Evaluation of Genetic Diversity and Parasite-Mediated Selection of MHC Class I Genes in Emberiza godlewskii (Passeriformes: Emberizidae)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. DNA and RNA Extraction
2.3. PCR Amplification and NGS Sequencing
2.4. MHC Allele Genotyping and Classification
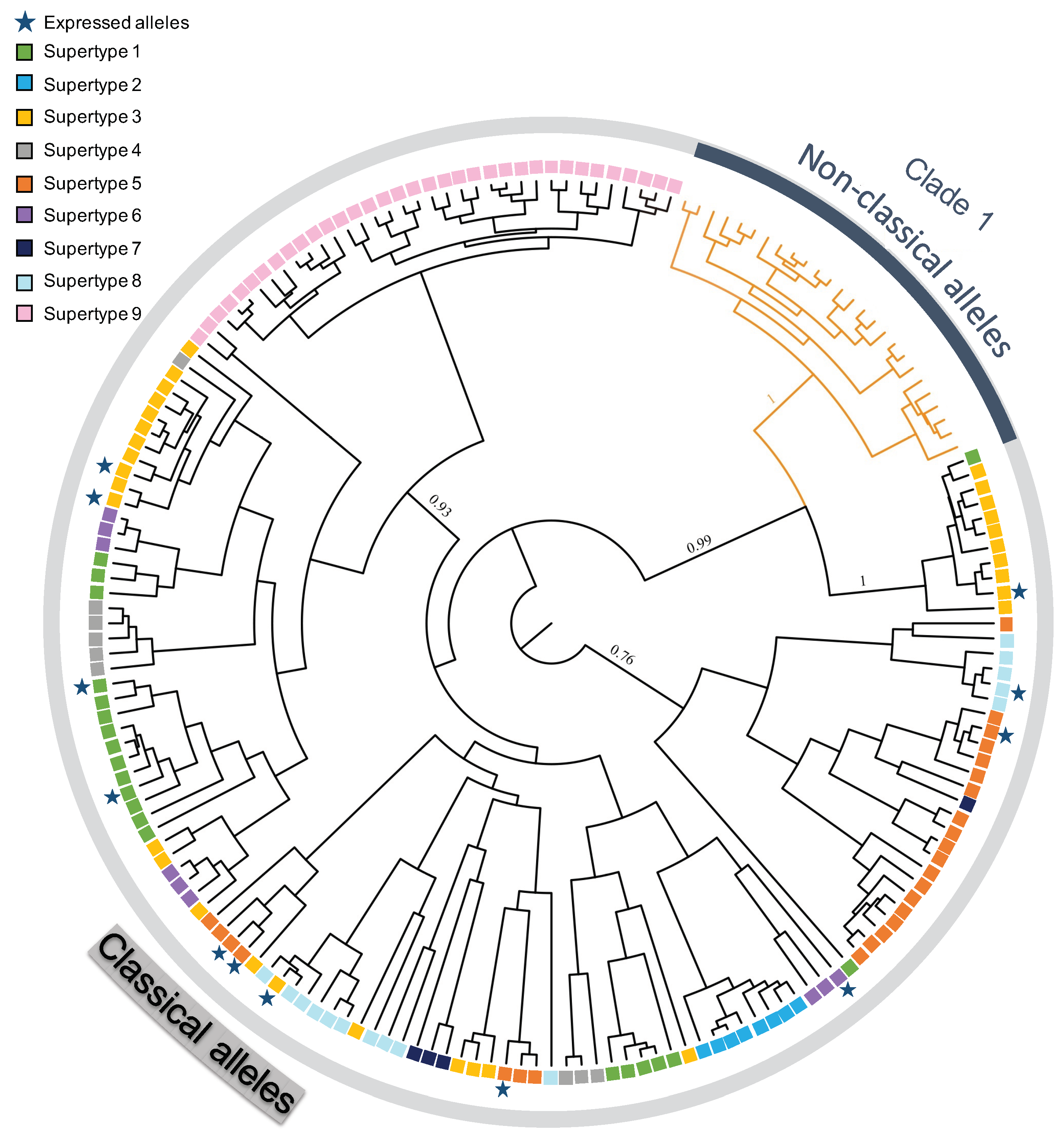
2.5. Functional Allele Identification and Phylogenetic Analysis
2.6. Historical Selection on MHC Alleles
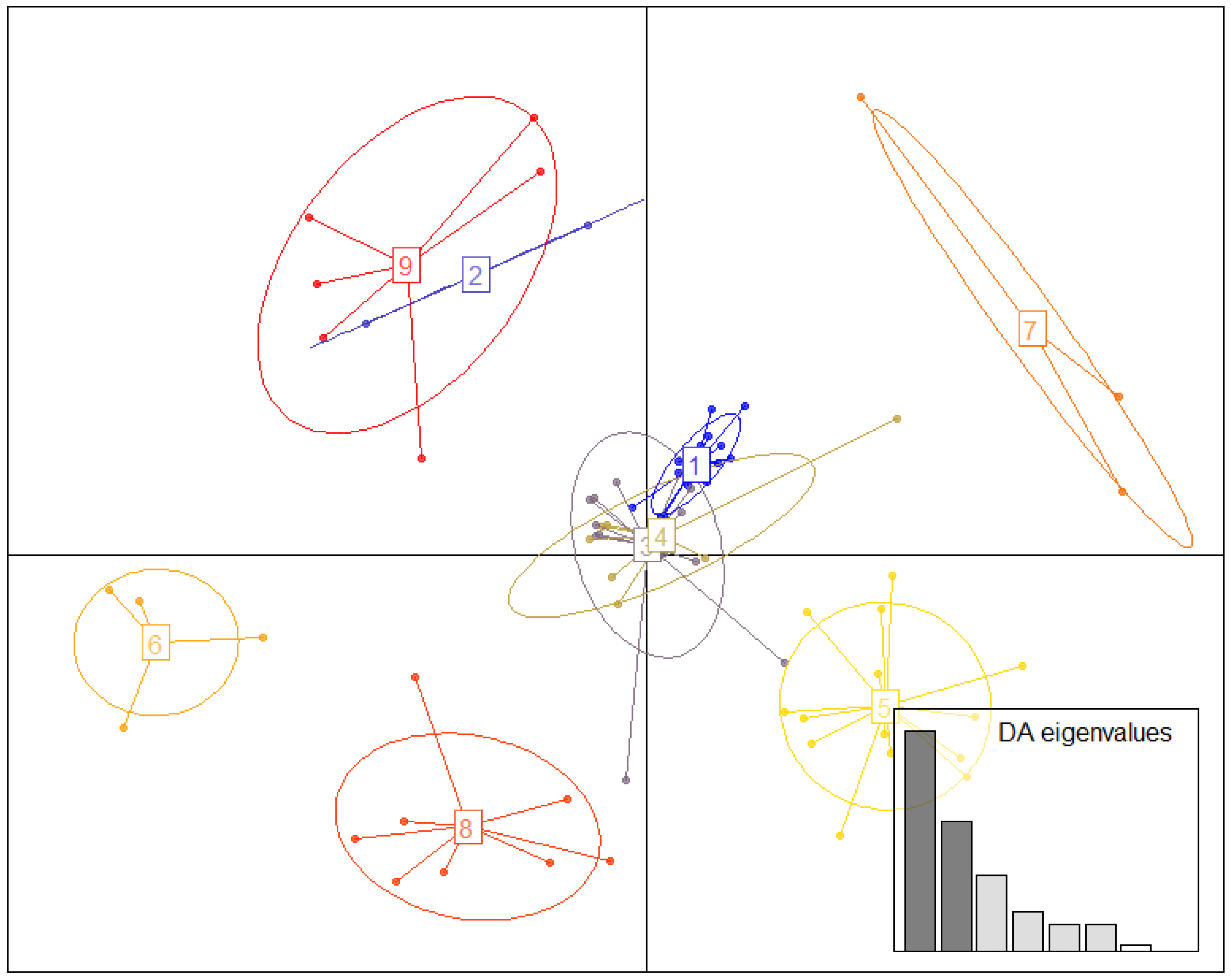
2.7. Supertype Identification
2.8. Avian Malaria Identification
2.9. Association between MHC Variation and Malaria Parasites
3. Results
3.1. MHC Allele Diversity and Classification
3.2. Historical Selection Analysis
3.3. Association between MHC Variation and Malaria Parasites
4. Discussion
4.1. MHC Variation in Godlewski’s Bunting
4.2. Classical and Non-Classical MHC Alleles
4.3. Selection on MHC Genes in Godlewski’s Buntings
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pilosof, S.; Fortuna, M.A.; Cosson, J.F.; Galan, M.; Kittipong, C.; Ribas, A.; Segal, E.; Krasnov, B.R.; Morand, S.; Bascompte, J. Host–parasite network structure is associated with community-level immunogenetic diversity. Nat. Commun. 2014, 5, 5172. [Google Scholar] [CrossRef] [PubMed]
- Bernatchez, L.; Landry, C. MHC studies in nonmodel vertebrates: What have we learned about natural selection in 15 years? J. Evol. Biol. 2003, 16, 363–377. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.J.; Reale, D.; Clements, M.N.; Morrissey, M.M.; Postma, E.; Walling, C.A.; Kruuk, L.E.B.; Nussey, D.H. An ecologist’s guide to the animal model. J. Anim. Ecol. 2010, 79, 13–26. [Google Scholar] [CrossRef]
- Piertney, S.B.; Oliver, M.K. The evolutionary ecology of the major histocompatibility complex. Heredity 2006, 96, 7–21. [Google Scholar] [CrossRef]
- Edwards, S.V.; Hedrick, P.W. Evolution and ecology of MHC molecules: From genomics to sexual selection. Trends Ecol. Evol. 1998, 13, 305–311. [Google Scholar] [CrossRef]
- Spurgin, L.G.; Richardson, D.S. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc. R. Soc. B-Biol. Sci. 2010, 277, 979–988. [Google Scholar] [CrossRef]
- Radwan, J.; Babik, W.; Kaufman, J.; Lenz, T.L.; Winternitz, J. Advances in the Evolutionary Understanding of MHC Polymorphism. Trends Genet. 2020, 36, 298–311. [Google Scholar] [CrossRef]
- Takahata, N.; Nei, M. Allelic Genealogy under Overdominant and Frequency-Dependent Selection and Polymorphism of Major Histocompatibility Complex Loci. Genetics 1990, 124, 967–978. [Google Scholar] [CrossRef]
- Bodmer, W.F. Evolutionary Significance of the HL-A System. Nature 1972, 237, 139–145. [Google Scholar] [CrossRef]
- Slade, R.W.; Mccallum, H.I. Overdominant Vs Frequency-Dependent Selection at Mhc Loci. Genetics 1992, 132, 861–862. [Google Scholar] [CrossRef]
- Hedrick, P.W. Pathogen resistance and genetic variation at MHC loci. Evolution 2002, 56, 1902–1908. [Google Scholar] [CrossRef] [PubMed]
- Milinski, M. The major histocompatibility complex, sexual selection, and mate choice. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 159–186. [Google Scholar] [CrossRef]
- Sin, Y.W.; Annavi, G.; Newman, C.; Buesching, C.; Burke, T.; Macdonald, D.W.; Dugdale, H.L. MHC class II-assortative mate choice in European badgers (Meles meles). Mol. Ecol. 2015, 24, 3138–3150. [Google Scholar] [CrossRef] [PubMed]
- Huchard, E.; Knapp, L.A.; Wang, J.L.; Raymond, M.; Cowlishaw, G. MHC, mate choice and heterozygote advantage in a wild social primate. Mol. Ecol. 2010, 19, 2545–2561. [Google Scholar] [CrossRef]
- Paterson, S.; Pemberton, J.M. No evidence for major histocompatibility complex-dependent mating patterns in a free-living ruminant population. Proc. R. Soc. B-Biol. Sci. 1997, 264, 1813–1819. [Google Scholar] [CrossRef]
- Kaufman, J.; Milne, S.; Gobel, T.W.F.; Walker, B.A.; Jacob, J.P.; Auffray, C.; Zoorob, R.; Beck, S. The chicken B locus is a minimal essential major histocompatibility complex. Nature 1999, 401, 923–925. [Google Scholar] [CrossRef]
- Westerdahl, H. Passerine MHC: Genetic variation and disease resistance in the wild. J. Ornithol. 2007, 148, S469–S477. [Google Scholar] [CrossRef]
- Minias, P.; Pikus, E.; Whittingham, L.A.; Dunn, P.O. Evolution of Copy Number at the MHC Varies across the Avian Tree of Life. Genome Biol. Evol. 2019, 11, 17–28. [Google Scholar] [CrossRef]
- He, K.; Minias, P.; Dunn, P.O. Long-Read Genome Assemblies Reveal Extraordinary Variation in the Number and Structure of MHC Loci in Birds. Genome Biol. Evol. 2021, 13, evaa270. [Google Scholar] [CrossRef]
- O’Connor, E.A.; Strandh, M.; Hasselquist, D.; Nilsson, J.A.; Westerdahl, H. The evolution of highly variable immunity genes across a passerine bird radiation. Mol. Ecol. 2016, 25, 977–989. [Google Scholar] [CrossRef]
- Biedrzycka, A.; O’Connor, E.; Sebastian, A.; Migalska, M.; Radwan, J.; Zajac, T.; Bielanski, W.; Solarz, W.; Cmiel, A.; Westerdahl, H. Extreme MHC class I diversity in the sedge warbler (Acrocephalus schoenobaenus); selection patterns and allelic divergence suggest that different genes have different functions. BMC Evol. Biol. 2017, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Drews, A.; Strandh, M.; Raberg, L.; Westerdahl, H. Expression and phylogenetic analyses reveal paralogous lineages of putatively classical and non-classical MHC-I genes in three sparrow species (Passer). BMC Evol. Biol. 2017, 17, 152. [Google Scholar] [CrossRef] [PubMed]
- Babik, W.; Taberlet, P.; Ejsmond, M.J.; Radwan, J. New generation sequencers as a tool for genotyping of highly polymorphic multilocus MHC system. Mol. Ecol. Resour. 2009, 9, 713–719. [Google Scholar] [CrossRef] [PubMed]
- Babik, W. Methods for MHC genotyping in non-model vertebrates. Mol. Ecol. Resour. 2010, 10, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Zagalska-Neubauer, M.; Babik, W.; Stuglik, M.; Gustafsson, L.; Cichon, M.; Radwan, J. 454 sequencing reveals extreme complexity of the class II Major Histocompatibility Complex in the collared flycatcher. BMC Evol. Biol. 2010, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Promerova, M.; Babik, W.; Bryja, J.; Albrecht, T.; Stuglik, M.; Radwan, J. Evaluation of two approaches to genotyping major histocompatibility complex class I in a passerine-CE-SSCP and 454 pyrosequencing. Mol. Ecol. Resour. 2012, 12, 285–292. [Google Scholar] [CrossRef]
- Sepil, I.; Moghadam, H.K.; Huchard, E.; Sheldon, B.C. Characterization and 454 pyrosequencing of Major Histocompatibility Complex class I genes in the great tit reveal complexity in a passerine system. BMC Evol. Biol. 2012, 12, 1–19. [Google Scholar] [CrossRef]
- Dunn, P.O.; Bollmer, J.L.; Freeman-Gallant, C.R.; Whittingham, L.A. Mhc Variation Is Related to a Sexually Selected Ornament, Survival, and Parasite Resistance in Common Yellowthroats. Evolution 2013, 67, 679–687. [Google Scholar] [CrossRef]
- Jones, M.R.; Cheviron, Z.A.; Carling, M.D. Variation in positively selected major histocompatibility complex class I loci in rufous-collared sparrows (Zonotrichia capensis). Immunogenetics 2014, 66, 693–704. [Google Scholar] [CrossRef]
- Bensch, S.; Hellgren, O.; Perez-Tris, J. MalAvi: A public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol. Ecol. Resour. 2009, 9, 1353–1358. [Google Scholar] [CrossRef]
- Clark, N.J.; Clegg, S.M.; Lima, M.R. A review of global diversity in avian haemosporidians (Plasmodium and Haemoproteus: Haemosporida): New insights from molecular data. Int. J. Parasitol. 2014, 44, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Biedrzycka, A.; Bielanski, W.; Cmiel, A.; Solarz, W.; Zajac, T.; Migalska, M.; Sebastian, A.; Westerdahl, H.; Radwan, J. Blood parasites shape extreme major histocompatibility complex diversity in a migratory passerine. Mol. Ecol. 2018, 27, 2594–2603. [Google Scholar] [CrossRef] [PubMed]
- Sepil, I.; Lachish, S.; Hinks, A.E.; Sheldon, B.C. Mhc supertypes confer both qualitative and quantitative resistance to avian malaria infections in a wild bird population. Proc. R. Soc. B-Biol. Sci. 2013, 280, 20130134. [Google Scholar] [CrossRef] [PubMed]
- Westerdahl, H.; Waldenstrom, J.; Hansson, B.; Hasselquist, D.; von Schantz, T.; Bensch, S. Associations between malaria and MHC genes in a migratory songbird. Proc. Biol. Sci. 2005, 272, 1511–1518. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.R.; Cheviron, Z.A.; Carling, M.D. Spatially variable coevolution between a haemosporidian parasite and the MHC of a widely distributed passerine. Ecol. Evol. 2015, 5, 1045–1060. [Google Scholar] [CrossRef]
- Loiseau, C.; Zoorob, R.; Robert, A.; Chastel, O.; Julliard, R.; Sorci, G. Plasmodium relictum infection and MHC diversity in the house sparrow (Passer domesticus). Proc. Biol. Sci. 2011, 278, 1264–1272. [Google Scholar] [CrossRef]
- Liu, B.Y.; Deng, Z.Q.; Huang, W.; Dong, L.; Zhang, Y.Y. High prevalence and narrow host range of haemosporidian parasites in Godlewski’s bunting (Emberiza godlewskii) in northern China. Parasitol. Int. 2019, 69, 121–125. [Google Scholar] [CrossRef]
- Griggio, M.; Biard, C.; Penn, D.J.; Hoi, H. Female house sparrows “count on” male genes: Experimental evidence for MHC-dependent mate preference in birds. BMC Evol. Biol. 2011, 11, 1–7. [Google Scholar] [CrossRef]
- Alcaide, M.; Liu, M.; Edwards, S.V. Major histocompatibility complex class I evolution in songbirds: Universal primers, rapid evolution and base compositional shifts in exon 3. Peerj 2013, 1, e86. [Google Scholar] [CrossRef]
- Westerdahl, H.; Wittzell, H.; von Schantz, T.; Bensch, S. MHC class I typing in a songbird with numerous loci and high polymorphism using motif-specific PCR and DGGE. Heredity 2004, 92, 534–542. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, A.; Herdegen, M.; Migalska, M.; Radwan, J. amplisas: A web server for multilocus genotyping using next-generation amplicon sequencing data. Mol. Ecol. Resour. 2016, 16, 498–510. [Google Scholar] [CrossRef]
- Biedrzycka, A.; Sebastian, A.; Migalska, M.; Westerdahl, H.; Radwan, J. Testing genotyping strategies for ultra-deep sequencing of a co-amplifying gene family: MHC class I in a passerine bird. Mol. Ecol. Resour. 2017, 17, 642–655. [Google Scholar] [CrossRef]
- Lenz, T.L.; Becker, S. Simple approach to reduce PCR artefact formation leads to reliable genotyping of MHC and other highly polymorphic loci - Implications for evolutionary analysis. Gene 2008, 427, 117–123. [Google Scholar] [CrossRef]
- Nei, M.; Gu, X.; Sitnikova, T. Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc. Natl. Acad. Sci. USA 1997, 94, 7799–7806. [Google Scholar] [CrossRef]
- Yang, Z.H. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Yang, Z.; Wong, W.S.; Nielsen, R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005, 22, 1107–1118. [Google Scholar] [CrossRef]
- Reche, P.A.; Reinherz, E.L. Sequence variability analysis of human class I and class II MHC molecules: Functional and structural correlates of amino acid polymorphisms. J. Mol. Biol. 2003, 331, 623–641. [Google Scholar] [CrossRef]
- Furlong, R.F.; Yang, Z. Diversifying and purifying selection in the peptide binding region of DRB in mammals. J. Mol. Evol. 2008, 66, 384–394. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Doytchinova, I.A.; Flower, D.R. In silico identification of supertypes for class II MHCs. J. Immunol. 2005, 174, 7085–7095. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, M.; Eriksson, L.; Jonsson, J.; Sjostrom, M.; Wold, S. New chemical descriptors relevant for the design of biologically active peptides. A multivariate characterization of 87 amino acids. J. Med. Chem. 1998, 41, 2481–2491. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 1–15. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2013. [Google Scholar]
- Lighten, J.; van Oosterhout, C.; Bentzen, P. Critical review of NGS analyses for de novo genotyping multigene families. Mol. Ecol. 2014, 23, 3957–3972. [Google Scholar] [CrossRef]
- Karlsson, M.; Westerdahl, H. Characteristics of MHC Class I Genes in House Sparrows Passer domesticus as Revealed by Long cDNA Transcripts and Amplicon Sequencing. J. Mol. Evol. 2013, 77, 8–21. [Google Scholar] [CrossRef]
- Westerdahl, H.; Asghar, M.; Hasselquist, D.; Bensch, S. Quantitative disease resistance: To better understand parasite-mediated selection on major histocompatibility complex. Proc. R. Soc. B-Biol. Sci. 2012, 279, 577–584. [Google Scholar] [CrossRef]

| Range | Mean | Median | SD | Minimum Number of Loci | |
|---|---|---|---|---|---|
| Number of alleles per individual | 1–30 | 16.06 | 16 | 3.33 | 15 |
| Number of classical alleles per individual | 1–18 | 8.42 | 8 | 2.33 | 9 |
| Number of supertypes per individual | 1–8 | 4.96 | 5 | 1.26 | NA |
| Model | Parameter Estimates | Model Comparison | p(LTR) |
|---|---|---|---|
| M0 | 0 = 0.54248 | ||
| M1 | 0 = 0.08505 p0 = 0.63235 1=1.000 p1 = 0.36765 | ||
| M2 | 0 = 0.04618 p0 = 0.47820 1 =1.000 p1 = 0.36387 2 =3.37284 p2 = 0.15794 | M1 vs. M2 | <0.05 |
| M7 | p = 0.19197 q = 0.29164 | ||
| M8 | p0 = 0.83551 p = 0.24542 q = 0.54364 (p1 = 0.16449) = 2.78333 | M7 vs. M8 | <0.05 |
| Site | Model 2 | Model 8 | ||
|---|---|---|---|---|
| Pr ( > 1) | Post Mean ± SE | Pr ( > 1) | Post Mean ± SE | |
| 5 | 0.990 * | 3.479 ± 0.266 | 0.999 ** | 2.502 ± 0.069 |
| 6 | 0.980 * | 3.454 ± 0.363 | 0.998 ** | 2.499 ± 0.097 |
| 9 | 1.000 ** | 3.505 ± 0.095 | 1.000 ** | 2.503 ± 0.055 |
| 11 | 0.951 * | 3.380 ± 0.544 | 0.998 ** | 2.499 ± 0.095 |
| 34 | 0.799 | 2.999 ± 1.004 | 0.978 * | 2.464 ± 0.258 |
| 52 | 1.000 ** | 3.505 ± 0.095 | 1.000 ** | 2.503 ± 0.055 |
| 53 | 0.760 | 2.899 ± 1.068 | 1.000 ** | 2.502 ± 0.064 |
| 60 | 1.000 ** | 3.505 ± 0.095 | 1.000 ** | 2.503 ± 0.055 |
| 66 | 0.999 ** | 3.503 ± 0.112 | 1.000 ** | 2.503 ± 0.056 |
| 67 | 1.000 ** | 3.505 ± 0.096 | 1.000 ** | 2.503 ± 0.055 |
| Fixed Effects | Estimate | Standard Error | Z Value | p Value | Malaria Lineage |
|---|---|---|---|---|---|
| Intercept | 0.488 | 1.610 | 0.303 | 0.762 | ALARV04 |
| Supertype 1 | -0.367 | 0.262 | −1.403 | 0.161 | |
| Supertype 2 | -0.153 | 0.279 | −0.550 | 0.583 | |
| Supertype 3 | 1.211 | 0.593 | 2.044 | 0.041 | |
| Supertype 4 | 0.067 | 0.251 | 0.266 | 0.790 | |
| Supertype 5 | 0.034 | 0.260 | 0.132 | 0.895 | |
| Supertype 6 | −0.319 | 0.245 | −1.304 | 0.192 | |
| Supertype 7 | 0.074 | 0.542 | 0.137 | 0.891 | |
| Supertype 8 | −0.613 | 0.307 | −1.994 | 0.046 | |
| Supertype 9 | −0.119 | 1.574 | −0.076 | 0.940 | |
| Number of supertypes | −0.161 | 0.100 | −1.612 | 0.107 | |
| Intercept | −1.912 | 1.712 | −1.117 | 0.264 | EMSPO05 |
| Supertype 1 | 1.628 | 0.622 | 2.618 | 0.009 | |
| Supertype 2 | −0.002 | 0.412 | −0.006 | 0.995 | |
| Supertype 3 | −0.397 | 0.633 | −0.627 | 0.531 | |
| Supertype 4 | 0.250 | 0.374 | 0.668 | 0.504 | |
| Supertype 5 | 0.690 | 0.436 | 1.582 | 0.114 | |
| Supertype 6 | 0.475 | 0.367 | 1.295 | 0.195 | |
| Supertype 7 | 0.298 | 0.814 | 0.367 | 0.714 | |
| Supertype 8 | −0.136 | 0.446 | −0.305 | 0.761 | |
| Supertype 9 | −2.341 | 1.580 | −1.482 | 0.138 | |
| Number of supertypes | 0.107 | 0.148 | 0.723 | 0.470 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, W.; Wang, X.; Liu, B.; Lenz, T.L.; Peng, Y.; Dong, L.; Zhang, Y. Evaluation of Genetic Diversity and Parasite-Mediated Selection of MHC Class I Genes in Emberiza godlewskii (Passeriformes: Emberizidae). Diversity 2022, 14, 925. https://doi.org/10.3390/d14110925
Huang W, Wang X, Liu B, Lenz TL, Peng Y, Dong L, Zhang Y. Evaluation of Genetic Diversity and Parasite-Mediated Selection of MHC Class I Genes in Emberiza godlewskii (Passeriformes: Emberizidae). Diversity. 2022; 14(11):925. https://doi.org/10.3390/d14110925
Chicago/Turabian StyleHuang, Wei, Xinyi Wang, Boye Liu, Tobias L. Lenz, Yangyang Peng, Lu Dong, and Yanyun Zhang. 2022. "Evaluation of Genetic Diversity and Parasite-Mediated Selection of MHC Class I Genes in Emberiza godlewskii (Passeriformes: Emberizidae)" Diversity 14, no. 11: 925. https://doi.org/10.3390/d14110925
APA StyleHuang, W., Wang, X., Liu, B., Lenz, T. L., Peng, Y., Dong, L., & Zhang, Y. (2022). Evaluation of Genetic Diversity and Parasite-Mediated Selection of MHC Class I Genes in Emberiza godlewskii (Passeriformes: Emberizidae). Diversity, 14(11), 925. https://doi.org/10.3390/d14110925






