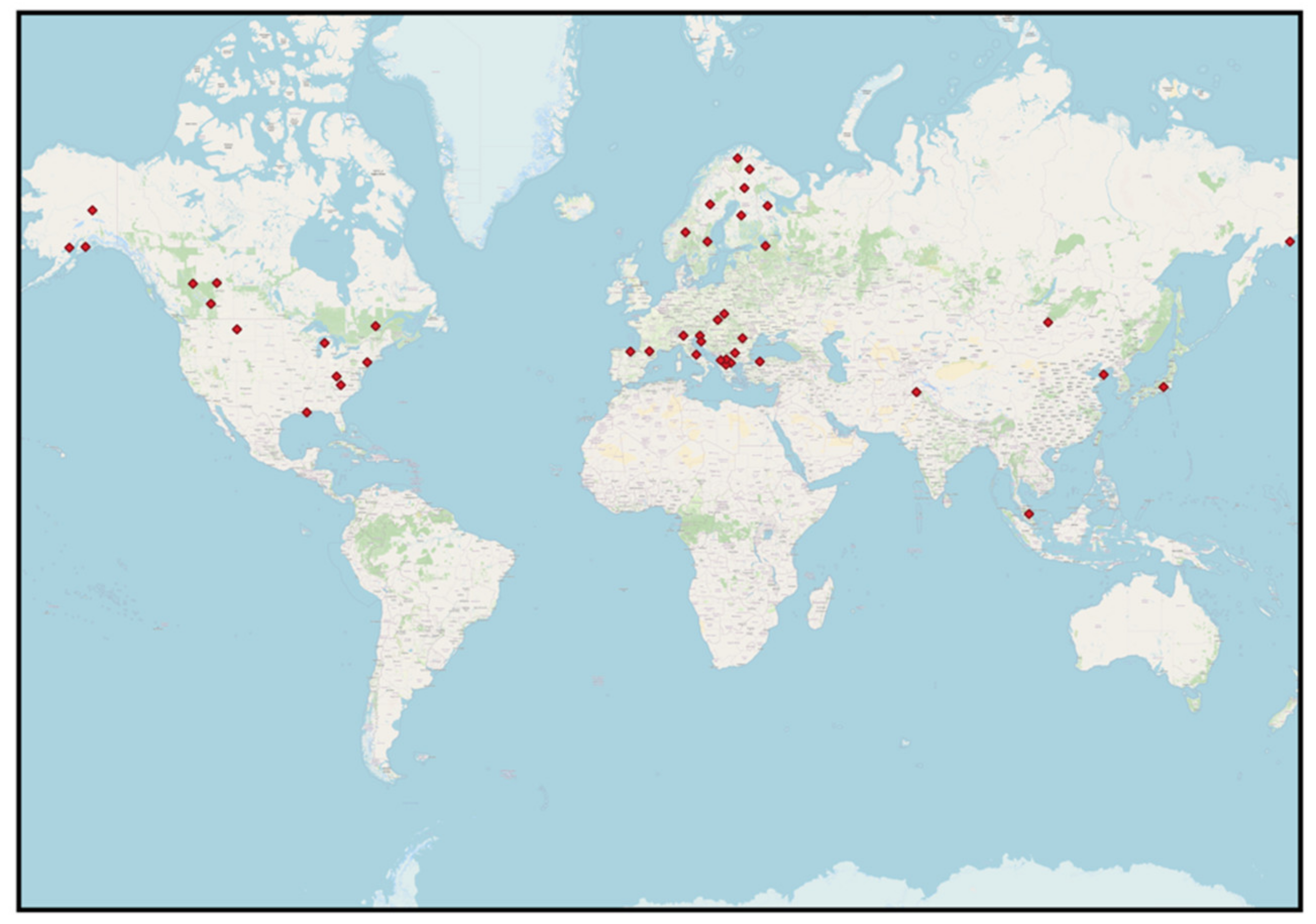
3.1. Sampling Scheme Including Location and Bear Population Size
3.2. Samples’ Collection and Storage
3.2.1. Faeces Collection
3.2.2. Hair Samples Collection
- Hair clumps: qualitative collection and storage.
- b.
- Attractants.
- c.
- Effectiveness of hair sampling.
4. Noninvasive Genetics in Bear Conservation and Management
5. Conclusions
- The Isohelix method to collect the samples needs to be tested in a local study and, if suitable, applied nationally, by considering the numerous advantages of this method and the high number of people involved in sample gathering, in addition to the shortcomings associated with the storage of such large quantities of samples.
- Trained dogs for faeces gathering should be used across the brown bear distribution, in parallel with hunting managers, foresters, and volunteers.
- Both faeces and hair samples should be collected using the systematic and opportunistic schemes, with a large focus on faeces.
- Samples following damage should be gathered without allowing much time to pass after the damaging event (these can be used for further forensic analysis).
- Sampling should be organised during autumn and winter because these seasons overlap with the hyperphagia behaviour, and it does not interfere with the cub’s period.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pimm, S.L.; Lawton, J.H. Planning for Biodiversity. Science 1998, 279, 2068–2069. [Google Scholar] [CrossRef]
- Zeng, H.; Sui, D.Z.; Wu, B. Human Disturbances on Landscapes in Protected Areas: A Case Study of the Wolong Nature Reserve. Ecol. Res. 2005, 20, 487–496. [Google Scholar] [CrossRef]
- Gaston, K.J.; Spicer, J.I. Biodiversity: An Introduction, 2nd ed.; Blackwell Publishing Ltd.: Malden, Massachusetts; Oxford, UK; Victoria, Australia, 2004; ISBN 1-4051-1857-1. [Google Scholar]
- Munguía, M.; Trejo, I.; González-Salazar, C.; Pérez-Maqueo, O. Human Impact Gradient on Mammalian Biodiversity. Glob. Ecol. Biogeogr. 2016, 6, 79–92. [Google Scholar] [CrossRef]
- Díaz, S.; Fargione, J.; Chapin, F.S.; Tilman, D. Biodiversity Loss Threatens Human Well-Being. PLoS Biol. 2006, 4, 277. [Google Scholar] [CrossRef]
- Weitzman, M. Diversity Functions. In Biodiversity Loss: Economic and Ecological Issues; Perrings, C., Maeler, K.G., Folke, C., Holling, C.S., Jansson, B.O., Eds.; Cambridge University Press: Cambridge, UK, 1995; pp. 21–43. [Google Scholar]
- Quratulann, S.; Muhammad Ehsan, M.; Rabia, E.; Sana, A. Review on Climate Change and Its Effect on Wildlife and Ecosystem. J. Environ. Biol. 2021, 008–014. [Google Scholar] [CrossRef]
- Pimm, S.L. Biodiversity: Climate Change or Habitat Loss—Which Will Kill More Species? Curr. Biol. 2008, 18, R117–R119. [Google Scholar] [CrossRef]
- Dinca, L.; Badea, O.; Guiman, G.; Braga, C.; Crisan, V.; Greavu, V.; Murariu, G.; Georgescu, L. Monitoring of Soil Moisture in Long-Term Ecological Research (LTER) Sites of Romanian Carpathians. Ann. For. Res. 2018, 61, 171–188. [Google Scholar] [CrossRef]
- Pearson, R.G.; Dawson, T.P. Predicting the Impacts of Climate Change on the Distribution of Species: Are Bioclimate Envelope Models Useful? Glob. Ecol. Biogeogr. 2003, 12, 361–371. [Google Scholar] [CrossRef]
- Araújo, M.B.; Rahbek, C. How Does Climate Change Affect Biodiversity? Science 2006, 313, 1396–1397. [Google Scholar] [CrossRef]
- Austin, M.P.; van Niel, K.P. Improving Species Distribution Models for Climate Change Studies: Variable Selection and Scale. J. Biogeogr. 2011, 38, 1–8. [Google Scholar] [CrossRef]
- Khalidah, K.N.; Wahdaniyah, S.; Kamarudin, N.; Lechner, A.M.; Azhar, B. Spared from Poaching and Natural Predation, Wild Boars Are Likely to Play the Role of Dominant Forest Species in Peninsular Malaysia. For. Ecol. Manag. 2021, 496, 119458. [Google Scholar] [CrossRef]
- Fischer, J.; Lindenmayer, D.B. Landscape Modification and Habitat Fragmentation: A Synthesis. Glob. Ecol. Biogeogr. 2007, 16, 265–280. [Google Scholar] [CrossRef]
- Scanes, C.G. Human Activity and Habitat Loss: Destruction, Fragmentation, and Degradation. Anim. Hum. Soc. 2018, 451–482. [Google Scholar] [CrossRef]
- Coulon, A.; Cosson, J.F.; Angibault, J.M.; Cargnelutti, B.; Galan, M.; Morellet, N.; Petit, E.; Aulagnier, S.; Hewison, A.J.M. Landscape Connectivity Influences Gene Flow in a Roe Deer Population Inhabiting a Fragmented Landscape: An Individual-Based Approach. Mol. Ecol. 2004, 13, 2841–2850. [Google Scholar] [CrossRef]
- Baguette, M.; Blanchet, S.; Legrand, D.; Stevens, V.M.; Turlure, C. Individual Dispersal, Landscape Connectivity and Ecological Networks. Biol. Rev. 2013, 88, 310–326. [Google Scholar] [CrossRef]
- Anderson, C.D.; Epperson, B.K.; Fortin, M.J.; Holderegger, R.; James, P.M.A.; Rosenberg, M.S.; Scribner, K.T.; Spear, S. Considering Spatial and Temporal Scale in Landscape-Genetic Studies of Gene Flow. Mol. Ecol. 2010, 19, 3565–3575. [Google Scholar] [CrossRef]
- Frid, A.; Dill, L. Human-Caused Disturbance Stimuli as a Form of Predation Risk. Ecol. Soc. 2002, 6. [Google Scholar] [CrossRef]
- Guanghun, J.; Jianzhang, M.; Minghai, Z. Spatial Distribution of Ungulate Responses to Habitat Factors in Wandashan Forest Region, Northeastern China. J. Wildl. Manag. 2006, 70, 1470–1476. [Google Scholar] [CrossRef]
- Paudel, P.K.; Kindlmann, P. Human Disturbance Is a Major Determinant of Wildlife Distribution in Himalayan Midhill Landscapes of Nepal. Anim. Conserv. 2012, 15, 283–293. [Google Scholar] [CrossRef]
- Tudose, N.C.; Cremades, R.; Broekman, A.; Sanchez-Plaza, A.; Mitter, H.; Marin, M. Mainstreaming the Nexus Approach in Climate Services Will Enable Coherent Local and Regional Climate Policies. Adv. Clim. Chang. Res. 2021, 12, 752–755. [Google Scholar] [CrossRef]
- Marin, M.; Clinciu, I.; Tudose, N.C.; Ungurean, C.; Adorjani, A.; Mihalache, A.L.; Davidescu, A.A.; Davidescu, Ș.O.; Dinca, L.; Cacovean, H. Assessing the Vulnerability of Water Resources in the Context of Climate Changes in a Small Forested Watershed Using SWAT: A Review. Environ. Res. 2020, 184, 109330. [Google Scholar] [CrossRef]
- Algotsson, E. Wildlife Conservation through People-Centred Approaches to Natural Resource Management Programmes and the Control of Wildlife Exploitation. Local Environ. 2006, 11, 79–93. [Google Scholar] [CrossRef]
- Hodgson, J.C.; Mott, R.; Baylis, S.M.; Pham, T.T.; Wotherspoon, S.; Kilpatrick, A.D.; Raja Segaran, R.; Reid, I.; Terauds, A.; Koh, L.P. Drones Count Wildlife More Accurately and Precisely than Humans. Methods Ecol. Evol. 2018, 9, 1160–1167. [Google Scholar] [CrossRef]
- García-Sánchez, M.P.; González-Ávila, S.; Solana-Gutiérrez, J.; Popa, M.; Jurj, R.; Ionescu, G.; Ionescu, O.; Fedorca, M.; Fedorca, A. Sex-Specific Connectivity Modelling for Brown Bear Conservation in the Carpathian Mountains. Landsc. Ecol. 2021. [Google Scholar] [CrossRef]
- Swenson, J.E.; Taberlet, P.; Bellemain, E. Genetics and Conservation of European Brown Bears Ursus Arctos. Mamm. Rev. 2011, 41, 87–98. [Google Scholar] [CrossRef]
- Antao, T.; Pérez-Figueroa, A.; Luikart, G. Early Detection of Population Declines: High Power of Genetic Monitoring Using Effective Population Size Estimators. Evol. Appl. 2011, 4, 144–154. [Google Scholar] [CrossRef]
- Bartoń, K.A.; Zwijacz-Kozica, T.; Zięba, F.; Sergiel, A.; Selva, N. Bears without Borders: Long-Distance Movement in Human-Dominated Landscapes. Glob. Ecol. Conserv. 2019, 17, e00541. [Google Scholar] [CrossRef]
- Epstein, Y.; Jos, J.; Vicente, J.; Opez-Bao, L.; Chapron, G.; Fischer, J. A Legal-Ecological Understanding of Favorable Conservation Status for Species in Europe. Conserv. Lett. 2016, 9, 81–88. [Google Scholar] [CrossRef]
- López-Bao, J.V.; Godinho, R.; Rocha, R.G.; Palomero, G.; Blanco, J.C.; Ballesteros, F.; Jiménez, J. Consistent Bear Population DNA-Based Estimates Regardless Molecular Markers Type. Biol. Conserv. 2020, 248. [Google Scholar] [CrossRef]
- Bischof, R.; Brøseth, H.; Gimenez, O. Wildlife in a Politically Divided World: Insularism Inflates Estimates of Brown Bear Abundance. Conserv. Lett. 2016, 9, 122–130. [Google Scholar] [CrossRef]
- De Barba, M.; Waits, L.P.; Genovesi, P.; Randi, E.; Chirichella, R.; Cetto, E. Comparing Opportunistic and Systematic Sampling Methods for Non-Invasive Genetic Monitoring of a Small Translocated Brown Bear Population. J. Appl. Ecol. 2010, 47, 172–181. [Google Scholar] [CrossRef]
- Boulanger, J.; Stenhouse, G.; Munro, R. Sources of Heterogeneity Bias When DNA Mark-Recapture Sampling Methods Are Applied to Grizzly Bear (Ursus Arctos) Populations. J. Mammal. 2004, 85, 618–624. [Google Scholar] [CrossRef]
- Boulanger, J.; Kendall, K.C.; Stetz, J.B.; Roon, D.A.; Waits, L.P.; Paetkau, D. Multiple Data Sources Improve DNA-Based Mark-Recapture Population Estimates of Grizzly Bears. Ecol. Appl. 2008, 18, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Sawaya, M.A.; Stetz, J.B.; Clevenger, A.P.; Gibeau, M.L.; Kalinowski, S.T. Estimating Grizzly and Black Bear Population Abundance and Trend in Banff National Park Using Noninvasive Genetic Sampling. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Latham, E.; Stetz, J.B.; Seryodkin, I.; Miquelle, D.; Gibeau, M.L. Non-Invasive Genetic Sampling of Brown Bears and Asiatic Black Bears in the Russian Far East: A Pilot Study. Ursus 2012, 23, 145–158. [Google Scholar] [CrossRef]
- Bischof, R.; Swenson, J.E. Linking Noninvasive Genetic Sampling and Traditional Monitoring to Aid Management of a Trans-Border Carnivore Population. Ecol. Appl. 2012, 22, 361–373. [Google Scholar] [CrossRef]
- Van Vliet, N.; Cornelis, D.; Beck, H.; Lindsey, P.A. Meat from the Wild: Extractive Uses of Wildlife and Alternatives for Sustainability. Curr. Trends Wildl. Res. 2016. [Google Scholar] [CrossRef]
- Albert, C.; Luque, G.M.; Courchamp, F. The Twenty Most Charismatic Species. PLoS ONE 2018, 13, e0199149. [Google Scholar] [CrossRef]
- Servheen, C.; Herrero, S.; Peyton, B. Bears. Status Survey and Conservation Action Plan; IUCN/SSC Bear and Polar Bear Specialist Groups, IUCN: Cland, Switzerland; Cambridge, UK; ISBN 2-8317-0462-6.
- Singh, N.J.; Danell, K.; Edenius, L.; Ericsson, G. Tackling the Motivation to Monitor: Success and Sustainability of a Participatory Monitoring Program. Ecol. Soc. 2014, 19. [Google Scholar] [CrossRef]
- Collins, D.M. Ursidae. In Fowler’s Zoo and Wild Animal Medicine; Elsevier: Amsterdam, The Netherlands, 2015; Volume 8, pp. 498–508. [Google Scholar]
- Kolchin, S.A.; Volkova, E.V.; Pokrovskaya, L.V.; Zavadskaya, A.V. Consequences of a Sockeye Salmon Shortage for the Brown Bear in the Basin of Lake Kurilskoe, Southern Kamchatka. Nat. Conserv. Res. 2021, 6, 53–65. [Google Scholar] [CrossRef]
- Ogurtsov, S.S. The Diet of the Brown Bear (Ursus Arctos) in the Central Forest Nature Reserve (West-European Russia), Based on Scat Analysis Data. Biology 2018, 45, 1039–1054. [Google Scholar] [CrossRef]
- Seryodkin, I.V.; Paczkowski, J.; Goodrich, J.M.; Petrunenko, Y.K. Locations of Dens with Respect to Space Use, Pre-and Post-Denning Movements of Brown Bears in the Russian Far East. Nat. Conserv. Res. 2021, 6, 97–109. [Google Scholar] [CrossRef]
- Chapron, G.; Kaczensky, P.; Linnell, J.D.C.; von Arx, M.; Huber, D.; Andrén, H.; Vicente López-Bao, J.; Adamec, M.; Álvares, F.; Anders, O.; et al. Recovery of Large Carnivores in Europe’s Modern Human-Dominated Landscapes. Science 2014, 346, 1517–1519. [Google Scholar] [CrossRef]
- Trouwborst, A. Managing the Carnivore Comeback: International and EU Species Protection Law and the Return of Lynx, Wolf and Bear to Western Europe. J. Environ. Law 2010, 22, 347–372. [Google Scholar] [CrossRef]
- Darimont, C.T.; Hall, H.; Eckert, L.; Mihalik, I.; Artelle, K.; Treves, A.; Paquet, P.C. Large Carnivore Hunting and the Social License to Hunt. Conserv. Biol. 2021, 35, 1111–1119. [Google Scholar] [CrossRef]
- Ogurtsov, S.S. Brown Bear (Ursus Arctos) Ecological Niche and Habitat Suitability Modeling in the Southern Taiga Subzone Using the Method of GNESFA. Nat. Conserv. Res. 2020, 5, 86–113. [Google Scholar] [CrossRef]
- De Barba, M.; Waits, L.P.; Garton, E.O.; Genovesi, P.; Randi, E.; Mustoni, A.; Groff, C. The Power of Genetic Monitoring for Studying Demography, Ecology and Genetics of a Reintroduced Brown Bear Population. Mol. Ecol. 2010, 19, 3938–3951. [Google Scholar] [CrossRef]
- Management and Action Plan for the Bear Population in Romania. Available online: http://www.mmediu.ro/app/webroot/uploads/files/17Management_Action_Plan.pdf" (accessed on 29 December 2021).
- Cotovelea, A.; Sofletea, N.; Ionescu, G.; Ionescu, O. Genetic Approaches for Romanian Conservation. Bull. Trans. Univ. Bras. Ser. II 2013, 6, 18–26. [Google Scholar]
- Kohn, M.H.; Wayne, R.K. Facts from Feces Revisited. Trends Ecol. Evol. 1997, 12, 223–227. [Google Scholar] [CrossRef]
- Bellemain, E.; Swenson, J.E.; Tallmon, D.; Taberlet, P. Estimating Population Size of Elusive Animals with DNA from Hunter-Collected Feces: Four Methods for Brown Bears. Conserv. Biol. 2005, 150–161. [Google Scholar] [CrossRef]
- Tumendemberel, O.; Zedrosser, A.; Proctor, M.F.; Reynolds, H.V.; Adams, J.R.; Sullivan, J.M.; Jacobs, S.J.; Khorloojav, T.; Tserenbataa, T.; Batmunkh, M.; et al. Phylogeography, Genetic Diversity, and Connectivity of Brown Bear Populations in Central Asia. PLoS ONE 2019, 14, 1–23. [Google Scholar] [CrossRef]
- Pranckutė, R. Web of Science (Wos) and Scopus: The Titans of Bibliographic Information in Today’s Academic World. Publications 2021, 9, 12. [Google Scholar] [CrossRef]
- Bramer, W.M.; Rethlefsen, M.L.; Kleijnen, J.; Franco, O.H. Optimal Database Combinations for Literature Searches in Systematic Reviews: A Prospective Exploratory Study. Syst. Rev. 2017, 6, 1–12. [Google Scholar] [CrossRef]
- Skrbinšek, T. Effects of Different Environmental and Sampling Variables on the Genotyping Success in Field-Collected Scat Samples: A Brown Bear Case Study. Acta Biol. Slov. 2020, 63, 89–98. [Google Scholar]
- Schwartz, M.K.; McKelvey, K.S. Why Sampling Scheme Matters: The Effect of Sampling Scheme on Landscape Genetic Results. Conserv. Genet. 2008, 10, 441–452. [Google Scholar] [CrossRef]
- Tredick, C.A.; Vaughan, M.R.; Stauffer, D.F.; Simek, S.L.; Eason, T. Sub-Sampling Genetic Data to Estimate Black Bear Population Size: A Case Study. Ursus 2007, 18, 179–188. [Google Scholar] [CrossRef]
- Schregel, J.; Kopatz, A.; Hagen, S.B.; Broseth, H.; Smith, M.E.; Wikan, S.; Wartiainen, I.; Aspholm, P.E.; Aspi, J.; Swenson, J.E.; et al. Limited Gene Flow among Brown Bear Populations in Far Northern Europe? Genetic Analysis of the East-West Border Population in the Pasvik Valley. Mol. Ecol. 2012, 21, 3474–3488. [Google Scholar] [CrossRef]
- Tallmon, D.A.; Bellemain, E.; Swendon, J.E.; Taberlet, P. Genetic Monitoring of Scandinavian Brown Bear Effective Population Size and Immigration. J. Wildl. Manag. 2004, 68, 960–965. [Google Scholar] [CrossRef]
- Kopatz, A.; Kleven, O.; Kojola, I.; Aspi, J.; Norman, A.J.; Spong, G.; Gyllenstrand, N.; Dalén, L.; Fløystad, I.; Hagen, S.B.; et al. Restoration of Transborder Connectivity for Fennoscandian Brown Bears (Ursus Arctos). Biol. Conserv. 2021, 253, 11–25. [Google Scholar] [CrossRef]
- Schregel, J.; Kopatz, A.; Eiken, H.G.; Swenson, J.E.; Hagen, S.B. Sex-Specific Genetic Analysis Indicates Low Correlation between Demographic and Genetic Connectivity in the Scandinavian Brown Bear (Ursus Arctos). PLoS ONE 2017, 12, e0180701. [Google Scholar] [CrossRef]
- Schregel, J.; Remm, J.; Eiken, H.G.; Swenson, J.E.; Saarma, U.; Hagen, S.B. Multi-Level Patterns in Population Genetics: Variogram Series Detects a Hidden Isolation-by-Distance-Dominated Structure of Scandinavian Brown Bears Ursus Arctos. Methods Ecol. Evol. 2018, 9, 1324–1334. [Google Scholar] [CrossRef]
- Schregel, J.; Eiken, H.G.; Grøndahl, F.A.; Hailer, F.; Aspi, J.; Kojola, I.; Tirronen, K.; Danilov, P.; Rykov, A.; Poroshin, E.; et al. Y Chromosome Haplotype Distribution of Brown Bears (Ursus Arctos) in Northern Europe Provides Insight into Population History and Recovery. Mol. Ecol. 2015, 24, 6041–6060. [Google Scholar] [CrossRef] [PubMed]
- Solberg, K.H.; Bellemain, E.; Drageset, O.M.; Taberlet, P.; Swenson, J.E. An Evaluation of Field and Non-Invasive Genetic Methods to Estimate Brown Bear (Ursus Arctos) Population Size. Biol. Conserv. 2006, 128, 158–168. [Google Scholar] [CrossRef]
- Kindberg, J.; Swenson, J.E.; Ericsson, G.; Bellemain, E.; Miquel, C.; Taberlet, P. Estimating Population Size and Trends of the Swedish Brown Bear Ursus Arctos Population. Wildl. Biol. 2011, 17, 114–123. [Google Scholar] [CrossRef]
- Skrbinšek, T.; Jelenčič, M.; Waits, L.; Kos, I.; Trontelj, P. Highly Efficient Multiplex PCR of Noninvasive DNA Does Not Require Pre-Amplification. Mol. Ecol. Resour. 2010, 10, 495–501. [Google Scholar] [CrossRef]
- Final Report <LIFE13 NAT/SI/000550 Life DINALP BEAR>. Available online: https://dinalpbear.eu/wp-content/uploads/LIFE-DINALP-BEAR_final-report_web.pdf (accessed on 9 June 2021).
- Jerina, K.; Jonozovič, M.; Krofel, M.; Skrbinšek, T. Range and Local Population Densities of Brown Bear Ursus Arctos in Slovenia. Eur. J. Wildl. Res. 2013, 59, 459–467. [Google Scholar] [CrossRef]
- Skrbinšek, T.; Luštrik, R.; Majić-Skrbinšek, A.; Potočnik, H.; Kljun, F.; Jelenčič, M.; Kos, I.; Trontelj, P. From Science to Practice: Genetic Estimate of Brown Bear Population Size in Slovenia and How It Influenced Bear Management. Eur. J. Wildl. Res. 2019, 65. [Google Scholar] [CrossRef]
- Straka, M.; Paule, L.; Ionescu, O.; Štofík, J.; Adamec, M. Microsatellite Diversity and Structure of Carpathian Brown Bears (Ursus Arctos): Consequences of Human Caused Fragmentation. Conserv. Genet. 2012, 13, 153–164. [Google Scholar] [CrossRef]
- Janiga, M.; Fečková, M.; Korňan, J. Preliminary Results on Genetic Tracking of the Brown Bear ( Ursus Arctos ) Individuals in the Malá Fatra National Park ( Slovakia ). Oecologia 2006, 15, 24–26. [Google Scholar]
- Graban, J.; Kisková, J.; Pepich, P.; Rigg, R. Genetic Analysis for Geographic Isolation Comparison of Brown Bears Living in the Periphery of the Western Carpathians Mountains with Bears Living in Other Areas. Open J. Genet. 2013, 3, 174–182. [Google Scholar] [CrossRef][Green Version]
- Iosif, R.; Skrbinšek, T.; Jelenčič, M.; Boljte, B.; Konec, M.; Erich, M.; Sulică, B.; Moza, I.; Ungureanu, L.; Rohan, R.; et al. Report on Monitoring Brown Bears Using Non-Invasive DNA Sampling in the Romanian Carpathians Bear Report FOUNDATION CONSERVATION CARPATHIA. Available online: https://www.carpathia.org/wp-content/uploads/2021/10/FCC-Report-on-monitoring-brown-bear-using-non-invasive-DNA-sampling-in-the-Romanian-Carpathians.pdf (accessed on 29 December 2021).
- Berezowska-Cnota, T.; Luque-Márquez, I.; Elguero-Claramunt, I.; Bojarska, K.; Okarma, H.; Selva, N. Effectiveness of Different Types of Hair Traps for Brown Bear Research and Monitoring. PLoS ONE 2017, 12, e0186605. [Google Scholar] [CrossRef]
- Frosch, C.; Dutsov, A.; Zlatanova, D.; Valchev, K.; Reiners, T.E.; Steyer, K.; Pfenninger, M.; Nowak, C. Noninvasive Genetic Assessment of Brown Bear Population Structure in Bulgarian Mountain Regions. Mamm. Biol. 2014, 79, 268–276. [Google Scholar] [CrossRef]
- Pylidis, C.; Anijalg, P.; Saarma, U.; Dawson, D.A.; Karaiskou, N.; Butlin, R.; Mertzanis, Y.; Giannakopoulos, A.; Iliopoulos, Y.; Krupa, A.; et al. Multisource Noninvasive Genetics of Brown Bears (Ursus Arctos) in Greece Reveals a Highly Structured Population and a New Matrilineal Contact Zone in Southern Europe. Ecol. Evol. 2021, 11, 6427–6443. [Google Scholar] [CrossRef]
- Karamanlidis, A.A.; de Hernando, M.G.; Krambokoukis, L.; Gimenez, O. Evidence of a Large Carnivore Population Recovery: Counting Bears in Greece. J. Nat. Conserv. 2015, 27, 10–17. [Google Scholar] [CrossRef]
- Tsaparis, D.; Karaiskou, N.; Mertzanis, Y.; Triantafyllidis, A. Non-Invasive Genetic Study and Population Monitoring of the Brown Bear (Ursus Arctos) (Mammalia: Ursidae) in Kastoria Region–Greece. J. Nat. Hist. 2015, 49, 393–410. [Google Scholar] [CrossRef]
- Karamanlidis, A.A.; Drosopoulou, E.; de Hernando, M.G.; Georgiadis, L.; Krambokoukis, L.; Pllaha, S.; Zedrosser, A.; Scouras, Z. Noninvasive Genetic Studies of Brown Bears Using Power Poles. Eur. J. Wildl. Res. 2010, 56, 693–702. [Google Scholar] [CrossRef]
- Karamanlidis, A.A.; Stojanov, A.; de Gabriel Hernando, M.; Ivanov, G.; Kocijan, I.; Melovski, D.; Skrbinšek, T.; Zedrosser, A. Distribution and Genetic Status of Brown Bears in FYR Macedonia: Implications for Conservation. Acta Theriol. 2014, 59, 119–128. [Google Scholar] [CrossRef]
- Karamanlidis, A.A.; Pllaha, S.; Krambokoukis, L.; Shore, K.; Zedrosser, A. Preliminary Brown Bear Survey in Southeastern Albania. Ursus 2014, 25, 1–7. [Google Scholar] [CrossRef]
- Ciucci, P.; Gervasi, V.; Boitani, L.; Boulanger, J.; Paetkau, D.; Prive, R.; Tosoni, E. Estimating Abundance of the Remnant Apennine Brown Bear Population Using Multiple Noninvasive Genetic Data Sources. J. Mammal. 2015, 96, 206–220. [Google Scholar] [CrossRef]
- Lorenzini, R.; Posillico, M.; Lovari, S.; Petrella, A. Non-Invasive Genotyping of the Endangered Apennine Brown Bear: A Case Study Not to Let One’s Hair Down. Anim. Conserv. 2004, 7, 199–209. [Google Scholar] [CrossRef]
- Clevenger, A.P.; Purroy, F.J. Sign Surveys for Estimating Trend of a Remnant Brown Bear Ursus Arctos Population in Northern Spain. Wildl. Biol. 1996, 2, 275–281. [Google Scholar] [CrossRef]
- Pérez, T.; Vázquez, F.; Naves, J.; Fernández, A.; Corao, A.; Albornoz, J.; Domínguez, A. Non-Invasive Genetic Study of the Endangered Cantabrian Brown Bear (Ursus Arctos). Conserv. Genet. 2009, 10, 291–301. [Google Scholar] [CrossRef]
- Sentilles, J.; Vanpé, C.; Quenette, P.-Y. Benefits of Incorporating a Scat-Detection Dog into Wildlife Monitoring: A Case Study of Pyrenean Brown Bear. J. Vertebr. Biol. 2021, 69. [Google Scholar] [CrossRef]
- Phoebus, I.; Boulanger, J.; Eiken, H.G.; Fløystad, I.; Graham, K.; Hagen, S.B.; Sorensen, A.; Stenhouse, G. Comparison of Grizzly Bear Hair-Snag and Scat Sampling along Roads to Inform Wildlife Population Monitoring. Wildl. Biol. 2020, 2020. [Google Scholar] [CrossRef]
- Wasser, S.K.; Davenport, B.; Ramage, E.R.; Hunt, K.E.; Parker, M.; Clarke, C.; Stenhouse, G. Scat Detection Dogs in Wildlife Research and Management: Application to Grizzly and Black Bears in the Yellowhead Ecosystem, Alberta, Canada. Can. J. Zool. 2004, 82, 475–492. [Google Scholar] [CrossRef]
- Boulanger, J.; Proctor, M.; Himmer, S.; Stenhouse, G.; Paetkau, D.; Cranston, J. An Empirical Test of DNA Mark-Recapture Sampling Strategies for Grizzly Bears. Ursus 2006, 17, 149–158. [Google Scholar] [CrossRef]
- Woods, J.G.; Paetkau, D.; Lewis, D.; McLellan, B.N.; Proctor, M.; Strobeck, C. Genetic Tagging of Free-Ranging Black and Brown Bears. Wild. Soc. Bull. 1999, 27, 616–627. [Google Scholar]
- Roy, J.; Yannic, G.; Côté, S.D.; Bernatchez, L. Negative Density-Dependent Dispersal in the American Black Bear (Ursus Americanus) Revealed by Noninvasive Sampling and Genotyping. Ecol. Evol. 2012, 2, 525–537. [Google Scholar] [CrossRef]
- Wirsing, A.J.; Quinn, T.P.; Adams, J.R.; Waits, L.P. Optimizing Selection of Brown Bear Hair for Noninvasive Genetic Analysis. Wild. Soc. Bull. 2020, 44, 94–100. [Google Scholar] [CrossRef]
- Beier, L.R.; Lewis, S.B.; Flynn, R.W.; Pendleton, G.; Schumacher, V.T. A Single-Catch Snare to Collect Brown Bear Hair for Genetic Mark–Recapture Studies. Wild. Soc. Bull. 2005, 33, 766–773. [Google Scholar] [CrossRef]
- Robinson, S.J.; Waits, L.P.; Martin, I.D. Estimating Abundance of American Black Bears Using DNA-Based Capture-Mark-Recapture Models. Ursus 2009, 20, 1–11. [Google Scholar] [CrossRef]
- Wheat, R.E.; Allen, J.M.; Miller, S.D.L.; Wilmers, C.C.; Levi, T. Environmental DNA from Residual Saliva for Efficient Noninvasive Genetic Monitoring of Brown Bears (Ursus Arctos). PLoS ONE 2016, 11, e0165259. [Google Scholar] [CrossRef] [PubMed]
- Kendall, K.C.; Stetz, J.B.; Boulanger, J.; Macleod, A.C.; Paetkau, D.; White, G.C. Demography and Genetic Structure of a Recovering Grizzly Bear Population. J. Wildl. Manag. 2009, 73, 3–17. [Google Scholar] [CrossRef]
- Gardner, B.; Royle, J.A.; Wegan, M.T.; Rainbolt, R.E.; Curtis, P.D. Estimating Black Bear Density Using DNA Data From Hair Snares. J. Wildl. Manag. 2010, 74, 318–325. [Google Scholar] [CrossRef]
- Triant, D.A.; Pace, R.M.; Stine, M. Abundance, Genetic Diversity and Conservation of Louisiana Black Bears (Ursus Americanus Luteolus) as Detected through Noninvasive Sampling. Conserv. Genet. 2004, 5, 647–659. [Google Scholar] [CrossRef]
- Boersen, M.R.; Clark, J.D.; King, T.L. Estimating Black Bear Population Density and Genetic Diversity at Tensas River, Louisiana Using Microsatellite DNA Markers. Wildl. Soc. Bull. 2003, 31, 197–207. [Google Scholar] [CrossRef]
- Dreher, B.P.; Winterstein, S.R.; Scribner, K.T.; Lukacs, P.M.; Etter, D.R.; Rosa, G.J.M.; Lopez, V.A.; Libants, S.; Filcek, K.B. Noninvasive Estimation of Black Bear Abundance Incorporating Genotyping Errors and Harvested Bear. J. Wildl. Manag. 2007, 71, 2684–2693. [Google Scholar] [CrossRef]
- Murphy, S.M.; Cox, J.J.; Augustine, B.C.; Hast, J.T.; Guthrie, J.M.; Wright, J.; McDermott, J.; Maehr, S.C.; Plaxico, J.H. Characterizing Recolonization by a Reintroduced Bear Population Using Genetic Spatial Capture–Recapture. J. Wildl. Manag. 2016, 80, 1390–1407. [Google Scholar] [CrossRef]
- Sun, C.C.; Fuller, A.K.; Hare, M.P.; Hurst, J.E. Evaluating Population Expansion of Black Bears Using Spatial Capture-Recapture. J. Wildl. Manag. 2017, 81, 814–823. [Google Scholar] [CrossRef]
- Bellemain, E.; Nawaz, M.A.; Valentini, A.; Swenson, J.E.; Taberlet, P. Genetic Tracking of the Brown Bear in Northern Pakistan and Implications for Conservation. Biol. Conserv. 2007, 134, 537–547. [Google Scholar] [CrossRef]
- Mccarthy, T.M.; Waits, L.P.; Mijiddorj, B. Status of the Gobi Bear in Mongolia as Determined by Noninvasive Genetic Methods. Ursus 2009, 20, 30–38. [Google Scholar] [CrossRef]
- Ambarli, H.; Mengüllüoğlu, D.; Fickel, J.; Förster, D.W. Population Genetics of the Main Population of Brown Bears in Southwest Asia. PeerJ 2018, 2018, e5660. [Google Scholar] [CrossRef]
- Tee, T.L.; Lai, W.L.; Ju Wei, T.K.; Shern, O.Z.; van Manen, F.T.; Sharp, S.P.; Wong, S.; Chew, J.; Ratnayeke, S. An Evaluation of Noninvasive Sampling Techniques for Malayan Sun Bears. Ursus 2020, 2020, 1–12. [Google Scholar] [CrossRef]
- Saito, M.; Yamauchi, K.; Aoi, T. Individual Identification of Asiatic Black Bears Using Extracted DNA from Damaged Crops. Ursus 2008, 19, 162–167. [Google Scholar] [CrossRef]
- Andreassen, R.; Schregel, J.; Kopatz, A.; Tobiassen, C.; Knappskog, P.M.; Hagen, S.B.; Kleven, O.; Schneider, M.; Kojola, I.; Aspi, J.; et al. A Forensic DNA Profiling System for Northern European Brown Bears (Ursus Arctos). Forensic Sci. Int. Genet. 2012, 6, 798–809. [Google Scholar] [CrossRef]
- Skrbinšek, T.; Jelenčič, M.; Boljte, B.; Konec, M.; Erich, M.; Iosif, R.; Moza, I.; Promberger, B. Report on Analysis of Genetic Samples Collected in 2017–2018 on Brown Bears (Ursus Arctos), Eurasian Lynx (Lynx Lynx) and Grey Wolf (Canis Lupus) in a Pilot Area in Southern Carpathians, Romania. Available online: https://www.carpathia.org/wp-content/uploads/2019/09/FCC2017.2018.FinalReport.Ver1_.1.pdf (accessed on 29 December 2021).
- Lynam, A.J.; Rabinowitz, A.; Myint, T.; Maung, M.; Latt, K.T.; Po, S.H.T. Estimating Abundance with Sparse Data: Tigers in Northern Myanmar. Popul. Ecol. 2009, 51, 115–121. [Google Scholar] [CrossRef]
- Breck, S. Sampling Rare or Elusive Species: Concepts, Designs, and Techniques for Estimating Population Parameters by William L. Thompson. Wildl. Soc. Bull. 2006, 897–998. [Google Scholar] [CrossRef]
- Wold, K.; Wirsing, A.J.; Quinn, T.P. Do Brown Bears Ursus Arctos Avoid Barbed Wires Deployed to Obtain Hair Samples? A Videographic Assessment. Wildl. Biol. 2020, 2020. [Google Scholar] [CrossRef]
- Karamanlidis, A.A.; Youlatos, D.; Sgardelis, S.; Scouras, Z. Using Sign at Power Poles to Document Presence of Bears in Greece. Ursus 2007, 18, 54–61. [Google Scholar] [CrossRef]
- Waits, L.P.; Paetkau, D. Noninvasive Genetic Sampling Tools for Wildlife Biologists: A Review of Applications and Recommendations for Accurate Data Collection. J. Wildl. Manag. 2005, 69, 1419–1433. [Google Scholar] [CrossRef]
- De Barba, M.; Miquel, C.; Lobréaux, S.; Quenette, P.Y.; Swenson, J.E.; Taberlet, P. High-Throughput Microsatellite Genotyping in Ecology: Improved Accuracy, Efficiency, Standardization and Success with Low-Quantity and Degraded DNA. Mol. Ecol. Resour. 2016, 17, 492–507. [Google Scholar] [CrossRef]
- Nsubuga, A.M.; Robbins, M.M.; Roeder, A.D.; Morin, P.A.; Boesch, C.; Vigilant, L. Factors Affecting the Amount of Genomic DNA Extracted from Ape Faeces and the Identification of an Improved Sample Storage Method. Mol. Ecol. 2004, 13, 2089–2094. [Google Scholar] [CrossRef]
- Sentilles, J.; Delrieu, N.; Quenette, P.Y. Un Chien Pour La Détection de Fèces: Premiers Résultats Pour Le Suivi de l Ours Brun Dans Les Pyrénées. Faune Sauvag. 2016, 312, 22–26. [Google Scholar]
- Stephanie, B.; Jenny, K.; Helen, S.; Nils, B.; Kathryn, J.J.; Etienne, F.; Akomo, O.; Rob Ogden, R.M. Improving Cost-Efficiency of Faecal Genotyping_ New Tools for Elephant Species. PLoS ONE 2019. [Google Scholar] [CrossRef]
- Petchey, A.; Gray, A.; Andrén, C.; Skelton, T.; Kubicki, B.; Allen, C.; Jehle, R. Characterisation of 9 Polymorphic Microsatellite Markers for the Critically Endangered Lemur Leaf Frog Agalychnis Lemur. Conserv. Genet. Res. 2014, 6, 971–973. [Google Scholar] [CrossRef]
- Werhahn, G.; Senn, H.; Kaden, J.; Joshi, J.; Bhattarai, S.; Kusi, N.; Sillero-Zubiri, C.; Macdonald, D.W. Phylogenetic Evidence for the Ancient Himalayan Wolf: Towards a Clarification of Its Taxonomic Status Based on Genetic Sampling from Western Nepal. R. Soc. Open Sci. 2017, 4, 170186. [Google Scholar] [CrossRef]
- Murphy, M.A.; Waits, L.P.; Kendall, K.C. The Influence of Diet on Faecal DNA Amplification and Sex Identification in Brown Bears (Ursus Arctos). Mol. Ecol. 2003, 12, 2261–2265. [Google Scholar] [CrossRef]
- Ross, S.; Costanzi, J.M.; al Jahdhami, M.; al Rawahi, H.; Ghazali, M.; Senn, H. First Evaluation of the Population Structure, Genetic Diversity and Landscape Connectivity of the Endangered Arabian Tahr. Mamm. Biol. 2020, 100, 659–673. [Google Scholar] [CrossRef]
- Planella, A.; Jiménez, J.; Palomero, G.; Ballesteros, F.; Blanco, J.C.; López-Bao, J.V. Integrating Critical Periods for Bear Cub Survival into Temporal Regulations of Human Activities. Biol. Conserv. 2019, 236, 489–495. [Google Scholar] [CrossRef]
- Huber, S.; Bruns, U.; Arnold, W. Sex Determination of Red Deer Using Polymerase Chain Reaction of DNA from Feces on JSTOR. Wildl. Soc. Bull. 2002, 30, 208–212. [Google Scholar]
- Murphy, M.A.; Kendall, K.C.; Robinson, A.; Waits, L.P. The Impact of Time and Field Conditions on Brown Bear (Ursus Arctos) Faecal DNA Amplification. Conserv. Genet. 2007, 8, 1219–1224. [Google Scholar] [CrossRef]
- Brinkman, T.J.; Schwartz, M.K.; Person, D.K.; Pilgrim, K.L.; Hundertmark, K.J. Effects of Time and Rainfall on PCR Success Using DNA Extracted from Deer Fecal Pellets. Conserv. Genet. 2009, 11, 1547–1552. [Google Scholar] [CrossRef]
- Kendall, K.C.; McKelvey, K.S. Hair Collection Methods. In Noninvasive Survey Methods for North American Carnivores; Island Press: Washington, DC, USA, 2008; pp. 135–176. ISBN 978-1-59726-119-7. [Google Scholar]
- Fusaro, J.L.; Conner, M.M.; Conover, M.R.; Taylor, T.J.; Kenyon, M.W. Best Management Practices in Counting Urban Black Bears. Hum. Wildl. Interact. 2017, 11, 64–77. [Google Scholar] [CrossRef]
- McLellan, M.L.; McLellan, B.N.; Sollmann, R.; Lamb, C.T.; Apps, C.D.; Wittmer, H.U. Divergent Population Trends Following the Cessation of Legal Grizzly Bear Hunting in Southwestern British Columbia, Canada. Biol. Conserv. 2019, 233, 247–254. [Google Scholar] [CrossRef]
- Gurney, S.M.; Smith, J.B.; Etter, D.R.; Williams, D.M. American Black Bears and Hair Snares: A Behavioral Analysis. Ursus 2020, 2020, 1–9. [Google Scholar] [CrossRef]
- Stetz, J.B.; Seitz, T.; Sawaya, M.A. Effects of Exposure on Genotyping Success Rates of Hair Samples from Brown and American Black Bears. J. Fish. Wildl. Manag. 2015, 6, 191–198. [Google Scholar] [CrossRef][Green Version]
- Sato, Y.; Kamiishi, C.; Tokaji, T.; Mori, M.; Koizumi, S.; Kobayashi, K.; Itoh, T.; Sonohara, W.; Takada, M.B.; Urata, T. Selection of Rub Trees by Brown Bears (Ursus Arctos) in Hokkaido, Japan. Acta Theriol. 2014, 59, 129–137. [Google Scholar] [CrossRef]
- Mowat, G.; Strobeck, C. Estimating Population Size of Grizzly Bears Using Hair Capture, DNA Profiling, and Mark-Recapture Analysis. J. Wildl. Manag. 2000, 64, 183. [Google Scholar] [CrossRef]
- Haroldson, M.A.; Gunther, K.A.; Reinhart, D.P.; Podruzny, S.R.; Cegelski, C.; Waits, L.; Wyman, T.; Smith, J. Changing Numbers of Spawning Cutthroat Trout in Tributary Streams of Yellowstone Lake and Estimates of Grizzly Bears Visiting Streams from DNA. Ursus 2005, 16, 167–180. [Google Scholar] [CrossRef]
- Evans, M.J.; Hawley, J.E.; Rego, P.W.; Rittenhouse, T.A.G. Hourly Movement Decisions Indicate How a Large Carnivore Inhabits Developed Landscapes. Oecologia 2018, 190, 11–23. [Google Scholar] [CrossRef]
- Paetkau, D.; Amstrup, S.C.; Born, E.W.; Calvert, W.; Derocher, A.E.; Garner, G.W.; Messier, F.; Stirling, I.; Taylor, M.K.; Wiig, O.; et al. Genetic Structure of the World’s Polar Bear Populations. Mol. Ecol. 1995, 9, 84. [Google Scholar] [CrossRef]
- Robinson, S.J.; Waits, L.P.; Martin, I.D. Evaluating Population Structure of Black Bears on the Kenai Peninsula Using Mitochondrial and Nuclear DNA Analyses. J. Mammal. 2007, 88, 1288–1299. [Google Scholar] [CrossRef]
- Shih, C.C.; Wu, S.L.; Hwang, M.H.; Lee, L.L. Evaluation on the Effects of Ageing Factor, Sampling and Preservation Methods on Asiatic Black Bear (Ursus Thibetanus) Noninvasive DNA Amplification. Taiwania 2017, 62, 363–370. [Google Scholar] [CrossRef]
- Skrbinšek, T. Collecting Lynx Noninvasive Genetic Samples: Instruction Manual for Field Personnel and Volunteers; Biotechnical Faculty, University of Ljubljana: Ljubljana, Slovenia, 2017. [Google Scholar]
- Puchkovskiy, S.V. Selectivity of Tree Species as Activity Target of Brown Bear in Taiga. Contemp. Probl. Ecol. 2009, 2, 260–268. [Google Scholar] [CrossRef]
- Green, G.I.; Mattson, D.J. Tree Rubbing by Yellowstone Grizzly Bears Ursus Arctos. Wildl. Biol. 2003, 9, 1–9. [Google Scholar] [CrossRef]
- Clapham, M.; Nevin, O.T.; Ramsey, A.D.; Rosell, F. The Function of Strategic Tree Selectivity in the Chemical Signalling Ofbrown Bears. Anim. Behav. 2013, 85, 1351–1357. [Google Scholar] [CrossRef]
- Lamb, C.T.; Walsh, D.A.; Mowat, G. Factors Influencing Detection of Grizzly Bears at Genetic Sampling Sites. Ursus 2016, 27, 31–44. [Google Scholar] [CrossRef]
- Rogers, L.L. Effects of Food Supply and Kinship on Social Behavior, Movements, and Population Growth of Black Bears in Northeastern Minnesota. Wildl. Monogr. 1987, 97, 3–72. [Google Scholar]
- Gabrielsen, C.G.; Kovach, A.I.; Babbitt, K.J.; McDowell, W.H. Limited Effects of Suburbanization on the Genetic Structure of an Abundant Vernal Pool-Breeding Amphibian. Conserv. Genet. 2013, 14, 1083–1097. [Google Scholar] [CrossRef]
- Gorospe, K.D.; Karl, S.A. Genetic Relatedness Does Not Retain Spatial Pattern across Multiple Spatial Scales: Dispersal and Colonization in the Coral, Pocillopora Damicornis. Mol. Ecol. 2013, 22, 3721–3736. [Google Scholar] [CrossRef]
- Keller, D.; Holderegger, R.; van Strien, M.J. Spatial Scale Affects Landscape Genetic Analysis of a Wetland Grasshopper. Mol. Ecol. 2013, 22, 2467–2482. [Google Scholar] [CrossRef]
- Nellemann, C.; Støen, O.G.; Kindberg, J.; Swenson, J.E.; Vistnes, I.; Ericsson, G.; Katajisto, J.; Kaltenborn, B.P.; Martin, J.; Ordiz, A. Terrain Use by an Expanding Brown Bear Population in Relation to Age, Recreational Resorts and Human Settlements. Biol. Conserv. 2007, 138, 157–165. [Google Scholar] [CrossRef]
- Piédallu, B.; Quenette, P.-Y.; Jordana, I.A.; Bombillon, N.; Gastineau, A.; Jato, R.; Miquel, C.; Muñoz, P.; Palazón, S.; Solà de la Torre, J.; et al. Better Together: A Transboundary Approach to Brown Bear Monitoring in the Pyrenees. bioRxiv 2016, 075663. [Google Scholar] [CrossRef]
- Matosiuk, M.; Śmietana, W.; Czajkowska, M.; Paule, L.; Štofik, J.; Krajmerová, D.; Bashta, A.T.; Jakimiuk, S.; Ratkiewicz, M. Genetic Differentiation and Asymmetric Gene Flow among Carpathian Brown Bear (Ursus Arctos) Populations—Implications for Conservation of Transboundary Populations. Ecol. Evol. 2019, 9, 1501–1511. [Google Scholar] [CrossRef]
- Zingstra, H.; Kovachev, A.; Kitnaes, K.; Tzonev, R.; Dimova, D.; Tzvetkov, P. Guidelines for Assessing Favorable Conservation Status of Natura 2000 Species and Habitat Types in Bulgaria 2009. Available online: https://library.wur.nl/WebQuery/wurpubs/fulltext/247891 (accessed on 29 December 2021).
- Taberlet, P.; Swenson, J.E.; Sandegren, F.; Bjarvall, A. Localization of a Contact Zone between Two Highly Divergent Mitochondrial DNA Lineages of the Brown Bear Ursus Arctos in Scandinavia. Conserv. Biol. 1995, 9, 1255–1261. [Google Scholar] [CrossRef]
- Bidon, T.; Janke, A.; Fain, S.R.; Eiken, H.G.; Hagen, S.B.; Saarma, U.; Hallström, B.M.; Lecomte, N.; Hailer, F. Brown and Polar Bear Y Chromosomes Reveal Extensive Male-Biased Gene Flow within Brother Lineages. Mol. Biol. Evol. 2014, 31, 1353–1363. [Google Scholar] [CrossRef]
- Støen, O.G.; Zedrosser, A.; Sæbø, S.; Swenson, J.E. Inversely Density-Dependent Natal Dispersal in Brown Bears Ursus Arctos. Oecologia 2006, 148, 356–364. [Google Scholar] [CrossRef]
- Bischof, R.; Milleret, C.; Dupont, P.; Chipperfield, J.; Tourani, M.; Ordiz, A.; de Valpine, P.; Turek, D.; Andrew Royle, J.; Gimenez, O.; et al. Estimating and Forecasting Spatial Population Dynamics of Apex Predators Using Transnational Genetic Monitoring. Proc. Natl. Acad. Sci. USA 2020, 117, 30531–30538. [Google Scholar] [CrossRef]
- Mills, L.S.; Citta, J.J.; Lair, K.P.; Schwartz, M.K.; Tallmon, D.A. Estimating Animal Abundance Using Noninvasive DNA Sampling: Promise and Pitfalls. Ecol. Appl. 2000, 10, 283. [Google Scholar] [CrossRef]
- Bellott, D.W.; Hughes, J.F.; Skaletsky, H.; Brown, L.G.; Pyntikova, T.; Cho, T.-J.; Koutseva, N.; Zaghlul, S.; Graves, T.; Rock, S.; et al. Mammalian Y Chromosomes Retain Widely Expressed Dosage-Sensitive Regulators. Nature 2014, 508, 494–499. [Google Scholar] [CrossRef]
- Hollerbach, L.; Heurich, M.; Reiners, T.E.; Nowak, C. Detection Dogs Allow for Systematic Non-Invasive Collection of DNA Samples from Eurasian Lynx. Mamm. Biol. 2018, 90, 42–46. [Google Scholar] [CrossRef]
- Gonzalez, E.G.; Blanco, J.C.; Ballesteros, F.; Alcaraz, L.; Palomero, G.; Doadrio, I. Genetic and Demographic Recovery of an Isolated Population of Brown Bear Ursus Arctos L., 1758. PeerJ 2016, 2016, e1928. [Google Scholar] [CrossRef] [PubMed]
- Goossens, B.; Waits, L.P.; Taberlet, P. Plucked Hair Samples as a Source of DNA: Reliability of Dinucleotide Microsatellite Genotyping. Mol. Ecol. 1998, 7, 1237–1241. [Google Scholar] [CrossRef] [PubMed]
- Ebert, C.; Knauer, F.; Storch, I.; Hohmann, U. Individual Heterogeneity as a Pitfall in Population Estimates Based on Non-Invasive Genetic Sampling: A Review and Recommendations. Wildl. Biol. 2010, 16, 225–240. [Google Scholar] [CrossRef]
| Category | Peer- Reviewed | Guidelines | Reports | Action Plans |
|---|---|---|---|---|
| N | 140 | 3 | 3 | 2 |
| Total | 148 | |||
| Location | Population | Sampling Scheme *1 | N Samples *2 | Temporal Extent | Study |
|---|---|---|---|---|---|
| Europe | |||||
| Northern Europe (E-W) | Scandinavian brown bear | M, SS | 3365 F, H, T | 2001, 2002, 2004, 2006 | [62] |
| Northern Europe (Se, Norw, Fi, Karelia) | Scandinavian brown bear | Dataset obtained from [63] | 2005–2017 | [64] | |
| Northern Europe | Scandinavian brown bear | Dataset obtained from [65] | [66] | ||
| Northern Europe | Scandinavian brown bear | Dataset obtained from regional monitoring programs | 2006–2013 | [65] | |
| Northern Europe | Scandinavian brown bear | Dataset obtained from regional monitoring programs | 2006–2012 | [67] | |
| Sweden | Scandinavian brown bear | Dataset obtained from [55] | 2001–2002 | [68] | |
| Sweden | Scandinavian brown bear | OS | 1904 F | 2001–2002 | [63] |
| Sweden | Scandinavian brown bear | OS | 5185 F | [69] | |
| Slovenia (south) | Dinaric brown bear | Dataset obtained from a pilot study | 2004–2007 | [59] | |
| Slovenia | Dinaric brown bear | SS | 1053 F | 2007–2008 | [70] |
| M, SS, OS | 4687 | 2015 | [71] | ||
| Slovenia | Dinaric brown bear | Dataset obtained from regional and national studies | 2007 | [72] | |
| Slovenia | Dinaric brown bear | CM, OS, SS | 2007 | [73] | |
| Carpathian brown bear | M, OS | 339 T, B, F, H, bones | 2004–2009 | [74] | |
| Slovakia | Carpathian brown bear | 76 H, F | 2005–2006 | [75] | |
| Slovakia | Carpathian brown bear | 140 F, H | 2007–2008, 2010 | [76] | |
| Romania | Carpathian brown bear | HT, OS, SS | 1426 F, H | 2017–2018 | [77] |
| Poland | Carpathian brown bear | HT, SS | 858 H | 2010 | [78] |
| Bulgaria | Eastern Balkan brown bear | HT, CM, OS, M | 355 F, H | 2004–2008, 2009–2012 | [79] |
| Greece | Eastern Balkan brown bear | HT, TS, CM, M, SS | 382 H, F, B | 2006–2010 | [80] |
| Greece | Eastern Balkan brown bear | HT, SS | 860 H | 2007–2010 | [81] |
| Greece (Kastoria) | Eastern Balkan brown bear | HT, OS | 232 H, F, B | 2011 | [82] |
| GR, FYROM, ALB | Eastern Balkan brown bear | HT, SS | 191 H | [83] | |
| FYR Macedonia | Eastern Balkan brown bear | HT, OC, SS | 106 H | 2008–2009 | [84] |
| Albania | Eastern Balkan brown bear | HT, M, TS, SS | 12 H | 2008–2009 | [85] |
| HT, OC, OS, SS | 643 H | 2011 | [86] | ||
| Italy | Apennine Brown Bear | TS, M, OS | 80 H, F, T | 1991–2002 | [87] |
| HT, TS, OS | 1164 F, H | 2003–2004 | [33] | ||
| Italy (Alps) | Alp Brown Bear | HT, TS, CM, M, OS, SS | 2781 F, H | 2002, 2003–2008 | [51] |
| Spain | TS, SS | 96 F | 1990–1992 | [88] | |
| Spain | CM, M, OS, SS | 133 F, H, B, T | 2004–2006 | [89] | |
| Spain | Cantabrian Brown Bear | TS, SS | 151 F, H | 2017 | [31] |
| France | Pyrenean Brown Bear | TS, OS, SS | 153 F | 2014–2019 | [90] |
| North America | |||||
| Alberta, Canada | Grizzly | HT, TS, OS, SS | 183 F, 958 H | 2016 | [91] |
| TS, SS | 880 F | 1999, 2001 | [92] | ||
| Alberta, Canada | Grizzly | HT, SS | 3363 H | 2004 | [93] |
| BNP, Canada | Grizzly and American black bear | HT, CM | 6236 H, T | 2006–2008 | [36] |
| BC, Canada | American black and brown bears | HT, SS | 447 H | 1995 | [94] |
| Quebec, Canada | American black bear | HT, SS | 411 H | 2005 | [95] |
| Alaska | Brown bear | HT, SS | 2245 H | 2014 - 2017 | [96] |
| Alaska | Grizzly bear | HT, SS | 466 H | 2002–2003 | [97] |
| HT, SS | 345 H | 2003–2005 | [98] | ||
| Alaska | Brown bear | TS, OS, SS | 428 F, saliva | 2014 | [99] |
| Montana, USA | Grizzly bear | HT, SS | 33741 H | 2004 | [100] |
| Northern New York, USA | American black bear | HT, SS | 2006 | [101] | |
| Louisiana, USA | Louisiana black bear | HT, SS | 922 H | 1999 | [102] |
| Louisiana, USA | Louisiana black bear | OS | 448 H | 1999 | [103] |
| NLP, Michigan, USA | American black bear | HT, SS | 1564 H, T | 2003 | [104] |
| Kentucky–Virginia, USA | American black bear | HT, SS | 1503 H | 2012–2013 | [105] |
| New York, USA | American black bear | HT, SS | 1985 H | 2012 | [106] |
| North Carolina, USA | American black bear | HT, SS | 468 H | 2001–2002 | [61] |
| Asia | |||||
| Pakistan | Brown bear | TS, SS | 136F | 2004 | [107] |
| HT, OS | 272 H | 2008 | [37] | ||
| Mongolia | Gobi bear | HT | 200 H | 1996–1998 | [108] |
| GKM, Turkey | Brown bear | CM, M, OS | 154 H, T | 2008–2014 | [109] |
| Malaysia | Malayan sun bear | HT | 69 H | 2017, 2019 | [110] |
| Japan | Asiatic black bear | OC | 99 corn-bite samples | 2004 | [111] |
| Hair Trap | Specifications | Bait and/or Lure | Location | Study |
|---|---|---|---|---|
| Hair corral | At least a single strand of barbed wire stretched around 4 or more trees at 50–55 cm above ground | yes | Italy, Poland, Malaysia, Turkey, California, Michigan, Montana, Alberta, BC, Quebec (Canada) | [33,35,36,78,86,94,109,110,132,133,134] |
| Adhesive rub stations | Tree trunk or wooden blocks wrapped with duct tape | yes | Malaysia | [110] |
| Power poles | Covered with barbed wire | yes, not on purpose | Greece, Albania, FYR Macedonia, Turkey, Montana | [81,82,83,85,109,117,135] |
| Natural rubs (bear rub trees) | equipped hair snagging devices (e.g., barbed wire) | no | Italy, Greece, Bulgaria, Romania, Poland, Alberta, Montana, BC, California, Alaska, Mongolia, Japan, Russian Far East, | [37,77,78,108,132,133,135,136] |
| Path traps | Barbed wire installed across known bear travel routes or at feeding routes | no | Italy, Poland, Alaska, Yellowstone Lake, | [37,77,78,79,108,133] |
| Modified hair snares | Barbed wire constructed in such way, that allows the bear to escape but keeps hair samples while doing it, and it disables after the process | no | Southeast Alaska | [97] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baciu, I.; Fedorca, A.; Ionescu, G. Noninvasive Genetics Knowledge from the Brown Bear Populations to Assist Biodiversity Conservation. Diversity 2022, 14, 121. https://doi.org/10.3390/d14020121
Baciu I, Fedorca A, Ionescu G. Noninvasive Genetics Knowledge from the Brown Bear Populations to Assist Biodiversity Conservation. Diversity. 2022; 14(2):121. https://doi.org/10.3390/d14020121
Chicago/Turabian StyleBaciu, Iulia, Ancuta Fedorca, and Georgeta Ionescu. 2022. "Noninvasive Genetics Knowledge from the Brown Bear Populations to Assist Biodiversity Conservation" Diversity 14, no. 2: 121. https://doi.org/10.3390/d14020121
APA StyleBaciu, I., Fedorca, A., & Ionescu, G. (2022). Noninvasive Genetics Knowledge from the Brown Bear Populations to Assist Biodiversity Conservation. Diversity, 14(2), 121. https://doi.org/10.3390/d14020121







