Abstract
The life cycles and biodiversity of Pacific coast gastropods were analyzed by videomicroscopy and DNA barcoding of individuals collected from tide pools and in plankton nets from a variety of shore stations. In many species (Families Calyptraeidae, Cerithiopsidae, Strombidae, Vermetidae, Columbellidae, Nassariidae, Olivellidae, Hermaeidae, Onchidorididae, Gastropteridae, Haminoeidae), the free-swimming veligers were recovered from plankton collections; in Roperia poulsoni (family Muricidae) veligers were usually recovered from egg sacs where they had been retained although some escapees were found in plankton collections; in Pteropurpura festiva (family Muricidae) free-living veligers were also found; and in Atlanta californiensis (family Atlantidae) both veligers and adults were obtained from plankton collections making this a holoplanktonic species. The results confirm that DNA barcoding based on COI gene sequencing is a useful strategy to match life-cycle stages within species as well as to identify species and to document the level of biodiversity within the gastropods.
Keywords:
Mollusks; gastropods; Zooplankton; plankton; COI mitochondrial gene; Pacific Ocean; larvae; DNA barcoding 1. Introduction
Gastropods (snails and slugs) occur in saltwater, freshwater, and terrestrial environments. They are the most diverse class in the phylum Mollusca, containing about 476 families with 65,000–80,000 living species. The class is thought to be second only to the insects in overall species number [1].
The fertilized egg of gastropods hatches directly into a spherical or pear-shaped free-swimming larval stage called the trochophore, carrying a ring of cilia [2]. The ciliary girdle then expands into large, heavily ciliated lobes, giving rise to a larval stage called the veliger. Later the larva undergoes torsion, a 180° twisting that brings the posterior part of the body to an anterior position behind the head. Torsion is unique to the gastropods.
The veliger has a shell (secreted by the dorsal shell gland), a foot, and a velum, which is a lobed, ciliated structure used for swimming and feeding [2]. In most cases, the veliger eventually sinks to the seabed, loses its velum, and completes its metamorphosis into a juvenile or adult with typical snail-like morphology (a heteroplanktonic life cycle). In some cases, the adult is also planktonic, making the life cycle holoplanktonic
Analyzing the development of gastropod larvae can be done by rearing and documenting individuals, but here we show that a simpler and more efficient method is to gather individuals and identify them by sequencing their DNA barcode, which is a portion of the cytochrome c oxidase I (COI or COX1) gene, found in mitochondrial DNA [3,4]. We have previously used this approach to document the life cycles of cnidarians [5] and crustaceans [6].
The class Gastropoda includes both shelled and unshelled species. The marine shelled species include whelks, abalone, conches, periwinkles, turbans, cowries, limpets, chitons, and others. In most cases the one-piece shell is coiled in both larval and adult stages, but in the limpets it is coiled only in the larval stage. Gastropods are distinguished by their asymmetrical anatomy, in which most of the organs are more developed on one side of the body than the other. They typically have a distinct head carrying two or four sensory tentacles bearing eyes. Their ventral foot is the basis for the name gastropod (“stomach-foot”). Many species have an operculum which allows closing of the shell. In the following we use the taxonomy established by the World Register of Marine Species [7].
2. Materials and Methods
Zooplankton was collected under Scientific Collecting Permit SC-12162 from the California Department of Fish and Wildlife. Collections were made from 16 sites between Newport Beach and Dana Point, Orange County California, as well as one off Santa Barbara and two from Baja California (Table 1 and Figure 1).

Table 1.
Collection locations.
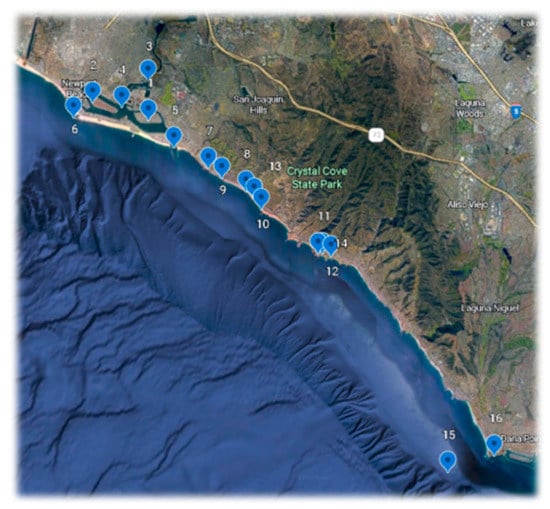
Figure 1.
Map of collecting sites listed in Table 1.
Field work. In the following account, only the localities outside of Orange County are identified specifically.
Shore-based collections were made with a 150 μm mesh net (aperture 30 cm) attached to a rope, with a 50 mL collection tube at the base. They were made from public docks using repeated horizontal sweeps near the surface and diagonal sweeps down to about 5 m. depth. About 5–10 sweeps of a total of about 35 m. usually yielded sufficient specimens, but no attempt was made to monitor collections quantitatively.
Ocean collection #15 was made with a 250 μm. mesh net attached to a 35 m. rope. The net (aperture 30 cm) was towed behind the vessel, just below the surface, for a period of 7 min at the slowest possible speed. Deployment and retrieval extended the total tow period to 10 min.
Laboratory analysis. Plankton collections were brought to the laboratory at the University of California, Irvine and examined under a dissecting microscope with lateral light and a dark background. Each specimen of interest was removed using a Pasteur Pipette, transferred to a depression slide, and recorded by video microscopy using a Zeiss microscope with a dark-field condenser, fitted with a phototube attached to a Nikon D5100 single-lens reflex camera. The most informative frames were taken from the videos and used in the figures for this paper. Each plankton specimen was preserved in 90% ethanol in a well of a 96-well microplate.
Each adult specimen was photographed in place, removed physically from its location, brought to the laboratory, and examined under the dissecting microscope. Using the microwave method [8] live adults were quickly heated sufficient to kill the animals and firm up their tissues; the steam from inside the shell forcing the body from the shell for easy removal. Multiple tissue samples were then removed using dissecting tools and transferred to the microplates. If available, typically three individuals per species were sacrificed, though occasionally more were used due to our inability to positively identify some species using field characteristics. Many of the species that also have a benthic stage in the life cycle have already been listed by the Southern California Association of Marine Invertebrate Taxonomists (SCAMIT) [9].
Filled plates were sent to the Canadian Centre for DNA Barcoding at the University of Guelph, 50 Stone Road East, Guelph, ON, N1G2W1, Canada for DNA extraction using an in-house protocol (http://ccdb.ca/resources/ accessed on 1 October 2022), and bidirectional sequencing of the standard 648-bp “DNA barcode” [3,4] in the COI mitochondrial gene. All samples except three were run with cocktail primers C_GasF1_t1 + GasR1_t1; Gast14_A01, Gast14_A02, and Gast14_A03 were run with both C_GasF1_t1 + GasR1_t1 and ZplankF1_t1 + ZplankR1_t1.
The procedure usually produced a DNA barcode of 658 nucleotides, and only those containing > 300 nucleotides were included in the sequence analysis. Groups of specimens with identical or almost identical DNA barcodes were assigned BIN numbers. They were compared with all barcode records on BOLD (10,580,183 Sequences) including the Public Record Barcode Database (2,529,561 Sequences/153,565 Species/66,474 Interim Species) using the Bold Aligner (Amino Acid Based HMM). The identification system on BOLD delivers a species identification if the query sequence shows less than 1% divergence to a reference sequence.
Images of all specimens as well as the DNA barcode sequences are in the public domain under code GASSC at the Canadian Centre for DNA Barcoding (http://www.boldsystems.org/index.php/MAS_Management_DataConsole?codes=GASSC accessed on 1 October 2022). In this paper, we concentrate on those species where we have found pelagic larval stages.
Conceptualization, Project administration, Data curation, Formal analysis: Peter Bryant; Investigation, Methodology: Timothy Arehart, Peter Bryant; Writing—original draft, Peter Bryant; Writing—review and editing, Peter Bryant and Timothy Arehart. Collections were made and analyzed with the assistance of Undergraduate students Taylor Sais, Alicia Navarro, Debbie Chung, Lesly Ortiz, and Bita Rostam.
3. Results
This project GASSC (Gastropoda of Southern California) included 1238 specimens of which 1235 provided images. 680 specimens provided a COI-5P sequence and 589 of these were Barcode-compliant, falling into 143 BINs containing 127 species (See Supplementary Materials). Our data (Figure 2) show a much larger range of interspecific divergences (seen in the graph of ”Distance to Nearest Neighbour”) compared to intraspecific divergences in this DNA sequence for gastropods suggesting the existence of a “DNA barcode gap”. Within species in this set of samples, the mean% divergence in sequence was 0.66 +/− 0.0 S.E. Within the largest set of conspecifics (Crepidula onyx, n = 54) the mean% divergence in sequence was 0.69 +/− 0.0 S.E., maximum 2.49%, minimum 0.0%.
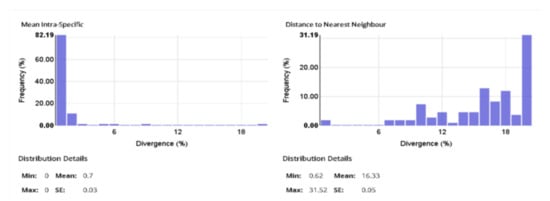
Figure 2.
Sequence divergence in the COI Barcode for intraspecific and interspecific comparisons using the data included in this publication. Distance Model: Kimura 2 Parameter; Alignment: BOLD Aligner; Length filter >/= 300 bp; Excluded: contaminants and misidentifications, records with stop codons.
Subclass caenogastropoda
Order Littorinimorpha: Sea Snails
Family Atlantidae Wiegmann and Ruthe, 1832
A family of microscopic (<1 cm shell diameter), holoplanktonic gastropods [1]. They have a transparent, coiled shell into which their bodies can be retracted, and an operculum that is used to close off the opening. The larval stage is a veliger in which the velum is initially small and bilobed, but with growth it develops three lobes (Figure 3). The larval shell and operculum are retained in the adult.
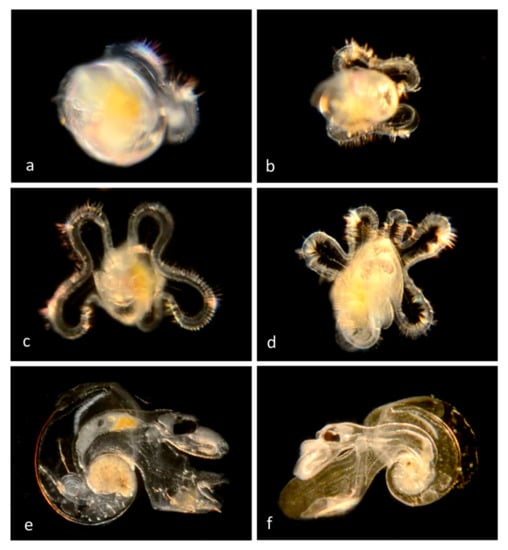
Figure 3.
Family Atlantidae: Sea snail, Atlanta californiensis (BINACV0973; n = 19). (a) Early veliger BIOUG01205-B08 from off Dana Point. (b) Later veliger with bilobed velum CCDB-24236-G01 from off Newport Pier. (c) Veliger with bilobed velum CCDB 31597 E12 from Newport Harbor entrance. (d) Later veliger with 6-lobed velum CCDB-24236-H05 from Newport Harbor. (e) Adult CCDB 31597 E09 from Newport Harbor entrance. (f) Adult CCDB-24722-D06 from La Profunda, Baja California.
Family Calyptraeidae Lamarck, 1809, the Slipper Shells
The shell is quite flat and has an internal shelf-like half shell, so that it resembles a traditional bedroom slipper; hence the name “slipper shell” or “slipper limpet” (although these are not true limpets). During mating they pile on top of each other to form a tower called a mating chain, in which all individuals start as males but the basal member transforms into a female. Females produce eggs that are fertilized internally, and developing embryos are held beneath the mother’s shell until they hatch into microscopic trochophore larvae before developing into swimming larvae (veligers) carrying shells, thousands of which disperse and later metamorphose into juveniles on the ocean floor.We have examined individuals of Crepidula onyx (Figure 4) Crepidula naticarum (Figure 5) and Crepidula huerta (Figure 6). In some species (e.g., Figure 6a and Figure 7b) the vela are decorated with yellow spots, the nature and function of which is unknown.
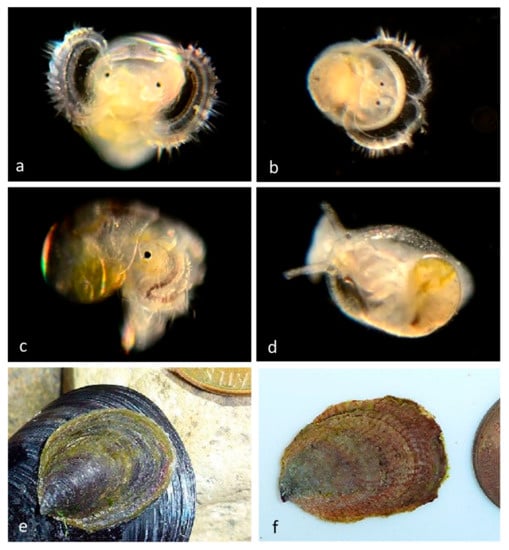
Figure 4.
Family Calyptraeidae. Onyx slipper snail, Crepidula onyx. (a) Veliger BIOUG01229-H01 from off Newport Pier. (b) Veliger BIOUG01229-B01 from off Newport Pier. (c) Late veliger CCDB 31597 F09 from Newport Harbor entrance. (d) Juvenile BIOUG01205-D02 from off Newport Pier. (e) Adult CCDB-32336 D06 from Balboa at Coral. (f) Adult CCDB-32336 E12 from Dana Point south shore.
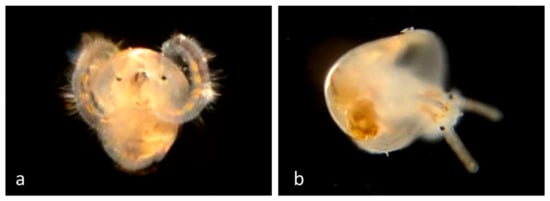
Figure 5.
Family Calyptraeidae. Slipper snail, Crepidula naticarum. (a) Veliger CCDB-24722-H04 from Newport Beach Harbor entrance. (b) Juvenile CCDB-24722-H09 from Newport Beach Harbor entrance.
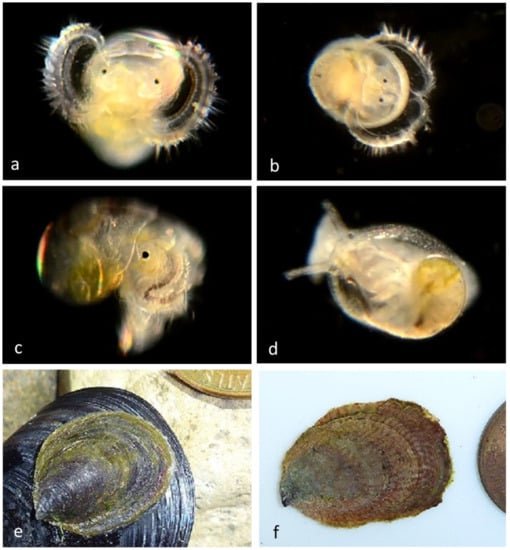
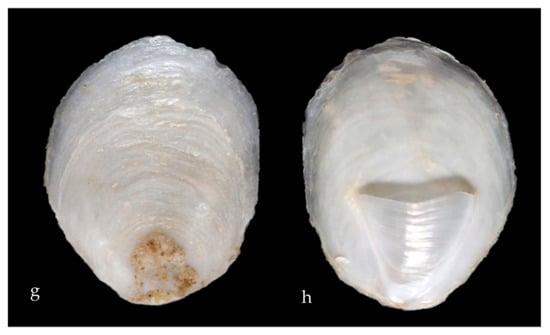
Figure 6.
Family Calyptraeidae. Slipper snail, Crepidula huertae (=Crepidula cf. perforans). (a) Veliger CCDB 31597 G03 from Newport Harbor entrance. (b) Adult CCDB-32336 D10 from inside the empty shell of a Bubble Snail (Bulla gouldiana) from Balboa at Coral. (c) Adult CCDB-32336 E01, 2&3 inside the empty shell of the same Bubble Snail (Bulla gouldiana). (d) Adult CCDB-32336 D10 and CCDB-32336 E01, 2&3 ventral views, live animals (no DNA sequences obtained). (e) CCDB-32336 D10 shell—ventral view. (f) CCDB-32336 E01, 2&3 shell—ventral view. (g,h), adult FMNH 282243. Subtidal in hermit crab shells, from Naples Reef, near Santa Barbara, Santa Barbara County, California. Length 30 mm [10]. We have found many stages of a different species of Slipper snail, Crepipatella lingulata. (Figure 6) which also has yellow spots on the vela.
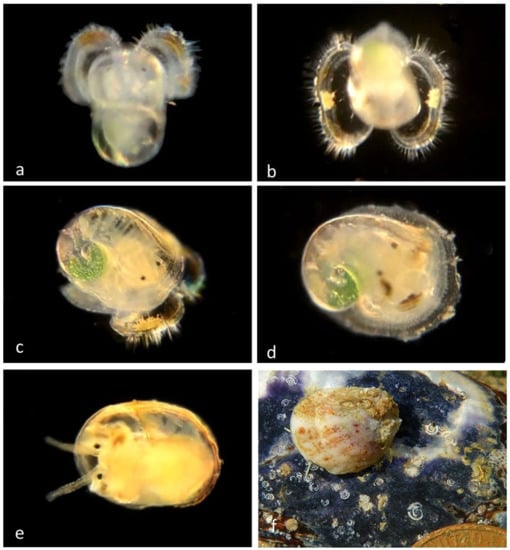
Figure 7.
Family Calyptraeidae. Slipper snail, Crepipatella lingulata. (BIN AAF2329; n = 27 veligers, 4 pre-adults, 2 adults) (a) Veliger CCDB-24236-E04 from Little Treasure Cove, Crystal Cove State Park. (b) Veliger CCDB 31597 F05 from Newport Beach harbor entrance. (c) Late veliger BIOUG01229-F06 from Newport Beach harbor entrance. (d) Juvenile CCDB-24722-H11 from Newport Beach harbor entrance. (e) Juvenile BIOUG01229-F02 from Newport Beach harbor entrance, (f) Adult CCDB-32336 G10, dorsal, from Shaw’s Cove south, Laguna Beach.
Our adult specimens of Crepidula huertae, (Figure 6) found inside the empty shell of a Bubble Snail (Bulla gouldiana) from Balboa at Coral (Newport Harbor), supported by the presence of DNA-sequenced veligers at the Harbor Entrance, extend the host and geographic range of this species, which was otherwise known from hermit crab shells at Naples Reef, near Santa Barbara, Santa Barbara County, California [10].
Family Cerithiopsidae H. Adams & A. Adams, 1853: The Cerithiopsids
A family of very small gastropods (Figure 8) with high spires and multiple whorls.
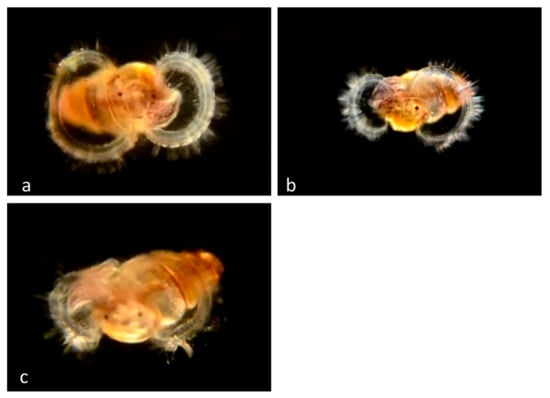
Figure 8.
Family Cerithiopsidae. Cerithiopsis sp.? (a) Veliger BIOUG01205-H03 from off Newport Pier. (b) Veliger CCDB-24236-B10 from off Newport Pier. (c) Veliger CCDB-24722-D04 from La Profunda, Baja California.
Family Strombidae Rafinesque, 1815: The true Conchs
Medium to very large snails (Figure 9), with eyes on long stalks. The shell has a long, narrow aperture with an anterior indentation that accommodates one of the eye stalks.
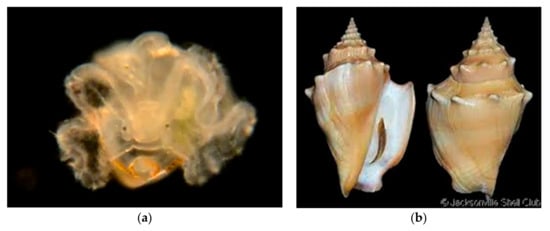
Figure 9.
Family Strombidae. Eastern Pacific fighting conch, Strombus gracilior. (a) Veliger, CCDB-24722-E09 from Bahia de Los Angeles, Baja California. (b) Adult, from Hardy’s Internet Guide to Marine Gastropods (This species is not recorded from Orange County).
Family Vermetidae Rafinesque, 1815: Worm Snails
Small to medium-sized snails (Figure 10), with very irregular elongated tubular shells often forming large clumps. Some species have opercula at the ends of the tubes, while others do not.
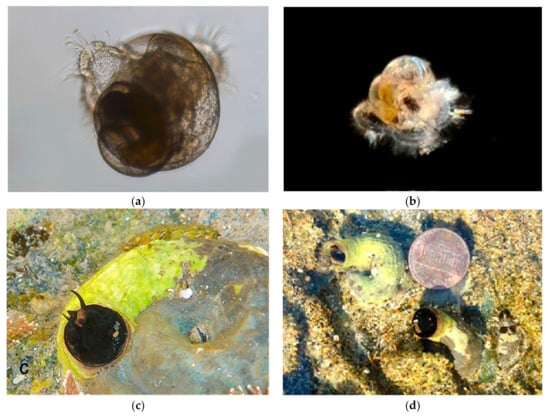
Figure 10.
Family Vermitidae. Scaly Worm Shell, Thylacodes squamigerus. (a) Veliger, BIOUG01205-B01. Off Crystal Cove State Park, Laguna Beach, Orange County, CA. (b) Veliger, BIOUG01229-D05. Off Newport Pier, Orange County, CA. (c) Adult CCDB 31729 A11 from Twin Points, Laguna Beach. (d) Adult CCDB-24002-C03 from Shaw’s Cove, Laguna Beach.
Subclass heterobranchia
The veliger has a shell, but this is lost during metamorphosis into the adult.
Family Hermaeidae H. Adams & A. Adams 1854
Small sea slugs (Figure 11) with cerata containing branches of the digestive gland, and rhinophores with a recessed tip.
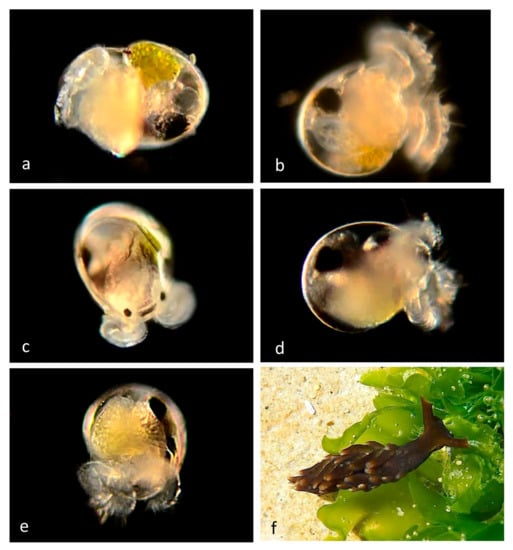
Figure 11.
Family Hermaeidae. Aplysiopsis enteromorphae. (a) Veliger larva BIOUG01229-H05 from Newport Pier. (b) Veliger larva CCDB-32329 H01 from Balboa at Coral. (c) Veliger larva BIOUG01205-H12 from Via Lido, Lido Island. (d) Late veliger BIOUG01229-H03 from Balboa at Coral. (e) Late veliger BIOUG01229-H04 from off Newport Pier. (f) Adult (no DNA barcode) from Rocky Bight, Crystal Cove State Park, Orange County, CA.
Family Onchidorididae Gray, 1827
Dorid nudibranchs (Figure 12), of which we have found only one pelagic larva.
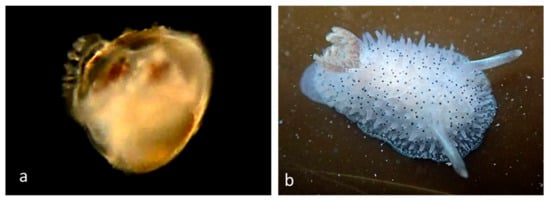
Figure 12.
Family Onchidorididae. Black-tipped Spiny Doris, Acanthodoris rhodoceras. (a) Larva CCDB 31597 D11 from Balboa at Coral. (b) Juvenile from Cabrillo Beach, Los Angeles County, CA. (http://nathistoc.bio.uci.edu/Molluscs/Acanthodoris%20rhodoceras/index.html accessed on 1 October 2022).
Order Neogastropoda: Sea Snails
Characterized by a long incurrent siphon and accompanying siphonal structure on the base of the shell.
Family Columbellidae Swainson, 1840: The Dove Snails
Minute to small snails (Figure 13) with a thick shell with a narrow opening. The foot is narrow, and the siphon is very long.

Figure 13.
Family Columbellidae. Carinate dove shell, Alia carinata (BIN AAZ4577). (a) Veliger BIOUG01229-G06 from Newport Beach, Harbor entrance. (b) Late veliger BIOUG01229-C06 from Pacific Ocean from Crystal Cove State Park. (c) Adult CCDB-24002-H02 from Dana Point rock field. (d) Adult CCDB 31729 D02 from Twin Points, Crystal Cove State Park.
Family Nassariidae Iredale, 1916 (1835): The Dog Whelks
Snails with rounded shells (Figure 14), a high spire, an oval aperture, and a siphonal notch.

Figure 14.
Family Nassariidae. Western mud nassa, Nassarius tiarula. (BIN ACV0694; n = 1 veliger, 7 adults). (a) Veliger BIOUG01205-G06 from Via Lido and Genoa, Lido Island. (b) Adult CCDB32329 G01 from Balboa at Coral (mud grab). (c) Adult CCDB-32329 F07 from Crew Dock, Back Bay Science Center (mud grab). (d) Adult CCDB-32329 F06 from Crew Dock, Back Bay Science (mud grab).
Family Muricidae Rafinesque, 1815
Within the family Muricidae, our collections include 27 adults but no larvae of Nucella ostrina (BIN AAA4209), 26 adults but no larvae of Acanthinucella spirata, 18 adults but no larvae of Ceratostoma nuttalli and 2 adults but no larvae of Mexacanthina lugubris. This is consistent with the finding that in these species of Muricidae the equivalent of the veliger stage occurs within the egg capsule, and the individuals hatch as juveniles which are not represented in our collections because of the limitations of our collection methods. In other family members Poulson’s Dwarf Triton, Roperia poulsoni, and Festive Murex, Pteropurpura festiva, individuals escape as veligers (Figure 15 and Figure 16).

Figure 15.
Family Muricidae. Poulson’s Dwarf Triton, Roperia poulsoni (BIN ADG1012, n = 6 adults, 2 veligers, 1 eggs). (a) Eggs CCDB 31597 C05 from Ocean off Dana Point. (b) Egg cases containing veligers, Gast14_A01—A03. Reef Point, Crystal Cove State Beach, Orange County, CA. 5/17/2021. (c) escaped veligers. (d) Veliger BIOUG01229-H10 from Newport Harbor entrance. (e) Adult CCDB 31729 B12 from Crescent Beach. (f) Adult CCDB 31729 B08 from Crescent Beach.
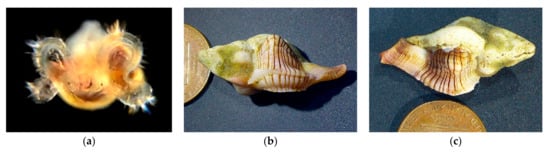
Figure 16.
Family Muricidae. Festive Murex, Pteropurpura festiva (BIN ADF9780, n = 6 veligers, 3 adults). (a) Veliger BIOUG01229-H09 from Newport Beach, Harbor entrance; (b) Adult (lateral) CCDB-36354 G07 from Reef Point, Crystal Cove State Park; (c) Adult (lateral) CCDB-36354 G09 from Pelican Point, Crystal Cove State Park.
Family Olivellidae Troschel, 1869: The Dwarf Olives
A family of small predatory snails (Figure 17) with smooth, shiny, elongated shells.
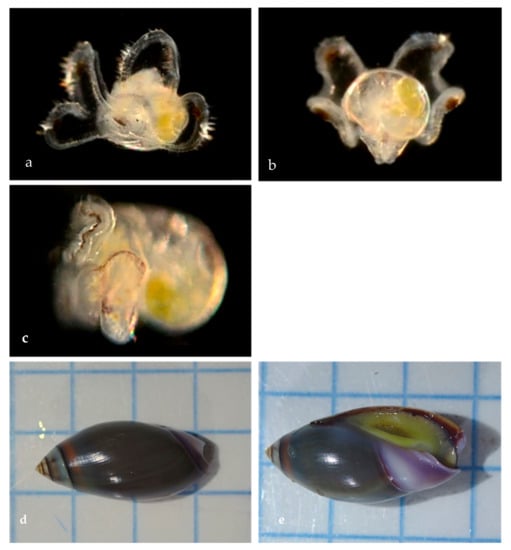
Figure 17.
Family Olivellidae. Purple dwarf olive, Callianax biplicata (BINACB8152; n = 8 veligers + 2 adults pending). (a) Veliger CCDB-24722-H05 from Newport Beach Harbor entrance. (b) Veliger CCDB-24722-H07 from Newport Beach Harbor entrance. (c) Veliger CCDB 31597 F10 from Ocean off Newport Beach. (d) Adult, dorsal CCDB-32329 G10 from Balboa at Coral. (e) Adult, ventral CCDB-32329 G09 from Balboa at Coral. Background ¼ inch squares.
Order Cephalaspidea: Sea Slugs and Bubble Snails
Family Gastropteridae Swainson, 1840 [1] Bat-Winged Slugs
Adults have no shell, or an internal reduced shell (Figure 18). They have outgrowths from the mantle wall called parapodia, used in swimming.
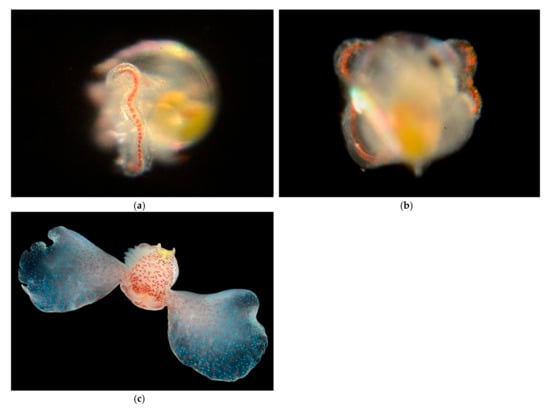
Figure 18.
Family Gastropteridae. Pacific Batwing Seaslug Gastropteron pacificum. (a,b) Veliger CCDB 31729 A07 from Newport Beach Harbor entrance. (a) lateral, (b) dorsal. (c) adult © Gustav Paulay. https://cdn.floridamuseum.ufl.edu/IZ/04ff1938-4872-41fd-9a4a-270ef0c5cc7e/ (accessed on 1 January 2022).
Family Haminoeidae Pilsbry, 1895
The shell (Figure 19) is partially or completely enfolded by lateral fleshy parapodial lobes. Represented in our collection by the Blister Glassy-bubble, Haminoea virescens (Figure 19) and the White Bubble snail, Haminoea vesicula (Figure 20).
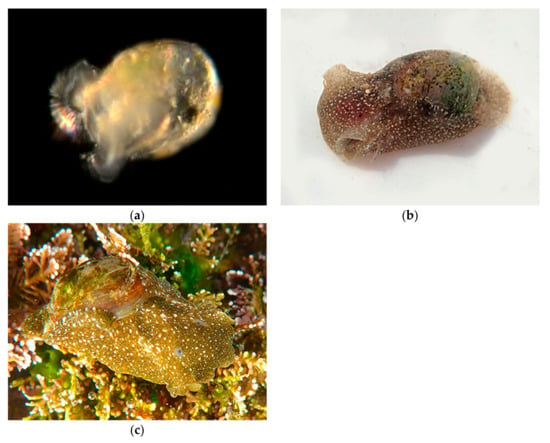
Figure 19.
Family Haminoeidae. Blister Glassy-bubble, Haminoea virescens. (a), Veliger CCDB-24722-G10 from Balboa at Coral. (b), Adult CCDB-24002-D05 from Shaw’s Cove south. (c), Adult CCDB-31729 B02 from Twin Points, Crystal Cove State Park.
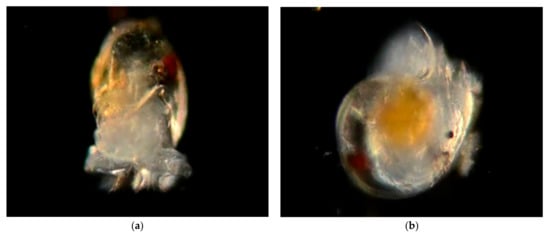
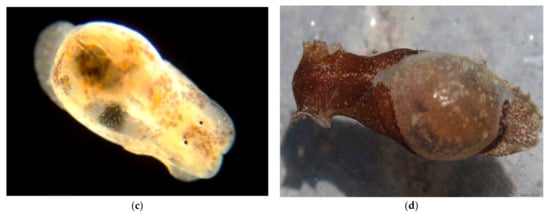
Figure 20.
Family Haminoeidae. White Bubble snail, Haminoea vesicula. (a), Veliger CCDB-24722-G12 from off Newport Pier. (b) Veliger CCDB-24002-A06 from Balboa at Coral. (c) Juvenile BIOUG01205-H09 from Balboa at Coral. (d) Adult from Bodega Head, CA, photo courtesy of Jackie Sones.
4. Discussion
Our results confirm that DNA barcoding using the COI barcode [3,4,11] is a useful strategy to match life-cycle stages within species as well as to identify species and to document the level of biodiversity within a taxon, in this case the gastropods. Our data also show the expected “DNA barcode gap”; i.e., a much larger range of interspecific divergences versus intraspecific divergences in this DNA sequence for gastropods (Figure 1). DNA barcoding has often revealed unexpected species diversity in many taxa [11,12] and the present study leads to the same conclusion for gastropods. The data will contribute to the development of a DNA barcode reference library, which will allow the rapid and convenient identification of individual gastropod individuals and parts collected at any developmental stage.
In this study we have begun to compile a collection of images of veligers, which are identified to the species level by matching of their DNA barcodes to those of morphologically recognized adults. These images confirm that the respective species have free-swimming veligers, and show that there is considerable morphological diversity, with some veligers having two velar lobes, some having four velar lobes, and some having yellow- or red-spotted vela. However, as expected from studies of larval stages in general, it is difficult to identify many species from the morphology of veligers.
The DNA sequence differences in the COI barcode are, of course, not responsible for the morphological differences we have observed between specimens in separate taxa. However, the DNA barcode differences that have evolved between morphologically distinct organisms can be used to examine the degree of relatedness between them. When the DNA sequence data are organized into a taxonomic tree (see Taxon Tree in Supplementary Materials), the results are generally consistent with the taxonomic tree according to conventional morphological methods. This can be explored by cladistic analysis, in which the taxonomic tree is examined for “DNA clades”—groups of species that are uniquely and exclusively related by DNA sequence. According to this analysis, all four species of Lottia, three species of Crepidula, two of Cerithidea, two of Nassarius, three of Littorina, two of Nuttalina, two of Epitonium, four of Tegula, three of Doriopsilla, two of Ancula, two of Felimare, two of Corambe, and two of Aplysia form clades, although the Lottia and Tegula clades include some different species.
DNA barcode reference libraries can lead to the development of more global sequencing strategies including metabarcoding and parallel sequencing of complex bulk samples including “environmental DNA”, which are being developed for monitoring ecosystem health. For example, in a recent study of plankton communities in the Baltic Sea, five nonindigenous species were discovered, and four of these were identified exclusively by metabarcoding [13]. Our work illustrates the need for more larval/adult matching to build sequence libraries specifically for meta- and eDNA barcoding.
Supplementary Materials
The following supporting information can be downloaded at: http://www.boldsystems.org/index.php/MAS_Management_DataConsole?codes=GASSC (accessed on 1 January 2022) (1238 records selected) Distance Model: Kimura 2 Parameter Marker: COI-5P Labels: Taxon, Sample ID, Life Stage, Barcode Cluster (BIN) Colorization: Barcode Cluster (BIN) Alignment: BOLD Aligner (Amino Acid based HMM) Filters Applied: ≥300 bp only, exclude records flagged as misidentifications, records with stop codons, contaminants. Sequence Count: 647 sequences; 104 Species; 84 Genera; 59 Families; 76 Unidentified; 143 BINs. For specimens that were large enough to allow separation of multiple samples (usually three samples to allow for sequencing failures) all of the sequences obtained are included in the tree; when full-length barcodes were obtained they always matched perfectly.
Author Contributions
Conceptualization, Project administration, Data curation, Formal analysis: P.J.B.; Investigation, Methodology: T.A., P.J.B.; Writing—original draft, P.J.B.; Writing—review and editing, P.J.B. and T.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The DNA barcodes generated during and/or analyzed during the current study are available at the Centre for Biodiversity Genomics, Biodiversity Institute of Ontario under the project GASSC: Gastropods of Southern California: http://www.boldsystems.org/index.php/MAS_Management_DataConsole?codes=GASSC (accessed on 1 October 2022).
Acknowledgments
Collections were made and analyzed with the assistance of Undergraduate students Taylor Sais, Alicia Navarro, Debbie Chung, Lesly Ortiz, and Bita Rostam.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bouchet, P.; Rocroi, J.P.; Hausdorf, B.; Kaim, A.; Kano, Y.; Nützel, A.; Parkhaev, P.; Schrödl, M.; Strong, E.E. Revised Classification, Nomenclator and Typification of Gastropod and Monoplacophoran Families. Malacologia 2017, 61, 1–526. [Google Scholar] [CrossRef]
- Boss, K.J. SHELLFISH|Characteristics of Molluscs. In Encyclopedia of Food Sciences and Nutrition; Academic Press: Cambridge, MA, USA, 2003; pp. 5216–5221. [Google Scholar]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; DeWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Hubert, N.; Hanner, R. DNA Barcoding, species delineation and taxonomy: A historical perspective. DNA Barcodes 2016, 3, 44–58. [Google Scholar] [CrossRef]
- Bryant, P.J.; Arehart, T.E. Diversity and life-cycle analysis of Pacific Ocean zooplankton by videomicroscopy and DNA barcoding: Hydrozoa. PLoS ONE 2019, 14, e0218848. [Google Scholar] [CrossRef] [PubMed]
- Bryant, P.; Arehart, T. Diversity and life-cycle analysis of Pacific Ocean zooplankton by video microscopy and DNA barcoding: Crustacea. J. Aquac. Mar. Biol. 2021, 10, 108–136. [Google Scholar]
- WoRMS Editorial Board World Register of Marine Species. Available online: https://www.marinespeces.org (accessed on 27 August 2022).
- Galindo, L.A.; Puillandre, N.; Strong, E.E.; Bouchet, P. Using microwaves to prepare gastropods for DNA barcoding. Mol. Ecol. Resour. 2014, 14, 700–705. [Google Scholar] [CrossRef] [PubMed]
- Barwick, K.; Cadien, D.B.; Lovell, L.L. (Eds.) A Taxonomic Listing of Benthic Macro-and Megainvertebrates, 12th ed.; The Southern California Association of Marine Invertebrate Taxonomists: Los Angeles, CA, USA, 2018. [Google Scholar]
- Collin, R. Calyptraeidae from the northeast Pacific (Gastropoda: Caenogastropoda). Zoosymposia 2019, 13, 107–130. [Google Scholar] [CrossRef]
- Bucklin, A.; Steinke, D.; Blanco-Bercial, L. DNA Barcoding of Marine Metazoa. Ann. Rev. Mar. Sci. 2011, 3, 471–508. [Google Scholar] [CrossRef] [PubMed]
- Waugh, J. DNA barcoding in animal species: Progress, potential and pitfalls. Bioessays 2007, 29, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Zaiko, A.; Samuiloviene, A.; Ardura, A.; Garcia-Vazquez, E. Metabarcoding approach for nonindigenous species surveillance in marine coastal waters. Mar. Pollut. Bull. 2015, 100, 53–59. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).