Phylogeography of Hypomasticus copelandii (Teleostei, Anostomidae) Reveals Distinct Genetic Lineages along Atlantic Coastal Drainages of Eastern Brazil
Abstract
:1. Introduction
2. Materials and Methods
2.1. Taxon Sampling, DNA Extraction, and Sequencing
2.2. Alignment and Phylogenetic Analyses
2.3. Species Delimitation Analyses
2.4. Divergence Time Estimates
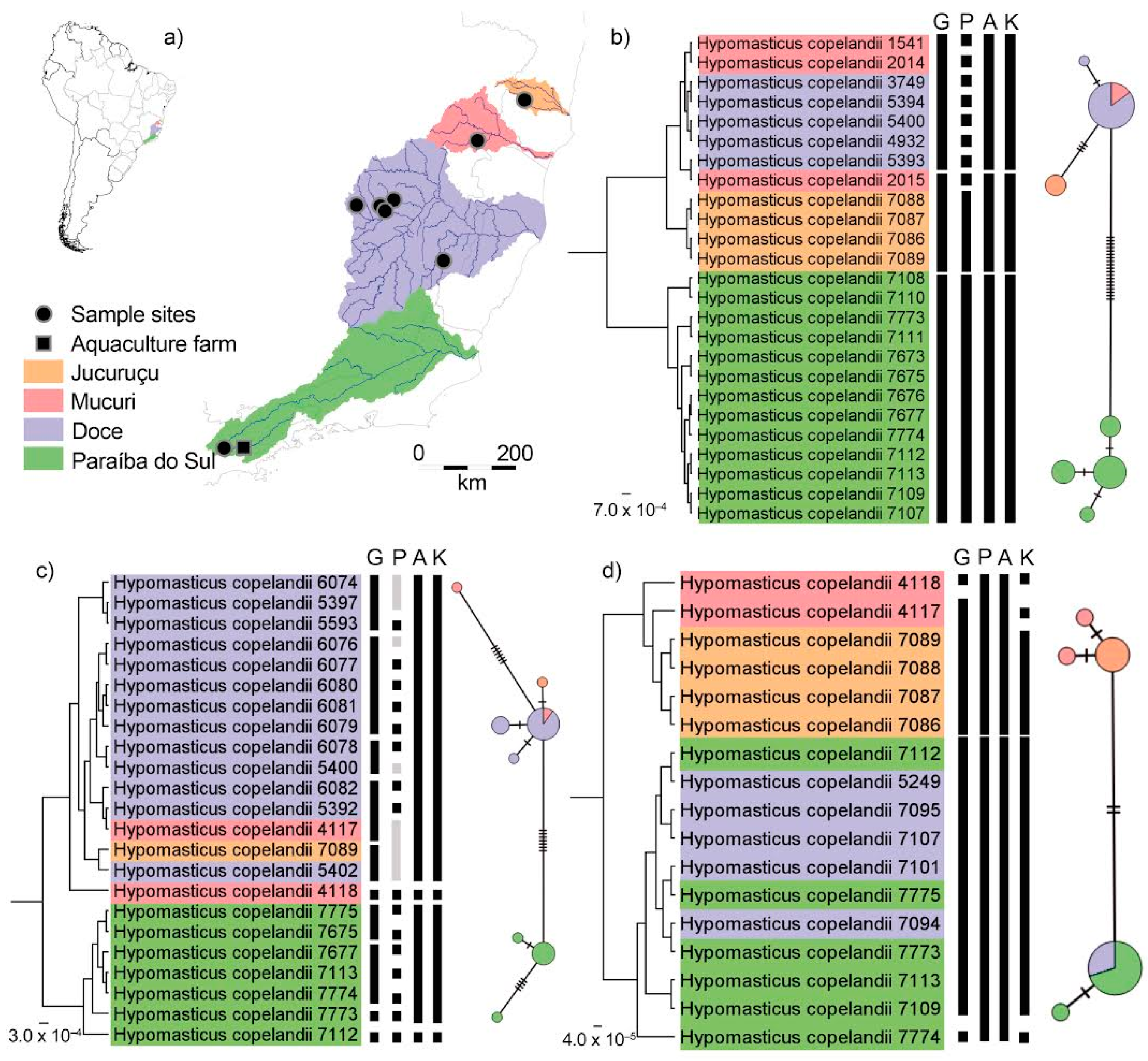
3. Results
3.1. Genetic Distances
3.2. Species Delimitation
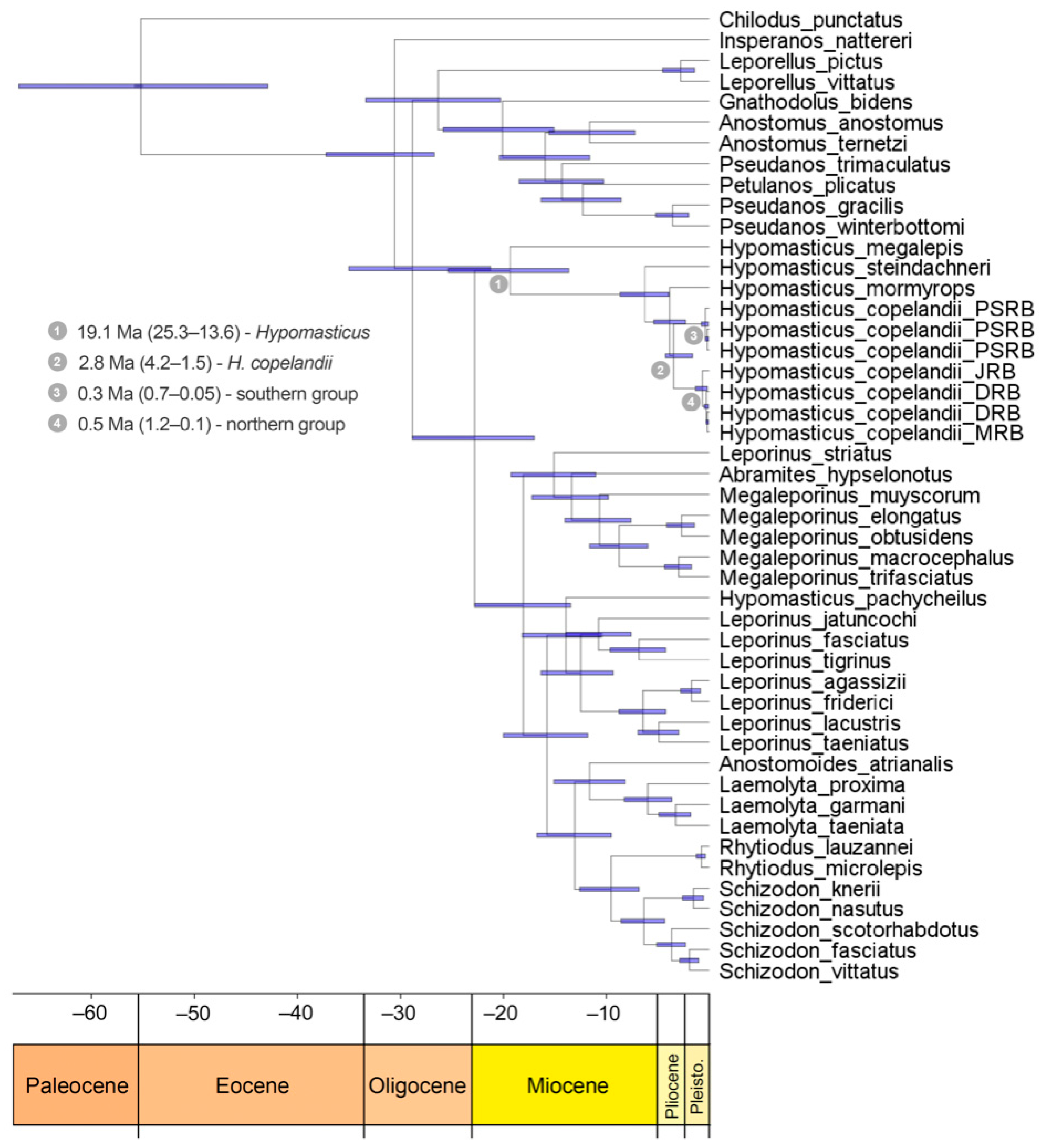
3.3. Timing of Diversification of Hypomasticus Copelandii
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Albert, J.S.; Reis, R.E. Historical Biogeography of Neotropical Freshwater Fishes; University of California Press: Oakland, CA, USA, 2011; p. 388. [Google Scholar]
- Albert, J.S.; Tagliacollo, V.A.; Dagosta, F. Diversification of neotropical freshwater fishes. Annu. Rev. Ecol. Evol. Syst. 2020, 51, 27–53. [Google Scholar] [CrossRef]
- Hubert, N.; Duponchelle, F.; Nuñez, J.; Rivera, R.; Bonhomme, F.; Renno, J.F. Isolation by distance and Pleistocene expansion of the lowland populations of the white piranha Serrasalmus rhombeus. Mol. Ecol. 2007, 16, 2488–2503. [Google Scholar] [CrossRef]
- Lundberg, J.G.; Marshall, L.G.; Guerrero, J.; Horton, B.K.; Malabarba, M.C.; Wesselingh, F. The stage for Neotropical fish diversification: A history of tropical South American rivers. In Phylogeny and Classification of Neotropical Fishes; Malabarba, L.R., Reis, R.E., Vari, R.P., Lucena, Z.M.S., Lucena, C.A.S., Eds.; Edipucrs: Porto Alegre, Brazil, 1998; pp. 13–48. [Google Scholar]
- Melo, B.F.; Sidlauskas, B.L.; Near, T.J.; Roxo, F.F.; Ghezelayagh, A.; Ochoa, L.E.; Stiassny, M.L.J.; Arroyave, J.; Chang, J.; Faircloth, B.C.; et al. Accelerated diversification explains the exceptional species richness of Tropical characoid fishes. Syst. Biol. 2021. [Google Scholar] [CrossRef]
- Ribeiro, A.C. Tectonic history and the biogeography of the freshwater fishes from the coastal drainages of eastern Brazil: An example of faunal evolution associated with a divergent continental margin. Neotrop. Ichthyol. 2006, 4, 225–246. [Google Scholar] [CrossRef]
- Ruokolainen, K.; Moulatlet, G.M.; Zuquim, G.; Hoorn, C.; Tuomisto, H. Geologically recent rearrangements in central Amazonian river network and their importance for the riverine barrier hypothesis. Front. Biogeogr. 2019, 11, e45046e. [Google Scholar] [CrossRef] [Green Version]
- Thomaz, A.T.; Malabarba, L.R.; Bonatto, S.L.; Knowles, L.L. Testing the effect of palaeodrainages versus habitat stability on genetic divergence in riverine systems: Study of a Neotropical fish of the Brazilian coastal Atlantic Forest. J. Biogeogr. 2015, 42, 2389–2401. [Google Scholar] [CrossRef]
- Thomaz, A. Riverscape Genetics: Insights into the Drivers of Divergence in Coastal Brazilian Fishes. Ph.D. Thesis, University of Michigan, Ann Harbor, MI, USA, 2017. [Google Scholar]
- Thomaz, A.T.; Lacey Knowles, L. Flowing into the unknown: Inferred paleodrainages for studying the ichthyofauna of brazilian coastal rivers. Neotrop. Ichthyol. 2018, 16, e180019. [Google Scholar] [CrossRef]
- Dagosta, F.C.P.; de Pinna, M.; Peres, C.A.; Tagliacollo, V.A. Existing protected areas provide a poor safety-net for threatened Amazonian fish species. Aquat. Conserv. Mar. Freshw. Ecosyst. 2020, 31, 1167–1189. [Google Scholar] [CrossRef]
- Fernandes, G.W.; Goulart, F.F.; Ranieri, B.D.; Coelho, M.S.; Dales, K.; Boesche, N.; Bustamante, M.; Carvalho, F.A.; Carvalho, D.C.; Dirzo, R.; et al. Deep into the mud: Ecological and socio-economic impacts of the dam breach in Mariana, Brazil. Nat. Conserv. 2016, 14, 35–45. [Google Scholar] [CrossRef]
- Ota, R.R.; Message, H.J.; da Graça, W.J.; Pavanelli, C.S. Neotropical Siluriformes as a model for insights on determining biodiversity of animal groups. PLoS ONE 2015, 10, e0132913. [Google Scholar] [CrossRef] [Green Version]
- Reis, R.E.; Albert, J.S.; Di Dario, F.; Mincarone, M.M.; Petry, P.; Rocha, L.A. Fish biodiversity and conservation in South America. J. Fish Biol. 2016, 89, 12–47. [Google Scholar] [CrossRef] [Green Version]
- Melo, B.F.; Ochoa, L.E.; Vari, R.P.; Oliveira, C. Cryptic species in the Neotropical fish genus Curimatopsis (Teleostei, Characiformes). Zool. Scr. 2016, 45, 650–658. [Google Scholar] [CrossRef]
- Ochoa, L.E.; Melo, B.F.; García-Melo, J.E.; Maldonado-Ocampo, J.A.; Souza, C.S.; Albornoz-Garzón, J.G.; Conde-Saldaña, C.C.; Villa-Navarro, F.; Ortega-Lara, A.; Oliveira, C. Species delimitation reveals an underestimated diversity of Andean catfishes of the family Astroblepidae (Teleostei: Siluriformes). Neotrop. Ichthyol. 2020, 18, 1–19. [Google Scholar] [CrossRef]
- Britz, R.; Hundsdörfer, A.; Fritz, U. Funding, training, permits—The three big challenges of taxonomy. Megataxa 2020, 1, 49–52. [Google Scholar] [CrossRef] [Green Version]
- Ely, C.V.; Bordignon, S.A.d.L.; Trevisan, R.; Boldrini, I.I. Implications of poor taxonomy in conservation. J. Nat. Conserv. 2017, 36, 10–13. [Google Scholar]
- Pereira, L.H.G.; Maia, G.M.G.; Hanner, R.; Foresti, F.; Oliveira, C. DNA barcodes discriminate freshwater fishes from the Paraíba do sul River Basin, São Paulo, Brazil. Mitochondrial Dna 2011, 22 (Suppl. 1), 71–79. [Google Scholar] [CrossRef]
- Pereira, L.H.G.; Hanner, R.; Foresti, F.; Oliveira, C. Can DNA barcoding accurately discriminate megadiverse Neotropical freshwater fish fauna? BMC Genet. 2013, 14, 20. [Google Scholar] [CrossRef] [Green Version]
- Calegari, B.B.; Vari, R.P.; Reis, R.E. Phylogenetic systematics of the driftwood catfishes (Siluriformes: Auchenipteridae): A combined morphological and molecular analysis. Zool. J. Linn. Soc. 2019, 187, 661–773. [Google Scholar] [CrossRef]
- Ramirez, J.L.; Carvalho-Costa, L.F.; Venere, P.C.; Carvalho, D.C.; Troy, W.P.; Galetti, P.M. Testing monophyly of the freshwater fish Leporinus (Characiformes, Anostomidae) through molecular analysis. J. Fish Biol. 2016, 88, 1204–1214. [Google Scholar] [CrossRef]
- Silva, G.S.C.; Melo, B.F.; Oliveira, C.; Benine, R.C. Revision of the South American genus Tetragonopterus Cuvier, 1816 (Teleostei: Characidae) with description of four new species. Zootaxa 2016, 4200, 1–46. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; DeWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [Green Version]
- Machado, V.N.; Collins, R.A.; Ota, R.P.; Andrade, M.C.; Farias, I.P.; Hrbek, T. One thousand DNA barcodes of piranhas and pacus reveal geographic structure and unrecognised diversity in the Amazon. Sci. Rep. 2018, 8, 8387. [Google Scholar] [CrossRef]
- Rossini, B.C.; Oliveira, C.A.M.; de Melo, F.A.G.; de Araújo Bertaco, V.; de Astarloa, J.M.D.; Rosso, J.J.; Foresti, F.; Oliveira, C. Highlighting Astyanax species diversity through DNA Barcoding. PLoS ONE 2016, 11, e0167203. [Google Scholar] [CrossRef]
- Britski, H.A.; Melo, B.F.; Vari, R.P.; Oliveira, C. Revalidation and redescription of Steindachnerina nigrotaenia and redescription of S. insculpta (Characiformes: Curimatidae). Neotrop. Ichthyol. 2019, 17, e180076. [Google Scholar] [CrossRef]
- Carvalho, D.C.; Oliveira, D.A.A.; Beheregaray, L.B.; Torres, R.A. Hidden genetic diversity and distinct evolutionarily significant units in an commercially important Neotropical apex predator, the catfish Pseudoplatystoma corruscans. Conserv. Genet. 2012, 13, 1671–1675. [Google Scholar] [CrossRef]
- Pires, A.A.; Ramirez, J.L.; Galetti, P.M., Jr.; Troy, W.P.; Freitas, P.D. Molecular analysis reveals hidden diversity in Zungaro (Siluriformes: Pimelodidade): A genus of giant South American catfish. Genetica 2017, 145, 335–340. [Google Scholar] [CrossRef]
- Ramirez, J.L.; Birindelli, J.L.; Carvalho, D.C.; Affonso, P.R.A.M.; Venere, P.C.; Ortega, H.; Carrillo-Avila, M.; Rodríguez-Pulido, J.A.; Galetti, P.M., Jr. Revealing hidden diversity of the underestimated Neotropical ichthyofauna: DNA Barcoding in the recently described genus Megaleporinus (Characiformes: Anostomidae). Front. Genet. 2017, 8, 149. [Google Scholar] [CrossRef] [Green Version]
- Gomes, L.C.; Pessali, T.C.; Sales, N.G.; Pompeu, P.S.; Carvalho, D.C. Integrative taxonomy detects cryptic and overlooked fish species in a neotropical river basin. Genetica 2015, 143, 581–588. [Google Scholar] [CrossRef]
- Pugedo, M.L.; de Andrade Neto, F.R.; Pessali, T.C.; Birindelli, J.L.O.; Carvalho, D.C. Integrative taxonomy supports new candidate fish species in a poorly studied neotropical region: The Jequitinhonha River Basin. Genetica 2016, 144, 341–349. [Google Scholar] [CrossRef]
- Ramirez, J.L.; Galetti, P.M., Jr. DNA barcode and evolutionary relationship within Laemolyta Cope 1872 (Characiformes: Anostomidae) through molecular analyses. Mol. Phylogenet. Evol. 2015, 93, 77–82. [Google Scholar] [CrossRef]
- Silva-Santos, R.; Ramirez, J.L.; Galetti, P.M., Jr.; Freitas, P.D. Molecular evidences of a hidden complex scenario in Leporinus cf. friderici. Front. Genet. 2018, 9, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fricke, R.; Eschmeyer, W.N.; Fong, J.D. CAS—Eschmeyer’s Catalog of Fishes—Genera/Species by Family/Subfamily. Available online: https://researcharchive.calacademy.org/research/ichthyology/catalog/SpeciesByFamily.asp (accessed on 24 June 2020).
- Garavello, J.C.; Britski, H.A. Family Anostomidae (Headstanders). Check List Freshw. Fishes South Cent. Am. 2003, 1, 71–84. [Google Scholar]
- Sidlauskas, B.L.; Birindelli, J.L.O. Family Anostomidae—toothed headstanders. In Field Guide to the Fishes of the Amazon, Orinoco & Guianas; Princeton University Press: Princeton, NJ, USA, 2017; pp. 82–89. [Google Scholar]
- Ramirez, J.L.; Birindelli, J.L.O.; Galetti, P.M. A new genus of Anostomidae (Ostariophysi: Characiformes): Diversity, phylogeny and biogeography based on cytogenetic, molecular and morphological data. Mol. Phylogenet. Evol. 2017, 107, 308–323. [Google Scholar] [CrossRef]
- Sidlauskas, B.L.; Assega, F.M.; Melo, B.F.; Oliveira, C.; Birindelli, J.L.O. Total evidence phylogenetic analysis reveals polyphyly of Anostomoides and uncovers an unexpectedly ancient genus of Anostomidae fishes (Characiformes). Zool. J. Linn. Soc. 2021. [Google Scholar] [CrossRef]
- Sidlauskas, B.L.; Vari, R.P. Phylogenetic relationships within the South American fish family Anostomidae (Teleostei, Ostariophysi, Characiformes). Zool. J. Linn. Soc. 2008, 154, 70–210. [Google Scholar] [CrossRef] [Green Version]
- Burns, M.D.; Frable, B.W.; Sidlauskas, B.L. A new species of Leporinus (Characiformes: Anostomidae), from the Orinoco Basin, Venezuela. Copeia 2014, 2014, 206–214. [Google Scholar] [CrossRef]
- Britski, H.A.; Garavello, J.C. Sobre Leporinus octofasciatus Steindachner da bacia do Paraná (Pisces, Anostomidae). Papéis Avulsos Zool. 1978, 31, 237–250. [Google Scholar]
- Garavello, J.C.; Britski, H.A. Revisão Taxonomica do Gênero Leporinus Spix, 1829 (Ostariophysi, Anostomidae); Universidade de São Paulo: São Paulo, Brazil, 1979. [Google Scholar]
- Sidlauskas, B. Testing for unequal rates of morphological diversification in the absence of a detailed phylogeny: A case study from Characiform fishes. Evolution 2007, 61, 299–316. [Google Scholar] [CrossRef]
- Birindelli, J.L.O.; Melo, B.F.; Ribeiro-Silva, L.R.; Diniz, D.; Oliveira, C. A new species of Hypomasticus from Eastern Brazil based on morphological and molecular data (Characiformes, Anostomidae). Copeia 2020, 108, 416–425. [Google Scholar] [CrossRef]
- Steindachner, F. Die Süsswasserfische des südöstlichen Brasilien (II). Sitzungsberichte der Kaiserlichen Akademie der Wissenschaften. Math. Nat. Cl. 1875, 71, 211–245. [Google Scholar]
- Costa, A.P.R. Aspectos da biologia reprodutiva de fêmeas do Piau-vermelho Leporinus copelandii Steindachner, 1875 (Pisces, Anostomidae), na bacia do Baixo Rio Paraíba do Sul (RJ). Master’s Thesis, Centro de Ciências e Tecnologia Agropecuárias, Universidade Estadual do Norte Fluminense Darcy Ribeiro, Campos de Goytacazes, Brazil, 1999. [Google Scholar]
- Abell, R.; Thieme, M.L.; Revenga, C.; Bryer, M.; Kottelat, M.; Bogutskaya, N.; Coad, B.; Mandrak, N.; Balderas, S.C.; Bussing, W.; et al. Freshwater ecoregions of the world: A new map of biogeographic units for freshwater biodiversity conservation. BioScience 2008, 58, 403–414. [Google Scholar] [CrossRef] [Green Version]
- Escobar, H. Mud tsunami wreaks ecological havoc in Brazil. Science 2015, 350, 1138–1139. [Google Scholar] [CrossRef] [PubMed]
- Garcia, L.C.; Ribeiro, D.B.; Roque, F.d.O.; Ochoa-Quintero, J.M.; Laurance, W.F. Brazil’s worst mining disaster: Corporations must be compelled to pay the actual environmental costs. Ecol. Appl. 2017, 27, 5–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, L.E.d.O.; Correa, L.B.; Sá, F.; Neto, R.R.; Bernardino, A.F. The impacts of the Samarco mine tailing spill on the Rio Doce estuary, Eastern Brazil. Mar. Pollut. Bull. 2017, 120, 28–36. [Google Scholar] [CrossRef]
- Sarmento-Soares, L.M.; Rodrigues, L.N.; Martins-Pinheiro, R.F. Peixes do rio Doce segundo as coleções. Bol. Soc. Bras. Ictiol. 2017, 123, 9–25. [Google Scholar]
- Aljanabi, S.M.; Martinez, I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997, 25, 4692–4693. [Google Scholar] [CrossRef]
- Ivanova, N.V.; Zemlak, T.S.; Hanner, R.H.; Hebert, P.D.N. Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Notes 2007, 7, 544–548. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Møller, P.R.; Sigsgaard, E.E.; Knudsen, S.W.; Jørgensen, O.A.; Willerslev, E. Environmental DNA from seawater samples correlate with trawl catches of subarctic, deepwater fishes. PLoS ONE 2016, 11, e0165252. [Google Scholar] [CrossRef] [Green Version]
- Chow, S.; Hazama, K. Universal PCR primers for S7 ribosomal protein gene introns in fish. Mol. Ecol. 1998, 7, 1255–1256. [Google Scholar] [PubMed]
- Ratnasingham, S.; Hebert, P.D.N. Bold: The Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, X. DAMBE5: A comprehensive software package for Data Analysis in Molecular Biology and Evolution. Mol. Biol. Evol. 2013, 30, 1720–1728. [Google Scholar] [CrossRef] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE) Institute of Electrical and Electronics Engineers, New Orleans, LA, USA, 14 November 2010. [Google Scholar]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2021, 22, 2688–2690. [Google Scholar] [CrossRef]
- Fujisawa, T.; Barraclough, T.G. Delimiting species using single-locus data and the Generalized Mixed Yule Coalescent approach: A revised method and evaluation on simulated data sets. Syst. Biol. 2013, 62, 707–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef] [Green Version]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef]
- Ward, R.D. DNA barcode divergence among species and genera of birds and fishes. Mol. Ecol. Resour. 2009, 9, 1077–1085. [Google Scholar] [CrossRef]
- Milan, D.T.; Mendes, I.S.; Damasceno, J.S.; Teixeira, D.F.; Sales, N.G.; Carvalho, D.C. New 12S metabarcoding primers for enhanced Neotropical freshwater fish biodiversity assessment. Sci. Rep. 2020, 10, 17966. [Google Scholar] [CrossRef]
- Brown, S.D.J.; Collins, R.A.; Boyer, S.; Lefort, M.-C.; Malumbres-Olarte, J.; Vink, C.J.; Cruickshank, R.H. Spider: An R package for the analysis of species identity and evolution, with particular reference to DNA barcoding—BROWN—2012—Molecular Ecology Resources—Wiley Online Library. Mol. Ecol. Resour. 2012, 12, 562–565. [Google Scholar] [CrossRef] [PubMed]
- Team, R.C. R: A Language and Environment for Statistical Computing. 2013. Available online: http://r.meteo.uni.wroc.pl/web/packages/dplR/vignettes/intro-dplR.pdf (accessed on 24 June 2020).
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanfear, R.; Calcott, B.; Ho, S.Y.W.; Guindon, S. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [Green Version]
- Ameghino, F. Sur l’Arrhinolemur, mammifére aberrant du tertiare du Paraná. Comptes Rendus L’académie Sci. 1898, 127, 395–396. [Google Scholar]
- Bogan, S.; Sidlauskas, B.; Vari, R.P.; Agnolin, F. Arrhinolemur scalabrinii Ameghino, 1898, of the late Miocene: A taxonomic journey from the Mammalia to the Anostomidae (Ostariophysi: Characiformes). Neotrop. Ichthyol. 2021, 10, 555–560. [Google Scholar] [CrossRef] [Green Version]
- Günther, A. Diagnoses of some new freshwater fishes from Surinam and Brazil, in the collection of the British Museum. J. Nat. Hist. 1868, 1, 475–481. [Google Scholar] [CrossRef]
- Kner, R. Beiträge zur Familie der Characinen. Sitzungsberichte der Kaiserlichen Akademie der Wissenschaften. Math. Nat. Cl. 1858, 30, 75–80. [Google Scholar]
- Lofeu, L.; Anelli, V.; Straker, L.C.; Kohlsdorf, T. Developmental plasticity reveals hidden fish phenotypes and enables morphospace diversification. Evolution 2021, 75, 1170–1188. [Google Scholar] [CrossRef]
- Ramirez, J.L.; Santos, C.A.; Machado, C.B.; Oliveira, A.K.; Garavello, J.C.; Britski, A.H.; Galetti, P.M., Jr. Molecular phylogeny and species delimitation of the genus Schizodon (Characiformes, Anostomidae). Mol. Phylogenetics Evol. 2020, 153, 106959. [Google Scholar] [CrossRef] [PubMed]
- Burns, M.D.; Chatfield, M.; Birindelli, J.L.O.; Sidlauskas, B.L. Systematic assessment of the Leporinus desmotes species complex, with a description of two new species. Neotrop. Ichthyol. 2017, 15, e160166. [Google Scholar] [CrossRef] [Green Version]
- Camelier, P.; Zanata, A.M. Biogeography of freshwater fishes from the Northeastern Mata Atlântica freshwater ecoregion: Distribution, endemism, and area relationships. Neotrop. Ichthyol. 2015, 12, 683–698. [Google Scholar] [CrossRef] [Green Version]
- Quevedo, R.; Reis, R.E. Pogonopoma obscurum: A new species of Loricariid catfish (Siluriformes: Loricariidae) from Southern Brazil, with comments on the genus Pogonopoma. Copeia 2002, 2002, 402–410. [Google Scholar] [CrossRef]
- Pereira, T.L.; Santos, U.; Schaefer, C.E.; Souza, G.O.; Paiva, S.R.; Malabarba, L.R.; Schmidt, E.E.; Dergam, J.A. Dispersal and vicariance of Hoplias malabaricus (Bloch, 1794) (Teleostei, Erythrinidae) populations of the Brazilian continental margin. J. Biogeogr. 2013, 40, 905–914. [Google Scholar] [CrossRef]
- Argolo, L.A.; López-Fernández, H.; Batalha-Filho, H.; de Mello Affonso, P.R.A. Unraveling the systematics and evolution of the ‘Geophagus’ brasiliensis (Cichliformes: Cichlidae) species complex. Mol. Phylogenet. Evol. 2020, 50, 106855. [Google Scholar] [CrossRef] [PubMed]
- Wendt, E.W.; Silva, P.C.; Malabarba, L.R.; Carvalho, T.P. Phylogenetic relationships and historical biogeography of Oligosarcus (Teleostei: Characidae): Examining riverine landscape evolution in southeastern South America. Mol. Phylogenetics Evol. 2019, 140, 106604. [Google Scholar] [CrossRef]
- Avise, J.C. Mitochondrial DNA and the evolutionary genetics of higher animals. Philos. Transact. R. Soc. Lond. Ser. B Biol. Sci. 1986, 312, 325–342. [Google Scholar]
- Gompert, Z.; Forister, M.L.; Fordyce, J.A.; Nice, C.C. Widespread mito-nuclear discordance with evidence for introgressive hybridization and selective sweeps in Lycaeides. Mol. Ecol. 2008, 17, 5231–5244. [Google Scholar] [CrossRef]
- Ivanov, V.; Lee, K.M.; Mutanen, M. Mitonuclear discordance in wolf spiders: Genomic evidence for species integrity and introgression. Mol. Ecol. 2018, 27, 1681–1695. [Google Scholar] [CrossRef] [PubMed]
- Toews, D.P.; Brelsford, A. The biogeography of mitochondrial and nuclear discordance in animals. Mol. Ecol. 2012, 21, 3907–3930. [Google Scholar] [CrossRef]
- Sarmento-Soares, L.M.; Martins-Pinheiro, R.F. Glanidium botocudo, a new species from the rio Doce and rio Mucuri, Minas Gerais, Brazil (Siluriformes: Auchenipteridae) with comments on taxonomic position of Glanidium bockmanni Sarmento-Soares & Buckup. Neotrop. Ichthyol. 2013, 11, 265–274. [Google Scholar]
- Roxo, F.F.; Silva, G.S.C.; Zawadzki, C.H.; Oliveira, C. Neoplecostomus doceensis: A new loricariid species (Teleostei, Siluriformes) from the rio doce basin and comments about its putative origin. ZooKeys 2014, 440, 115–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, R.P.; Melo, B.F.; Yazbeck, G.M.; Oliveira, R.S.; Hilário, H.O.; Prosdocimi, F.; Carvalho, D.C. Diversification of Prochilodus in the eastern Brazilian Shield: Evidence from complete mitochondrial genomes (Teleostei, Prochilodontidae). J. Zool. Syst. Evol. Res. 2021, 59, 1053–1063. [Google Scholar] [CrossRef]
- Souza, C.S.; Silva, G.S.C.; Ochoa, L.E.; Roxo, F.F.; Costa-Silva, G.J.; Foresti, F.; Melo, B.F.; Oliveira, C. Molecular and morphological diversity in species of Kronichthys (Teleostei, Loricariidae) from Atlantic coastal rivers of Brazil. J. Fish Biol. 2021, 98, 668–679. [Google Scholar] [CrossRef]

| Taxon | Voucher | Tissues | Locality | Coordinates | Accession Numbers (COI, 12S, S7) |
|---|---|---|---|---|---|
| Hypomasticus mormyrops | 29,070 | LBP29070 | PSRB | 23S 22′ 26″ 46W 03′ 11″ | GU702177.1, -, - |
| Hypomasticus steindachneri | - | LGC2984 | Jequitinhonha RB | 16S 59′ 68″41W 59′ 82″ | - |
| Hypomasticus copelandii | - | LGC3749 | DRB | 18S 58′ 48″ 43W 26′ 11″ | MK770203, -, - |
| Hypomasticus copelandii | MCNIP-1398 | LGC4116 | MRB | 17S 35′ 824″ 40W 59′ 205″ | MUCU153-14, -, - |
| Hypomasticus copelandii | MCNIP-1398 | LGC4117 | MRB | 17S 35′ 824″ 40W 59′ 205″ | MUCU154-14, MH187580, MF850394 |
| Hypomasticus copelandii | MCNIP- 1398 | LGC4118 | MRB | 17S 35′ 824″ 40W 59′ 205″ | MUCU120-14, MH187587, F850395 |
| Hypomasticus copelandii | - | LGC5249 | DRB | 42W 42′ 26″ 18S 53′ 52″ | -, -, MF850383 |
| Hypomasticus copelandii | - | LGC5392 | DRB | 42W 40′ 59″ 18S 53′ 50″ | -, MH187599, - |
| Hypomasticus copelandii | - | LGC5393 | DRB | 18S 53′ 30″ 42W 41′ 44″ | MK770205, -, - |
| Hypomasticus copelandii | - | LGC5394 | DRB | 18S 53′ 30″ 42W 41′ 44″ | MK770206 -, - |
| Hypomasticus copelandii | - | LGC5397 | DRB | 42W 56′ 31″ 19S 00′ 23″ | -, MH187598, - |
| Hypomasticus copelandii | - | LGC5400 | DRB | 18S 59′ 18″ 42W 57′ 49″ | MK770207, MH187579, - |
| Hypomasticus copelandii | - | LGC5402 | DRB | 42W 57′ 24″ 18S 59′ 43″ | -, MH187597, - |
| Hypomasticus copelandii | - | LGC5593 | DRB | 19S 59′ 03″ 41W 43′ 03″ | MK770209, MH187586, - |
| Hypomasticus thayeri | - | LGC5784 | DRB | 18S 55′ 01″ 43W 27′ 42″ | MF850389 |
| Hypomasticus copelandii | - | LGC6074 | DRB | 42W 52′ 57″ 19S 03′ 41″ | -, MH187596, - |
| Hypomasticus copelandii | - | LGC6076 | DRB | 42W 52′ 34″ 19S 04′ 37″ | -, MH187595, - |
| Hypomasticus copelandii | - | LGC6077 | DRB | 42W 52′ 34″ 19S 04′ 37″ | -, MH187594, - |
| Hypomasticus copelandii | - | LGC6078 | DRB | 42W 57′ 49″ 18S 59′ 18″ | -, MH187593, - |
| Hypomasticus copelandii | - | LGC6079 | DRB | 42W 57′ 39″ 18S 59′ 38″ | -, MH187592, - |
| Hypomasticus copelandii | - | LGC6080 | DRB | 42W 56′ 31″ 19S 00′ 23″ | -, MH187591, - |
| Hypomasticus copelandii | - | LGC6081 | DRB | 42W 55′ 28″ 19S 03′ 04″ | -, MH187590, - |
| Hypomasticus copelandii | - | LGC6082 | DRB | 42W 41′ 44″ 18S 53′ 30″ | -, MH187589, - |
| Hypomasticus copelandii | - | LGC7086 | JRB | 44W 59′ 55″ 79S 51′ 27″ | MK770212, -, MF850392 |
| Hypomasticus copelandii | - | LGC7087 | JRB | 44W 59′ 55″ 79S 51′ 27″ | MK770213, -, MF850393 |
| Hypomasticus copelandii | - | LGC7088 | JRB | 44W 59′ 55″ 79S 51′ 27″ | MK770214, -, MF850390 |
| Hypomasticus copelandii | - | LGC7089 | JRB | 44W 59′ 55″ 79S 51′ 27″ | MK770215, MH187585, MF850391 |
| Hypomasticus copelandii | - | LGC7094 | DRB | 19S 44′ 58″ 41W 46′ 58″ | -, -, MF850384 |
| Hypomasticus copelandii | - | LGC7095 | DRB | 19S 44′ 58″ 41W 46′ 58″ | -, -, MF850385 |
| Hypomasticus copelandii | - | LGC7101 | DRB | 19S 44′ 58″ 41W 46′ 58″ | -, -, MF850381 |
| Hypomasticus copelandii | - | LGC7107 | PSRB | 23S 22′ 25″ 45W 39′ 59″ | MK770216, -, MF850386 |
| Hypomasticus copelandii | - | LGC7108 | PSRB | 23S 22′ 25″ 45W 39′ 59″ | MK770217, -, - |
| Hypomasticus copelandii | - | LGC7109 | PSRB | 23S 22′ 25″ 45W 39′ 59″ | MK770218, -, MF850387 |
| Hypomasticus copelandii | - | LGC7110 | PSRB | 23S 22′ 25″ 45W 39′ 59″ | MK770219, -, - |
| Hypomasticus copelandii | - | LGC7111 | PSRB | 23S 22′ 25″ 45W 39′ 59″ | MK770220, -, - |
| Hypomasticus copelandii | - | LGC7112 | PSRB | 23S 22′ 25″ 45W 39′ 59″ | MK770221, MH187578, MF850388 |
| Hypomasticus copelandii | - | LGC7113 | PSRB | 23S 22′ 25″ 45W 39′ 59″ | MK770222, MH187584, MF850382 |
| Hypomasticus copelandii | - | LGC7673 | PSRB | 23S 22′ 25″ 45W 39′ 59″ | MK770231, -, - |
| Hypomasticus copelandii | - | LGC7675 | PSRB | 23S 22′ 25″ 45W 39′ 59″ | MK770232, MH187583, - |
| Hypomasticus copelandii | - | LGC7676 | PSRB | 45E 49′ 01″ 36S 16′ 35″ | MK770233, -, - |
| Hypomasticus copelandii | - | LGC7677 | PSRB | 45E 49′ 01″ 36S 16′ 35″ | MK770234, MH187582, - |
| Hypomasticus copelandii | - | LGC7773 | PSRB | 23S 22′ 25″ 45W 39′ 59″ | MK770235, -, - |
| Hypomasticus copelandii | - | LGC7774 | PSRB | 23S 22′ 25″ 45W 39′ 59″ | MK770236, -, - |
| Hypomasticus copelandii | - | LGC7775 | DRB | 23S 22′ 25″ 45W 39′ 59″ | -, undeposited, undeposited |
| Lineages | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 1 Hypomasticus copelandii DRB | 0.000 | 0.000 | 0.003 | 0.008 | 0.008 |
| 2 Hypomasticus copelandii MRB | 0.000 | 0.000 | 0.003 | 0.008 | 0.008 |
| 3 Hypomasticus copelandii JRB | 0.005 | 0.005 | 0.000 | 0.008 | 0.008 |
| 4 Hypomasticus copelandii PSRB | 0.036 | 0.036 | 0.042 | 0.002 | 0.001 |
| 5 Hypomasticus copelandii EHAP | 0.035 | 0.035 | 0.041 | 0.002 | 0.001 |
| Lineages | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 1 Hypomasticus copelandii DRB | 0.001 | 0.003 | 0.002 | 0.006 | 0.006 |
| 2 Hypomasticus copelandii MRB | 0.007 | 0.013 | 0.004 | 0.007 | 0.007 |
| 3 Hypomasticus copelandii JRB | 0.003 | 0.009 | - | 0.006 | 0.006 |
| 4 Hypomasticus copelandii PSRB | 0.017 | 0.023 | 0.018 | 0.001 | 0.002 |
| 5 Hypomasticus copelandii EHAP | 0.021 | 0.027 | 0.023 | 0.005 | 0.001 |
| Lineages | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 1 Hypomasticus copelandii DRB | 0.000 | 0.003 | 0.002 | 0.001 | 0.000 |
| 2 Hypomasticus copelandii MRB | 0.005 | 0.003 | 0.001 | 0.003 | 0.003 |
| 3 Hypomasticus copelandii JRB | 0.003 | 0.002 | 0.000 | 0.002 | 0.002 |
| 4 Hypomasticus copelandii PSRB | 0.001 | 0.006 | 0.004 | 0.002 | 0.001 |
| 5 Hypomasticus copelandii EHAP | 0.000 | 0.005 | 0.003 | 0.001 | 0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendes, I.S.; Melo, B.F.; Damasceno, J.S.; Teixeira, D.F.; Carvalho, D.C. Phylogeography of Hypomasticus copelandii (Teleostei, Anostomidae) Reveals Distinct Genetic Lineages along Atlantic Coastal Drainages of Eastern Brazil. Diversity 2022, 14, 29. https://doi.org/10.3390/d14010029
Mendes IS, Melo BF, Damasceno JS, Teixeira DF, Carvalho DC. Phylogeography of Hypomasticus copelandii (Teleostei, Anostomidae) Reveals Distinct Genetic Lineages along Atlantic Coastal Drainages of Eastern Brazil. Diversity. 2022; 14(1):29. https://doi.org/10.3390/d14010029
Chicago/Turabian StyleMendes, Izabela S., Bruno F. Melo, Júnio S. Damasceno, Daniel F. Teixeira, and Daniel C. Carvalho. 2022. "Phylogeography of Hypomasticus copelandii (Teleostei, Anostomidae) Reveals Distinct Genetic Lineages along Atlantic Coastal Drainages of Eastern Brazil" Diversity 14, no. 1: 29. https://doi.org/10.3390/d14010029
APA StyleMendes, I. S., Melo, B. F., Damasceno, J. S., Teixeira, D. F., & Carvalho, D. C. (2022). Phylogeography of Hypomasticus copelandii (Teleostei, Anostomidae) Reveals Distinct Genetic Lineages along Atlantic Coastal Drainages of Eastern Brazil. Diversity, 14(1), 29. https://doi.org/10.3390/d14010029








