Genotypic and Phenotypic Selection of Newly Improved Putra Rice and the Correlations among Quantitative Traits
Abstract
1. Introduction
2. Materials and Methods
2.1. Breeding Materials and Breeding Scheme
2.2. Crossing Procedure for Gene Pyramiding
2.3. Molecular Genotyping Procedure
2.4. Phenotypic Assessment and Evaluation of Plants for Disease Resistance
2.5. Experiment Design and Statistical Analysis
3. Results
3.1. Genotypic Selection
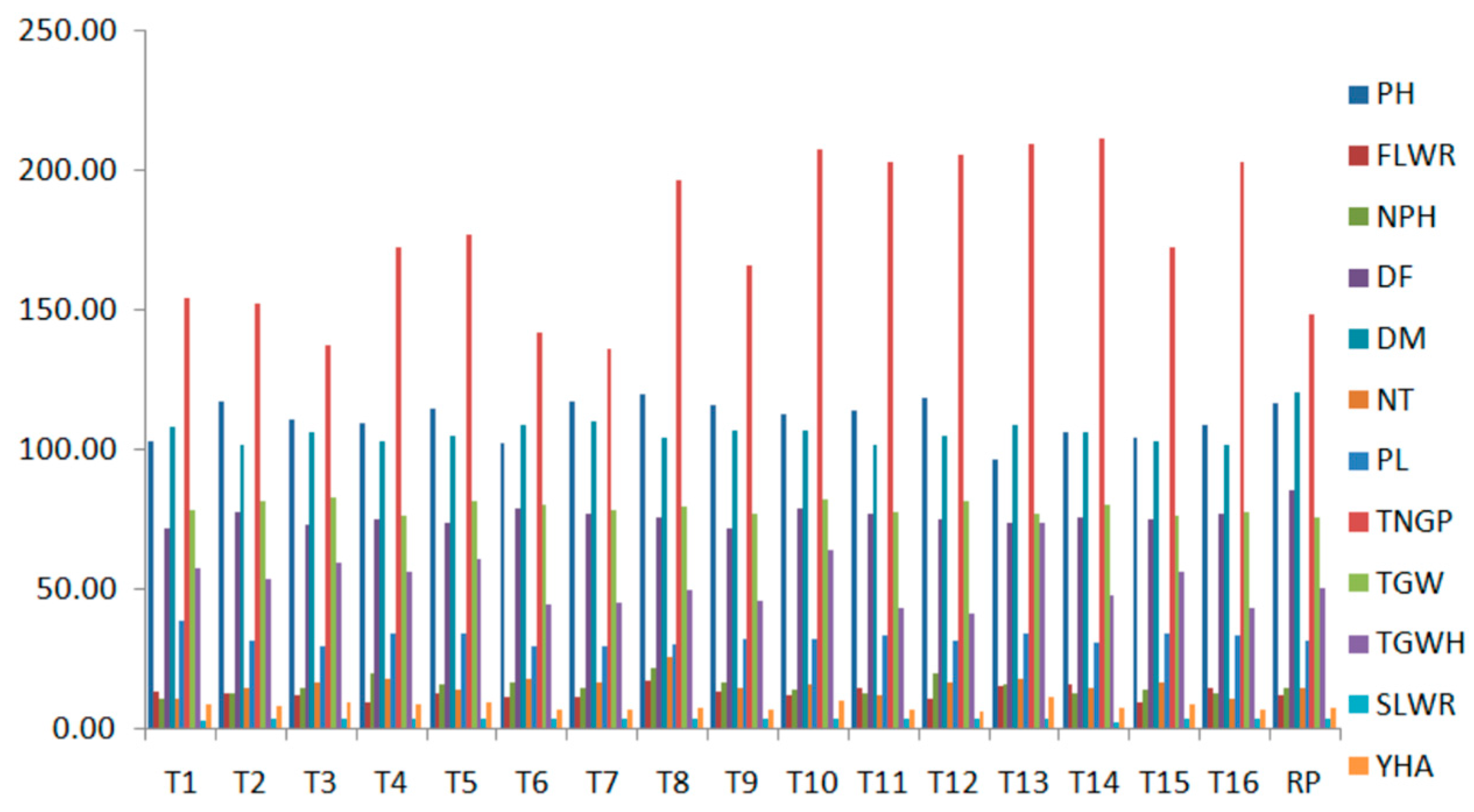
3.2. Phenotypic Selection
3.3. Trait Variation and Correlation
3.4. Correlation among the Quantitative Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chukwu, S.C.; Rafii, M.Y.; Ramlee, S.I.; Ismail, S.I.; Hasan, M.M.; Oladosu, Y.A.; Magaji, U.G.; Akos, I.; Olalekan, K.K. Bacterial leaf blight resistance in rice: A review of conventional breeding to molecular approach. Mol. Biol. Rep. 2019, 46, 1519–1532. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.K.; Chun-Lian, W.A.N.G.; Yuan-Jie, Y.U.; Liang, Y.T.; Kai-Jun, Z.H.A.O. Identification and mo-lecular mapping of Xa32 (t), a novel resistance gene for bacterial blight (Xanthomonas oryzaepv. oryzae) in rice. Acta Agron. Sin. 2009, 35, 1173–1180. [Google Scholar]
- Zhang, Q.; Wang, C.L.; Shi, A.N.; Bai, J.F.; Ling, S.C.; Li, D.Y.; Chen, C.B.; Pang, H.H. Evaluation of resistance to bacterial blight (Xan-thomonasoryzaepv. oryzae) in wild rice species. Sci. Agric. Sin. 1994, 27, 1–9. [Google Scholar]
- He, B.; Huang, X.; Li, D.; Jiang, C.; Yu, T.; Yin, F.; Li, W.; Cheng, Z. The cDNA cloning of a novel bacterial blight-resistance gene ME137. Acta Biochim. Biophys. Sin. 2013, 45, 422–424. [Google Scholar] [CrossRef][Green Version]
- Akos, I.S.; Rafii, M.Y.; Ismail, M.R.; Ramlee, S.I.; Shamsudin, N.A.A.; Ramli, A.; Chukwu, S.C.; Swaray, S.; Jalloh, M. Evaluation of Inherited Resistance Genes of Bacterial Leaf Blight, Blast and Drought Tolerance in Improved Rice Lines. Rice Sci. 2021, 28, 279–288. [Google Scholar] [CrossRef]
- Wu, J.; Yu, H.; Dai, H.; Mei, W.; Huang, X.; Zhu, S.; Peng, M. Metabolite profiles of rice cultivars containing bacterial blight-resistant genes are distinctive from susceptible rice. Acta Biochim. Biophys. Sin. 2012, 44, 650–659. [Google Scholar] [CrossRef]
- Halim, A.; Rafii, A.; Mohd, Y.; Mohamad, B.; Osman, Y.O.; Samuel, C.C. Ageing Effects, Generation Means, and Path Coefficient Analyses on High Kernel Elongation in MahsuriMutan and Basmati 370 Rice Populations. BioMed Res. Int. 2021, 2021, 8350136. [Google Scholar] [CrossRef]
- Chukwu, S.C.; Rafii, M.Y.; Ramlee, S.I.; Ismail, S.I.; Oladosu, Y.; Muhammad, I.I.; Musa, I.; Ahmed, M.; Jatto, M.I.; Yusuf, B.R. Recovery of recurrent parent genome in a marker-assisted backcrossing against rice blast and blight infections using functional markers and SSRs. Plants 2020, 9, 1411. [Google Scholar] [CrossRef]
- Zhou, Y.-L.; Uzokwe, V.N.; Zhang, C.-H.; Cheng, L.-R.; Wang, L.; Chen, K.; Gao, X.-Q.; Sun, Y.; Chen, J.-J.; Zhu, L.-H.; et al. Improvement of bacterial blight resistance of hybrid rice in China using the Xa23 gene derived from wild rice (Oryza rufipogon). Crop Prot. 2011, 30, 637–644. [Google Scholar] [CrossRef]
- Chukwu, S.C.; Rafii, M.Y.; Ramlee, S.I.; Ismail, S.I.; Oladosu, Y.; Kolapo, K.; Musa, I.; Halidu, J.; Muhammad, I.I.; Ahmed, M. Mark-er-assisted introgression of multiple resistance genes confers broad spectrum resistance against bacterial leaf blight and blast diseases in Putra-1 rice variety. Agronomy 2020, 10, 42. [Google Scholar] [CrossRef]
- Singh, V.K.; Singh, A.; Singh, S.P.; Ellur, R.K.; Choudhary, V.; Sarkel, S.; Singh, D.; Krishnan, S.G.; Nagarajan, M.; Vinod, K.K.; et al. Incorporation of blast resistance into “PRR78”, an elite Basmati rice restorer line, through marker assisted backcross breeding. Field Crops Res. 2012, 128, 8–16. [Google Scholar] [CrossRef]
- Miah, G.; Rafii, M.Y.; Ismail, M.R.; Puteh, A.B.; Rahim, H.A.; Islam, K.; Latif, M.A. A review of microsatellite markers and their ap-plications in rice breeding programs to improve blast disease resistance. Int. J. Mol. Sci. 2013, 14, 22499–22528. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. Isolation ofplant DNA from fresh tissue. Focus 1990, 12, 39–40. [Google Scholar]
- McCouch, S.R.; McNally, K.L.; Wang, W.; Sackville Hamilton, R. Genomics of gene banks: A case study in rice. Am. J. Bot. 2012, 99, 407–423. [Google Scholar] [CrossRef] [PubMed]
- Jena, K.K.; Mackill, D.J. Molecular Markers and Their Use in Marker-Assisted Selection in Rice. Crop Sci. 2008, 48, 1266–1276. [Google Scholar] [CrossRef]
- Divakara, B.N.; Upadhyaya, H.D.; Wani, S.P.; Gowda, C.L. Biology and genetic improvement of Jatropha curcas L.: A review. Appl. Energy 2010, 87, 732–742. [Google Scholar] [CrossRef]
- Sandhu, S.K.; Singh, G. Genomic-Assisted Breeding in Oilseed Brassicas. In Genomic-Assisted Breeding in Oilseed Brassicas; Springer: Cham, Switzerland, 2018; pp. 317–348. [Google Scholar] [CrossRef]
- Sabri, R.S.; Rafii, M.Y.; Ismail, M.R.; Yusuff, O.; Chukwu, S.C.; Hasan, N. Assessment of Agro-Morphologic Performance, Genetic Parameters and Clustering Pattern of Newly Developed Blast Resistant Rice Lines Tested in Four Environments. Agronomy 2020, 10, 1098. [Google Scholar] [CrossRef]
- IRRI. International Rice Research Institute. 2014. Available online: http://irri.org Practical Manual (accessed on 22 November 2019).
- Oladosu, Y.; Rafii, M.Y.; Magaji, U.; Abdullah, N.; Miah, G.; Chukwu, S.C.; Hussin, G.; Ramli, A.; Kareem, I. Genotypic and phe-notypic relationship among yield components in rice under tropical conditions. BioMed Res. Int. 2018, 15, 2018. [Google Scholar]
- van Berloo, R. GGT 2.0: Versatile Software for Visualization and Analysis of Genetic Data. J. Hered. 2008, 99, 232–236. [Google Scholar] [CrossRef]
- Hashim, N.; Rafii, M.Y.; Oladosu, Y.; Ismail, M.R.; Ramli, A.; Arolu, F.; Chukwu, S. Integrating multivariate and univariate statis-tical models to investigate genotype–environment interaction of advanced fragrant rice genotypes under rainfed condition. Sustainability 2021, 13, 4555. [Google Scholar] [CrossRef]
- Eslinger, J. The SAS Programmer’s PROC REPORT Handbook: Basic to Advanced Reporting Techniques; Sas Institute, Inc.: Cary, NC, USA, 2016; p. 23. [Google Scholar]
- Ahmed, M.A.; Rafii, M.Y.; Izzati, M.Z.N.A.; Khalilah, A.K.; Awad, E.A.; Kaka, U.; Chukwu, S.C.; Liang, J.B.; Sazili, A.Q. Biological additives improved qualities, in vitro gas production kinetics, digestibility, and rumen fermentation characteristics of different varieties of rice straw silage. Anim. Prod. Sci. 2022. [Google Scholar] [CrossRef]
- Yi, M.; Nwe, K.T.; Vanavichit, A.; Chai-Arree, W.; Toojinda, T. Marker assisted backcross breeding to improve cooking quality traits in Myanmar rice cultivar Manawthukha. Field Crop. Res. 2009, 113, 178–186. [Google Scholar] [CrossRef]
- Ribaut, J.M.; Jiang, C.; Hoisington, D.A. Simulation experiments on efficiencies of gene introgression by backcrossing. Crop Sci. 2002, 42, 557–565. [Google Scholar] [CrossRef]
- Frisch, M.; Bohn, M.; Melchinger, A.E. Comparison of Selection Strategies for Marker-Assisted Backcrossing of a Gene. Crop Sci. 1999, 39, 1295–1301. [Google Scholar] [CrossRef]
- Olalekan, K.K.; Rafii, M.Y.; Salleh, A.M.; Mohamed, M.T.; Ahmad, K.; Misran, A.; Abro, T.F.; Oladosu, Y.; Arolu, I.W.; Samuel, C.; et al. Analysis of recurrent parent genome recovery in marker-assisted backcross breeding programme in Watermelon. Int. J. Sci. Technol. Res. 2019, 8, 945–955. [Google Scholar]
- Oladosu, Y.; Rafii, M.Y.; Samuel, C.; Fatai, A.; Magaji, U.; Kareem, I.; Kamarudin, Z.S.; Muhammad, I.; Kolapo, K. Drought Resistance in Rice from Conventional to Molecular Breeding: A Review. Int. J. Mol. Sci. 2019, 20, 3519. [Google Scholar] [CrossRef]
- Oladosu, Y.; Rafii, M.Y.; Arolu, F.; Chukwu, S.C.; Muhammad, I.; Kareem, I.; Salisu, M.A.; Arolu, I.W. Submergence Tolerance in Rice: Review of Mechanism, Breeding and, Future Prospects. Sustainability 2020, 12, 1632. [Google Scholar] [CrossRef]
- Frisch, M.; Melchinger, A.E. Marker-assisted backcrossing for simultaneous introgression of two genes. Crop Sci. 2001, 41, 1716–1725. [Google Scholar] [CrossRef]
- Chukwu, S.C.; Rafii, M.Y.; Ramlee, S.I.; Ismail, S.I.; Oladosu, Y.; Okporie, E.; Onyishi, G.; Utobo, E.; Ekwu, L.; Swaray, S.; et al. Marker-assisted selection and gene pyramiding for resistance to bacterial leaf blight disease of rice (Oryza sativa L.). Biotechnol. Biotechnol. Equip. 2019, 33, 440–455. [Google Scholar] [CrossRef]
- Tanksley, S.D.; Young, N.D.; Paterson, A.H.; Bonierbale, M.W. RFLP Mapping in Plant Breeding: New Tools for an Old Science. Nat. Biotechnol. 1989, 7, 257–264. [Google Scholar] [CrossRef]
- Visscher, P.M.; Haley, C.S.; Thompson, R. Marker-Assisted Introgression in Backcross Breeding Programs. Genetics 1996, 144, 1923–1932. [Google Scholar] [CrossRef] [PubMed]
- Oladosu, Y.; Rafii, M.Y.; Arolu, F.; Chukwu, S.C.; Salisu, M.A.; Fagbohun, I.K.; Muftaudeen, T.K.; Swaray, S.; Haliru, B.S. Superabsorbent Polymer Hydrogels for Sustainable Agriculture: A Review. Horticulturae 2022, 8, 605. [Google Scholar] [CrossRef]
- Tanksley, S.D. Molecular markers in plant breeding. Plant Mol. Biol. Rep. 1983, 1, 3–8. [Google Scholar] [CrossRef]
- Miah, G.; Rafii, M.Y.; Ismail, M.R.; Puteh, A.B.; Rahim, H.A.; Latif, M.A. Recurrent parent genome recovery analysis in a mark-er-assisted backcrossing program of rice (Oryza sativa L.). Comptesrendusbiologies 2015, 338, 83–94. [Google Scholar]
- Melchinger, A.E. Use of Molecular Markers in Breeding for Oligogenic Disease Resistance. Plant Breed. 1990, 104, 1–19. [Google Scholar] [CrossRef]
- Rathour, R.; Chopra, M.; Sharma, T.R. Development and validation of microsatellite markers linked to the rice blast resistance gene Pi-z of Fukunishiki and Zenith. Euphytica 2008, 163, 275–282. [Google Scholar] [CrossRef]
- Fjellstrom, R.; McClung, A.M.; Shank, A.R. SSR Markers Closely Linked to the Pi-z Locus are Useful for Selection of Blast Resistance in a Broad Array of Rice Germplasm. Mol. Breed. 2006, 17, 149–157. [Google Scholar] [CrossRef]
- Mondal, U.; Khanom, M.; Hassan, L.; Begum, S. Foreground selection through SSRs markers for the development of salt tolerant rice variety. J. Bangladesh Agric. Univ. 2013, 11, 67–72. [Google Scholar] [CrossRef]
- Ebem, E.C.; Afuape, S.O.; Chukwu, S.C.; Ubi, B.E. Genotype × Environment Interaction and Stability Analysis for Root Yield in Sweet Potato [Ipomoea batatas (L.) Lam]. Front. Agron. 2021, 3, 1–14. [Google Scholar] [CrossRef]
- Young, N.D.; Tanksley, S.D. RFLP analysis of the size of chromosomal segments retained around the Tm-2 locus of tomato during backcross breeding. Theor. Appl. Genet. 1989, 77, 353–359. [Google Scholar] [CrossRef]
- Luu, M.C.; Luu, T.N.H.; Pham, T.M.H.; Vu, T.T.H.; Nguyen, Q.D.; Pham, T.M.; Vu, D.Q.; Abdelbagi, M.I.; Le, H.H. Application of marker assisted backcrossing to introgress the submergence tolerance QTL SUB1 into the Vietnam elite rice variety-AS996. Am. J. Plant Sci. 2012, 25, 2012. [Google Scholar]
- Semagn, K.; Ndjiondjopd, M.N.; Cissoko, M. Microsatellites and agronomic traits for assessing genetic relationships among 18 New Rice for Africa (NERICA) varieties. Afr. J. Biotechnol. 2006, 5, 42852. [Google Scholar]
- Hospital, F.; DeCoux, G. Popmin: A program for the numerical optimization of population sizes in marker-assisted backcross programs. J. Hered. 2002, 93, 383–384. [Google Scholar] [CrossRef] [PubMed]
- Basavaraj, S.H.; Singh, V.K.; Singh, A.; Singh, A.; Singh, A.; Anand, D.; Yadav, S.; Ellur, R.K.; Singh, D.; Krishnan, S.G.; et al. Marker-assisted improvement of bacterial blight resistance in parental lines of Pusa RH10, a superfine grain aromatic rice hybrid. Mol. Breed. 2010, 26, 293–305. [Google Scholar] [CrossRef]
- Singh, A.K.; Sarma, B.K.; Singh, P.K.; Nandan, R. Screening of rice (Oryza sativa L.) germplasms against Xanthomonas oryzaepv. oryzae. J. Eco-Friendly Agric. 2013, 8, 86–88. [Google Scholar]
- Gopalakrishnan, S.; Sharma, R.K.; Anand Rajkumar, K.; Joseph, M.; Singh, V.P.; Singh, A.K.; Bhat, K.V.; Singh, N.K.; Mohapatra, T. Integrating marker assisted background analysis with foreground selection for identification of superior bacterial blight re-sistant recombinants in Basmati rice. Plant Breed. 2008, 127, 131–139. [Google Scholar] [CrossRef]
- Chukwu, S.C.; Rafii, M.Y.; Ramlee, S.I.; Ismail, S.I.; Oladosu, Y.; Muhammad, I.I.; Ubi, B.E.; Nwokwu, G. Genetic analysis of microsat-ellites associated with resistance against bacterial leaf blight and blast diseases of rice (Oryza sativa L.). Biotechnol. Biotechnol. Equip. 2020, 34, 898–904. [Google Scholar] [CrossRef]
- Jeung, J.U.; Heu, S.G.; Shin, M.S.; Vera Cruz, C.M.; Jena, K.K. Dynamics of Xanthomonas oryzaepv.oryzae populations in Korea and their relationship to known bacterial blight resistance genes. Phytopathology 2006, 96, 867–875. [Google Scholar] [CrossRef] [PubMed]
- Khanh, T.D.; Linh, L.H.; Linh, T.H.; Ham, L.H.; Xuan, T.D. Rapid and high-precision marker assisted backcrossing to introgress the SUB1 QTL into the Vietnamese elite rice variety. J. Plant Breed. Crop. Sci. 2013, 5, 26–33. [Google Scholar] [CrossRef]
- Ashkani, S.; Rafii, M.Y.; Rusli, I.; Sariah, M.; Abdullah, S.N.A.; Rahim, H.A.; Latif, M.A. SSRs for Marker-Assisted Selection for Blast Resistance in Rice (Oryza sativa L.). Plant Mol. Biol. Rep. 2011, 30, 79–86. [Google Scholar] [CrossRef]
- Okporie, E.O.; Chukwu, S.C.; Onyishi, G.C. Phenotypic recurrent selection for increase yield and chemical constituents of maize (Zea mays L.). World Appl. Sci. J. 2013, 21, 994–999. [Google Scholar]
- Chukwu, S.C.; Ibeji, C.A.; Ogbu, C.; Oselebe, H.O.; Okporie, E.O.; Rafii, M.Y.; Oladosu, Y. Primordial Initiation, Yield and Yield Component Traits of Two Genotypes of Oyster Mushroom (Pleurotus spp.) as Affected by Various Rates of Lime. Sci. Rep. Springer Nat. 2022; in press. [Google Scholar]
- Kusutani, A.; Cui, J.; Toyota, M.; Asanuma, K.I. Studies on the varietal difference of harvest index in rice: Relationship between harvest index and morphological characteristics and leaf color. Jpn. J. Crop. Sci. 2000, 69, 359–364. [Google Scholar]
- Chukwu, S.C.; Ekwu, L.G.; Onyishi, G.C.; Okporie, E.O.; Obi, I.U. Correlation between agronomic and chemical characteristics of maize (Zea mays L.) genotypes after two years of mass selection. Int. J. Sci. Res. 2013, 4, 1708–1712. [Google Scholar]
- Sarif, H.M.; Rafii, M.Y.; Ramli, A.; Oladosu, Y.; Musa, H.M.; Rahim, H.A.; Zuki, Z.M.; Chukwu, S.C. Genetic diversity and variability among pigmented rice germplasm using molecular marker and morphological traits. Biotechnol. Biotechnol. Equip. 2020, 34, 747–762. [Google Scholar] [CrossRef]
- Ji, Z.-J.; Yang, S.-D.; Zeng, Y.-X.; Liang, Y.; Yang, C.-D.; Qian, Q. Pyramiding blast, bacterial blight and brown planthopper resistance genes in rice restorer lines. J. Integr. Agric. 2016, 15, 1432–1440. [Google Scholar] [CrossRef]
- Blair, M.W.; Garris, A.J.; Iyer, A.S.; Chapman, B.; Kresovich, S.; McCouch, S.R. High resolution genetic mapping and candidate gene identification at the xa5 locus for bacterial blight resistance in rice (Oryza sativa L.). Theor. Appl. Genet. 2003, 107, 62–73. [Google Scholar] [CrossRef]
- Zhang, G.Q.; Angeles, E.R.; Abenes, M.L.; Khush, G.S.; Huang, N. RAPD and RFLP mapping of the bacterial blight resistance gene xa-13 in rice. Theor. Appl. Genetics. 1996, 93, 65–70. [Google Scholar] [CrossRef]
- Singh, A.K.; Gopalakrishnan, S.; Singh, V.P.; Prabhu, K.V.; Mohapatra, T.; Singh, N.K.; Sharma, T.R.; Nagarajan, M.; Vinod, K.K.; Singh, D.; et al. Marker assisted selection: A paradigm shift in Basmati breeding. Indian J. Genet. Plant Breeding. 2011, 71, 120. [Google Scholar]
- Dokku, P.; Das, K.M.; Rao, G.J.N. Genetic enhancement of host plant-resistance of the Lalat cultivar of rice against bacterial blight employing marker-assisted selection. Biotechnol. Lett. 2013, 35, 1339–1348. [Google Scholar] [CrossRef]
- Singh, A.K.; Dharmraj, E.; Nayak, R.; Singh, P.K.; Singh, N.K. Identification of bacterial leaf blight resistance genes in wild rice of eastern India. Turk. J. Bot. 2015, 39, 1060–1066. [Google Scholar] [CrossRef]
- McCouch, S.R.; Sebastian, L.S.; Ikeda, R.; Huang, N.; Imbe, T.; Coffman, W.R. Molecular mapping of resistance to rice tungro spherical virus and green leafhopper. Phytopathol. 1996, 86, 25–30. [Google Scholar]
- Yoshimura, S.; Yoshimura, A.; Iwata, N.; McCouch, S.R.; Abenes, M.L.; Baraoidan, M.R.; Mew, T.W.; Nelson, R.J. Tagging and combin-ing bacterial blight resistance genes in rice using RAPD and RFLP markers. Mol. Breeding. 1995, 1, 375–387. [Google Scholar] [CrossRef]
- Ronald, P.C.; Albano, B.; Tabien, R.; Abenes, L.; Wu, K.S.; McCouch, S.; Tanksley, S.D. Genetic and physical analysis of the rice bacte-rial blight disease resistance locus, Xa21. Mol. Gen. Genet. MGG 1992, 236, 113–120. [Google Scholar] [CrossRef]
- Amgai, R.B.; Niroula, R.K.; Pantha, S.; Hamal, S.S.; Tamang, B.G.; Sah, B.P.; Bhatta, M.R. Marker Assisted Screening of Nepalese rice for bacterial leaf blight (BLB) resistance. Nepal J. Biotechnol. 2015, 3, 35–39. [Google Scholar] [CrossRef][Green Version]
- Deshmukh, U.C.; Verma, R.K.; Saxena, R.R.; Mohan, P.; Verulkar, S.B.; Chandra, D.U.; Kumar, V.R.; Pratibha, M. Marker assisted selection for bacterial leaf blight resistance in segregating populations of Karma Mahsuri. Vegetos 2017, 30, 55. [Google Scholar] [CrossRef]
- Sundaram, R.M.; Laha, G.S.; Viraktamath, B.C.; Sujatha, K.; Natarajkumar, P.; Hari, Y.; Srinivasa Rao, K.; Reddy, C.S.; Balachandran, S.M.; Madhav, M.S.; et al. Marker assisted breeding for development of bacterial blight resistant rice. In Genomics and Crop Improvement: Relevance and Reservations; Institute of Biotechnology, Acharya NG Ranga Agricultural University: Hyderabad, Indian, 2011; Volume 500, pp. 154–182. [Google Scholar]
- Rajpurohit, D.; Kumar, R.; Kumar, M.; Paul, P.; Awasthi, A.; Basha, P.O.; Puri, A.; Jhang, T.; Singh, K.; Dhaliwal, H.S. Pyramiding of two bacterial blight resistance and a semidwarfing gene in Type 3 Basmati using marker-assisted selection. Euphytica 2010, 178, 111–126. [Google Scholar] [CrossRef]
- Huang, N.; Angeles, E.R.; Domingo, J.; Magpantay, G.; Singh, S.; Zhang, G.; Kumaravadivel, N.; Bennett, J.; Khush, G.S. Pyramiding of bacterial blight resistance genes in rice: Marker-assisted selection using RFLP and PCR. Theor. Appl. Genet. 1997, 95, 313–320. [Google Scholar] [CrossRef]
- Bojun, M.; Wenming, W.; Bin, Z.; Yongli, Z.; Lihuang, Z.; Wenxue, Z. Studies of PCR marker for the rice bacterial blight resistance gene Xa-4. Yi Chuan Hered. 1999, 21, 9–12. [Google Scholar]
- Miah, G.; Rafii, M.Y.; Ismail, M.R.; Puteh, A.B.; Rahim, H.A.; Latif, M.A. Marker-assisted introgression of broad-spectrum blast resistance genes into the cultivated MR219 rice variety. J. Sci. Food Agric. 2017, 97, 2810–2818. [Google Scholar] [CrossRef]

| S/n | Marker | Gene | Chromo. | Primer Sequence (F) | Exp. (bp) | Putra1 (bp) | IRBB60 (bp) |
|---|---|---|---|---|---|---|---|
| Blast | |||||||
| 1. | RM6836 | Piz, Pi2, Pi9 | 6 | F: TGTTGCATATGGTGCTATTTGA R: GATACGGCTTCTAGGCCAAA | 240 | 244 | 218 |
| 2. | RM8225 | Piz | 6 | F: ATGCGTGTTCAGAAATTAGG R: TTGTTGTATACCTCATCGACAG | 221 | 268 | 246 |
| BLB | |||||||
| 3. | MP | Xa4 | 4 | F: ATCGATCGATCTTCACGAGG R: TCGTATAAAAGGCATTCGGG | 150 | 219 | 104 |
| 4. | RM13 | xa5 | 5 | F: TCCAACATGGCAAGAGAGAG R: GGTGGCATTCGATTCCAG | 141 | 187 | 162 |
| 5. | RM21 | xa5 | 11 | F: ACAGTATTCCGTAGGCACGG R: GCTCCATGAGGGTGGTAGAG | 157 | 164 | 154 |
| 6. | Xa13prom | xa13 | 8 | F: GGCCATGGCTCAGTGTTTAT R: GAGCTCCAGCTCTCCAAATG | 450 | 311 | 484 |
| 7. | Xa21FR | Xa21 | 11 | F: TCCAACATGGCAAGAGAGAG R: GGTGGCATTCGATTCCAG | 140 | 144 | 132 |
| 8. | pTA248 | Xa21 | 11 | F: AGACGCGGAAGGGTGGTTCCCGGA R: AGACGCGGTAATCGAAGATGAAA | 925 | 500 | 687 |
| S/n | Parameter | Code | Description |
|---|---|---|---|
| 1. | Plant height | PH | This was measured from the soil surface to the tip of the tallest flag leaf. The unit of measurement is centimetres (cm). |
| 2. | Days to flowering | DF | Counted from the days from planting until 50% flowering. The unit of measurement is days. |
| 3. | Days to maturity | DM | Counted from the days from planting until 80% of the grains became golden yellow. The unit of measurement is days. |
| 4. | Total number of productive tillers per plant | NT | Counted as all of the tillers on each plant bearing panicles with grains. The unit of measurement is number. |
| 5. | Panicle length | PL | Measured from the first node to the tip of the last spikelet (excluding awns). The unit of measurement is centimetres (cm). |
| 6. | Total number of filled grains per panicle | TNG/P | This was recorded as the total number of matured spikelets filled with grains per panicle. The unit of measurement is number. |
| 7. | Total number of unfilled grains per panicle | NUFG | Counted as the number of spikelets without seed or grain. The unit of measurement is number. |
| 8. | 1000-grain weight | 1000-GW | One thousand filled grains were counted and weighed. The unit of measurement is gramme (g). |
| 9. | Grain yield per plant | Y/P | All the grains harvested from each plant were weighed. The unit of measurement is gramme (g). |
| 10. | Seed length | SL | Ten grains were measured using a Vernier calliper (Mitutoyo, Japan) from the base of the lowermost sterile lemma to the tip of the fertile lemma or palea. The unit of measurement is millimetre (mm). |
| 11. | Seed width | SW | Ten grains were measured using a Vernier calliper (Mitutoyo, Japan) from the distance across the fertile lemma and palea at the widest point. The unit of measurement is millimetre (mm). |
| 12. | Seed length:width ratio | SLWR | This was recorded as seed length divided by the seed width. |
| 13. | Seed shape | SS | The seed shape was categorized using the record taken on seed length:width ratio. |
| 14. | Grain yield per hectare | GY/ha | Grain yield per hectare was calculated using the equivalence of the grain yield per plant with a spacing of 25 cm × 25 cm. |
| s/n | Improved Lines | Xa21FR (Xa21) | pTA248 (Xa21) | Xa13prom (xa13) | RM21 (xa5) | MP (Xa4) | RM6836 (Pi2, Pi9, PiZ) | RM8225 (Piz) |
|---|---|---|---|---|---|---|---|---|
| 1. | BC2F2–157 | -- | ++ | ++ | ++ | ++ | ++ | ++ |
| 2. | BC2F2–122 | -- | - | ++ | ++ | +- | ++ | ++ |
| 3. | BC2F2–9 | ++ | - | ++ | -- | ++ | ++ | ++ |
| 4. | BC2F2–196 | +- | ++ | +- | -- | ++ | ++ | ++ |
| 5. | BC2F2–120 | -- | -- | ++ | +- | ++ | ++ | ++ |
| 6. | BC2F2–208 | ++ | -- | +- | -- | ++ | ++ | ++ |
| 7. | BC2F2–155 | ++ | ++ | +- | -- | ++ | ++ | ++ |
| 8. | BC2F2–4 | ++ | - | +- | -- | ++ | ++ | ++ |
| 9. | BC2F2–109 | -- | - | -- | ++ | ++ | ++ | ++ |
| 10. | BC2F2–161 | - | - | - | ++ | ++ | ++ | ++ |
| 11. | BC2F2–144 | -- | - | ++ | -- | ++ | ++ | ++ |
| 12. | BC2F2–1 | -- | -- | ++ | -- | ++ | ++ | ++ |
| 13. | BC2F2–50 | -- | -- | ++ | -- | ++ | ++ | ++ |
| 14. | BC2F2–172 | - | -- | ++ | -- | ++ | ++ | ++ |
| 15. | BC2F2–166 | - | - | -- | ++ | ++ | ++ | ++ |
| 16. | BC2F2–14 | - | - | ++ | -- | ++ | ++ | ++ |
| Improved Lines | PH (cm) | FLWR | NP/H | DF | DM | NT | PL (cm) | TNG/P | 1000 GW (g) | TGW/H (g) | SLWR | Y/HA (t/ha) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 103.0 o | 13.5 e | 11.0 g | 72.0 a,b | 108.0 a,b,c | 11.0 h | 38.6 a | 154.0 f,g,h | 78.6 e | 57.4 e | 3.4 h | 9.2 b,c |
| 2 | 117.0 d | 13.2 g | 13.0 g,f | 78.0 a,b | 102.0 b,c | 15.0 e | 32.0 d,c | 152.0 g,h,i | 81.8 a,b | 53.6 g | 3.8 f | 8.6 c,d |
| 3 | 110.5 j | 12.2 j | 15.0 d,e,f | 73.0 a,b | 106.0 a,b,c | 17.0 c | 30.1 g,f | 137.0 i | 82.8 a | 59.3 d | 3.7 g | 9.5 b |
| 4 | 109.2 k | 9.7 o | 20.0 a | 75.0 b | 103.0 c | 18.0 b | 34.1 b | 172.0 d,e,f | 76.3 g | 56.3 f | 4.1 b | 9.0 c,b |
| 5 | 114.7 g | 13.1 h | 16.0 d,c | 74.0 a,b | 105.0 a,b,c | 14.0 f | 34.4 b | 177.0 d,e | 81.9 a,b | 60.7 c | 3.9 e | 9.7 b |
| 6 | 102.2 p | 11.5 m | 17.0 c,d,e | 79.0 a,b | 109.0 a,b,c | 18.0 b | 29.6 g | 142.0 h,i | 80.6 c | 44.5 k | 3.9 e | 7.1 f,g,h |
| 7 | 117.5 c | 11.7 l | 15.0 d,e,f | 77.0 a,b | 110.0 a,b,c | 17.0 c | 29.7 g | 136.0 i | 78.6 e | 45.1 j | 3.8 f | 7.2 e,f,g,h |
| 8 | 119.8 a | 17.6 a | 22.0 a,b | 76.0 a,b | 104.0 a,b,c | 26.0 a | 30.2 g,f | 196.0 b,c | 79.5 d | 49.9 h | 4.2 a | 8.0 d,e,f |
| 9 | 115.7 f | 13.3 f | 17.0 b,d,c | 72.0 a,b | 107.0 a,b | 15.0 e | 32.6 c | 166.0 e,f,g | 76.8 g | 46.1 j | 4.0 c | 7.4 e,f,g,h |
| 10 | 112.5 i | 12.1 k | 14.0 e,f | 79.0 a | 107.0 a,b,c | 16.0 d | 32.7 d,e | 207.0 a,b | 82.5 a | 64.0 b | 4.0 c | 10.2 b |
| 11 | 114.0 h | 15.0 d | 13.0 g,f | 77.0 a,b | 102.0 b,c | 12.0 g | 33.8 b | 203.0 a,b | 77.9 f,e | 43.6 l | 4.0 c | 7.0 e,f,g,h |
| 12 | 118.5 b | 11.1 n | 20.0 a,b,c | 75.0 a,b | 105.0 a,b,c | 17.0 c | 32.0 d,c | 205.0 a,b | 81.3 b,c | 41.5 m | 3.8 f | 6.6 h |
| 13 | 96.5 q | 15.3 c | 16.0 c,d,e | 74.0 b | 109.0 a,b,c | 18.0 b | 34.3 b | 209.0 a,b | 77.3 f | 73.8 a | 4.2 a | 11.8 a |
| 14 | 106.5 m | 16.3 b | 13.0 g,f | 76.0 a | 106.0 a,b,c | 15.0 e | 30.9 f,e | 211.0 a | 80.6 c | 48.2 i | 2.9 i | 7.7 e,f,g |
| 15 | 104.0 n | 9.7 o | 14.0 e,f | 75.0 a,b | 103.0 a,b,c | 17.0 c | 34.1 b | 172.0 d,e,f,g | 76.3 g | 56.3 f | 4.1 b | 9.0 b,c |
| 16 | 108.7 l | 15.0 d | 13.0 g,f | 77.0 a,b | 102.0 a,b,c | 11.0 h | 33.8 b | 203.0 d,c | 77.9 e,f | 43.6 l | 4.0 c | 7.0 g,h |
| Recurrent parent | 116.50 e | 12.56 i | 15.00 d,e,f | 85.67 a | 120.67 a | 15.00 e | 31.83 c,d,e | 148.00 f,g,h | 75.53 g | 50.41 h | 3.92 d | 8.07 ed |
| Reaction | Observed | Expected | Chi-Square (3:1) | p-Value |
|---|---|---|---|---|
| BLB | ||||
| Resistant | 170 | 165 | 0.15 | p > 0.05 |
| Susceptible | 44 | 55 | 2.22 | p > 0.05 |
| Total | 220 | 220 | 2.37 | p > 0.05 |
| Blast | ||||
| Resistant | 155 | 165 | 0.63 | p > 0.05 |
| Susceptible | 65 | 55 | 1.89 | p > 0.05 |
| Total | 220 | 220 | 2.53 | p > 0.05 |
| Xoo Pathotype P7.7 | Magnaportheoryzae Pathotype P7.2 | |||||
|---|---|---|---|---|---|---|
| Trait | BLD | BLT | %DLA | BLD | BLT | %DLA |
| BLD | 1.00 | 1.00 | ||||
| BLT | 0.99 ** | 1.00 | 0.99 ** | 1.00 | ||
| %DLA | 0.99 | 0.99 | 1.00 | 0.96 | 0.96 | 1.00 |
| PH | FLL | FLW | LA | LAI | FLWR | NPH | DF | DM | NL | NT | PL | TNGP | NUFG | NTGW | TGWH | SL | SW | SLWR | YHA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PH | 1.00 | 0.125 | 0.095 | 0.151 | 0.148 | 0.082 | −0.138 | −0.115 | 0.027 | −0.254 | −0.254 | 0.455 | 0.496 | −0.364 | −0.286 | 0.048 | −0.334 | −0.470 | 0.135 | 0.049 |
| FLL | 1.00 | 0.247 | 0.950 ** | 0.913 ** | 0.942 ** | −0.073 | −0.147 | −0.025 | −0.027 | −0.027 | 0.054 | 0.262 | −0.295 | −0.031 | 0.052 | −0.302 | −0.173 | −0.108 | 0.052 | |
| FLW | 1.00 | 0.533 * | 0.590 * | −0.091 | −0.216 | −0.187 | 0.340 | −0.216 | −0.216 | 0.107 | −0.612 * | −0.022 | 0.201 | −0.074 | 0.147 | 0.133 | −0.039 | −0.074 | ||
| LA | 1.00 | 0.988 ** | 0.791 ** | −0.122 | −0.212 | 0.067 | −0.078 | −0.078 | 0.113 | 0.032 | −0.281 | 0.008 | 0.027 | −0.214 | −0.118 | −0.096 | 0.028 | |||
| LAI | 1.00 | 0.732 ** | −0.013 | −0.268 | 0.087 | 0.022 | 0.022 | 0.175 | −0.014 | −0.279 | 0.024 | 0.132 | −0.176 | −0.143 | −0.061 | 0.133 | ||||
| FLWR | 1.00 | −0.009 | −0.070 | −0.122 | 0.036 | 0.036 | −0.009 | 0.478 | −0.282 | −0.078 | 0.073 | −0.369 | −0.213 | −0.115 | 0.073 | |||||
| NPH | 1.00 | −0.304 | 0.057 | 0.962 ** | 0.962 ** | 0.320 | 0.109 | −0.127 | 0.027 | 0.928 ** | 0.143 | −0.217 | 0.235 | 0.927** | ||||||
| DF | 1.00 | −0.108 | −0.225 | −0.225 | −0.489 | 0.095 | 0.152 | 0.324 | −0.231 | 0.007 | −0.145 | 0.150 | −0.231 | |||||||
| DM | 1.00 | −0.048 | −0.048 | −0.082 | −0.265 | −0.129 | −0.075 | 0.172 | −0.137 | −0.024 | −0.136 | 0.171 | ||||||||
| NL | 1.00 | 1.000 ** | 0.297 | 0.037 | −0.135 | 0.024 | 0.867 ** | 0.249 | −0.150 | 0.276 | 0.867 ** | |||||||||
| NT | 1.00 | 0.297 | 0.037 | −0.135 | 0.024 | 0.867 ** | 0.249 | −0.150 | 0.276 | 0.867 ** | ||||||||||
| PL | 1.00 | 0.175 | −0.516 * | −0.345 | 0.423 | −0.099 | −0.072 | −0.029 | 0.423 | |||||||||||
| TNGP | 1.00 | −0.029 | −0.069 | 0.151 | −0.390 | −0.387 | 0.050 | 0.151 | ||||||||||||
| NUFG | 1.00 | 0.437 | −0.297 | 0.383 | 0.191 | 0.197 | −0.297 | |||||||||||||
| NTGW | 1.00 | 0.069 | 0.138 | 0.514 | −0.329 | 0.069 | ||||||||||||||
| TGWH | 1.00 | 0.069 | −0.240 | 0.193 | 1.000 ** | |||||||||||||||
| SL | 1.00 | 0.316 | 0.622 ** | 0.069 | ||||||||||||||||
| SW | 1.00 | −0.532 * | −0.241 | |||||||||||||||||
| SLWR | 1.00 | 0.194 | ||||||||||||||||||
| YHA | 1.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chukwu, S.C.; Rafii, M.Y.; Oladosu, Y.; Okporie, E.O.; Akos, I.S.; Musa, I.; Swaray, S.; Jalloh, M.; Al-Mamun, M. Genotypic and Phenotypic Selection of Newly Improved Putra Rice and the Correlations among Quantitative Traits. Diversity 2022, 14, 812. https://doi.org/10.3390/d14100812
Chukwu SC, Rafii MY, Oladosu Y, Okporie EO, Akos IS, Musa I, Swaray S, Jalloh M, Al-Mamun M. Genotypic and Phenotypic Selection of Newly Improved Putra Rice and the Correlations among Quantitative Traits. Diversity. 2022; 14(10):812. https://doi.org/10.3390/d14100812
Chicago/Turabian StyleChukwu, Samuel C., Mohd Y. Rafii, Yusuff Oladosu, Emmanuel O. Okporie, Ibrahim S. Akos, Ibrahim Musa, Senesie Swaray, Momodu Jalloh, and Md. Al-Mamun. 2022. "Genotypic and Phenotypic Selection of Newly Improved Putra Rice and the Correlations among Quantitative Traits" Diversity 14, no. 10: 812. https://doi.org/10.3390/d14100812
APA StyleChukwu, S. C., Rafii, M. Y., Oladosu, Y., Okporie, E. O., Akos, I. S., Musa, I., Swaray, S., Jalloh, M., & Al-Mamun, M. (2022). Genotypic and Phenotypic Selection of Newly Improved Putra Rice and the Correlations among Quantitative Traits. Diversity, 14(10), 812. https://doi.org/10.3390/d14100812









