Abstract
Quinolizidine alkaloids (QAs) are synthesized by the genus Lupinus as a defense against herbivores. Synthesis of QAs in lupins is species- and organ-specific. Knowledge about their biosynthesis and their corresponding pathways are still fragmentary, in part because lupins of commercial importance were mainly investigated, representing a small sample of the chemodiversity of the genus. Here, we explore the use of three Mexican lupins: Lupinus aschenbornii, Lupinus montanus, and Lupinus bilineatus as a model to study the physiology of QA biosynthesis. The corresponding QA patterns cover widely and narrowly distributed tetracyclic QAs. Quinolizidine alkaloid patterns of seeds and plantlets at different developmental stages were determined by GLC–MS and compared to identify the onset of de novo QA synthesis and to gain insight into specific and common biosynthesis trends. Onset of de novo QA biosynthesis occurred after the metabolization of seed QA during germination and was species-specific, as expected. A common QA pattern, from which the diversity of QA observed in these species is generated, was not found; however, lupanine and 3β-lupanine were found in the three specieswhile sparteine was not found in Lupinus bilineatus, suggesting that this simplest tetracyclic QA is not the precursor of more complex QAs. Similar patterns of metabolization and biosynthesis of structurally related QAs were observed, suggesting a common regulation.
1. Introduction
Lupinus L. (Fabaceae) is a genus from the Mediterranean region (16 ± 5 million years BP), distributed in Europe, North Africa, and America [1], known for producing quinolizidine alkaloids (QAs) as chemical defenses against herbivores. Each lupin species produces a characteristic pattern of QAs with few major compounds and several others as traces [2,3,4]. The chemical properties and insecticidal, antimicrobial, and medicinal activities of QAs are of interest in the chemical, biotechnological, and pharmaceutical industries [5,6,7,8,9,10,11,12,13,14,15].
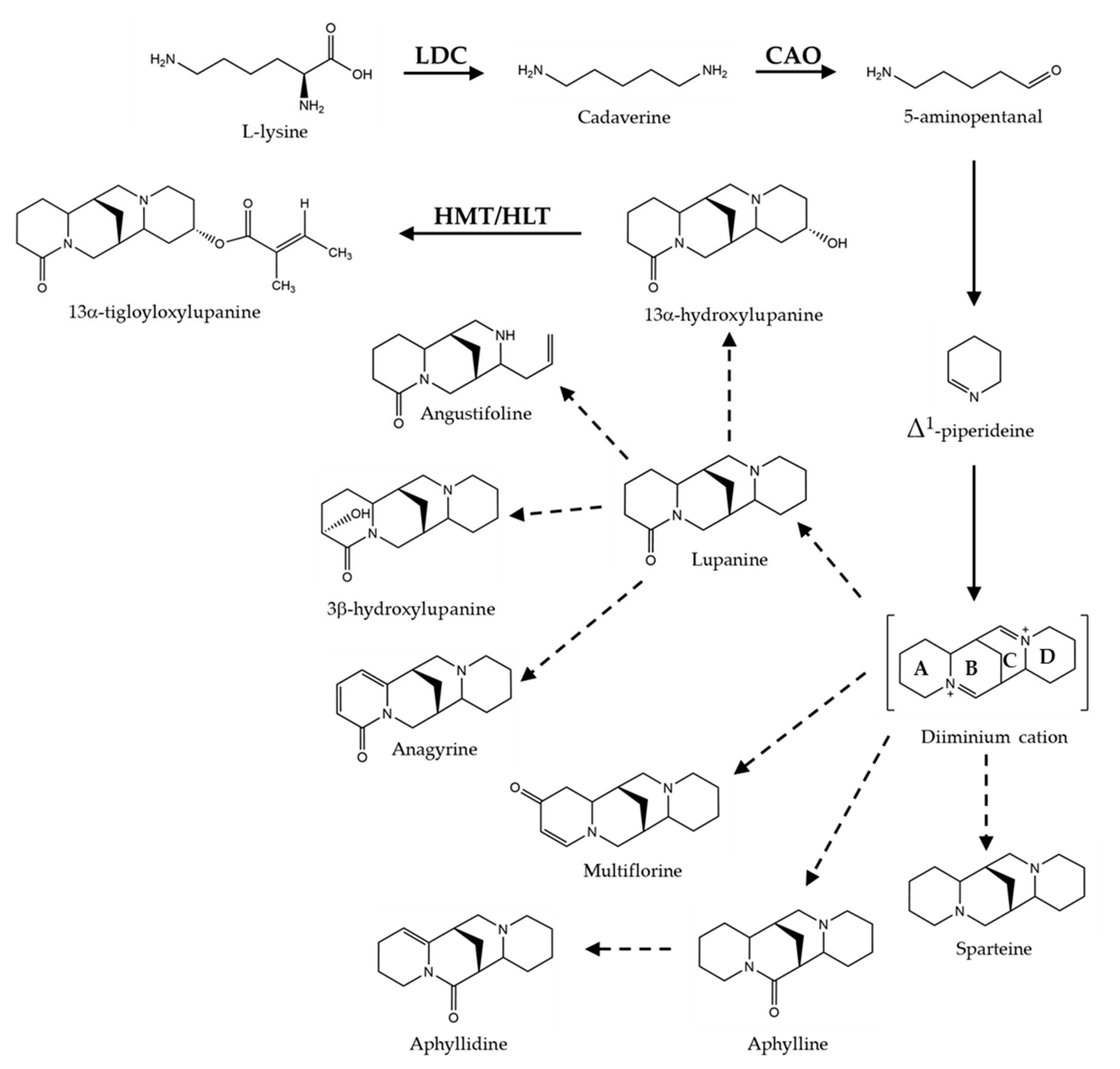
Biosynthesis of QAs is developmentally regulated and under environmental control [16,17,18]. QAs are synthesized from L-lysine, mainly in the chloroplasts of leaves [19]; biosynthesis in hypocotyls, stems, and pods also occurs, albeit at a lower extent [18,20,21]. QAs are transported from their place of synthesis to the whole plant via the phloem and stored in epidermal tissues and seeds, the latter as both a defense mechanism and as a source of nitrogen for the growth of the nascent plant [22]. During seed germination, QAs are metabolized and mobilized from cotyledons to the roots [22,23]. De novo biosynthesis of QAs activates during the early development of plantlets in a species-specific manner [22]. The first step of QA biosynthesis involves the action of lysine decarboxylase (LDC), which decarboxylates lysine to the diamine cadaverine (Figure 1) [24]. Cadaverine is then converted to 5-aminopentanal, putatively, by copper amino oxidase enzyme (CAO) or amino transferase [20,25]. The consensus is that 5-aminopentanal cyclizes spontaneously to produce Δ1-piperideine, the intermediate from which bicyclic (lupinine) and tetracyclic QA (sparteine, lupanine, and multiflorine) are formed; these are then converted to a vast diversity of related QAs through tailoring reactions, including oxidation, dehydrogenation, hydroxylation, acylation, and methylation. Tigloyl-CoA:13α-hydroxymultiflorine/13α-hydroxylupanine O-tigloyltransferase, (HMT/HLT) catalyzes the acylation of 13α-hydroxymultiflorine and 13α-hydroxylupanine to form 13α-tigloyloxymultiflorine and 13α-tigloyloxylupanine using tigloyl-CoA as acyl donor [26].
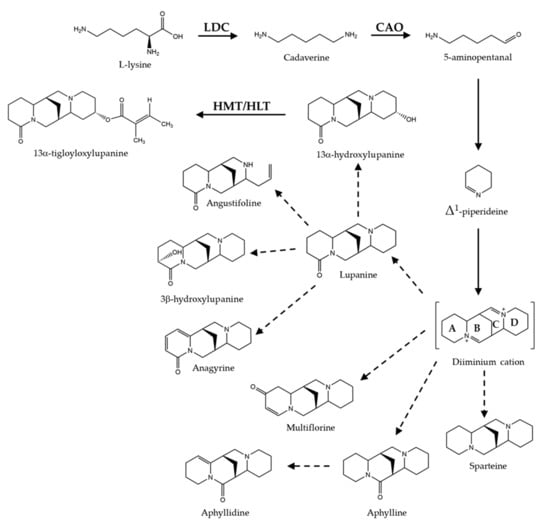
Figure 1.
Biosynthetic pathway of quinolizidine alkaloids. QAs are synthesized from lysine, which is converted to cadaverine and then to 5-aminopentanal that spontaneously cyclize to form Δ1-piperideine; the latter intermediate is the precursor of the diiminium cation, from which it is thought tetracyclic QA are derived. The four heterocyclic rings of the diiminium cation are indicated for the letters A, B, C, and D. Enzymes molecularly characterized are indicated: LDC, lysine decarboxylase; CAO, copper amino oxidase; HMT/HLT, tigloyl-CoA:13α-hydroxymultiflorine/13α-hydroxylupanine O-tigloyltransferase. Solid lines represent the known steps and dotted lines those on construction.
The fact that little is known about the rest of the enzymatic machinery involved in the pathway and its genetic regulation might soon change, considering the significant progress on the identification of candidate biosynthetic and regulatory genes achieved using transcriptomics and genomics in the last 4 years [17,25,27]. This advancement of knowledge has been achieved through research of high and low QA producing varieties of Lupinus angustifolius due to the strong commercial interest on generating lupin varieties with stable low or null QA content in seeds, which could be safely used for food and feed purposes. However, L. angustifolius represents a small sample of the chemodiversity of the genus. Studies to unravel QA biosynthesis involving more than one Lupinus species are a steppingstone towards better understanding of the poorly known mechanisms involved in the generation of the diversity of QA patterns in nature, enabling us to answer questions, such as, is there a precursor QA pattern for the diversification of QAs? If so, which QAs composed this starting pool?
Mexico is one of the three centers of diversification of the genus Lupinus in the American continent; the Rocky Mountains and the Andes are the other two [1,28]. There are approximately 65 lupin species in Mexico [29], distributed across the country from Baja California and Tamaulipas to Chiapas, along Sierra Madre Occidental, Sierra Madre Oriental, and particularly the Trans-Mexican Volcanic Belt [30]. Mexican lupins constitute a vast genetic pool and source of QAs of commercial importance [31,32]. Moreover, from a biological point of view, they represent an unexplored species diversity to study the biosynthesis of QA and the possible genetic changes that may have played a role in the adaptation of lupin species to the American habitats.
Our research group has characterized the QA patterns of wild Mexican lupins for over 20 years [31,33,34]. Results from this work have led us to the conclusion that Lupinus aschenbornii S. Schauer, Lupinus bilineatus Benth, and Lupinus montanus Kunth (Figure 2) constitute interesting models for the study of QA biosynthesis and its genetic regulation, due to the similarities and differences in the QA patterns they produce. Quinolizidine alkaloids from these species span a broad range of related molecules, including the main QAs produced by most lupin plants (sparteine, lupanine, and multiflorine) and those with restricted distribution, such as aphylline and aphyllidine, found only in some American species [2].

Figure 2.
Lupinus aschenbornii (A,B), Lupinus bilineatus (C,D), and Lupinus montanus (E,F); (A,B,E,F) growing at the Iztaccihuatl–Popocatepetl National Park and (C,D) growing at Chichinautzin Corridor.
Lupinus aschenbornii is a perennial species native to the Trans-Mexican Volcanic Belt; it is found in the high mountains of the states of Mexico, Michoacán, and Puebla, at altitudes ranging from 2800 to 4300 m above sea level (a.s.l.). This species produces the most diverse QA pattern among the Mexican species chemically characterized so far [30]. Up to 24 QAs have been identified in the leaves of L. aschenbornii, including sparteine, lupanine, 13α-hydroxylupanine, angustifoline, N-formylangustifoline, multiflorine, and 13α-tigloyloxylupanine, among other esters, which are the main QAs produced by this species [33,35]. Lupinus bilineatus is an annual, biennial, or short-lived perennial species that grows in the states of Aguascalientes, Morelos, Michoacán, and Mexico at 2780 to 2945 m a.s.l. [30]. It produces a different (and less diverse) QA pattern compared to L. aschenbornii. The main QAs synthesized by this species are aphylline, aphyllidine, lupanine, hydroxyaphylline, and hydroxyaphyllidine [Bermúdez-Torres, personal communication]. Lupinus montanus is a perennial polymorphic species [36]. It is the most widely distributed of the Mexican lupins as it grows in pine, oak, and alpine meadow forests at 2500 to 4100 m a.s.l. from Chihuahua to Guatemala [30]. Sparteine, lupanine, and aphylline are the main alkaloids produced by this species [31].
As we were aware of the potential of using L. aschenbornii, L. bilineatus, and L. montanus as a model to unravel QA biosynthesis, we embarked on the characterization of their QA patterns during germination and early plantlet development. Our aim was to identify the developmental stage at which de novo biosynthesis starts in each and the common pool of QA from which the diversity of QAs produced by these three species could be explained.
2. Materials and Methods
2.1. Seed Harvest
Seeds of L. aschenbornii, L. bilineatus, and L. montanus were collected from populations growing at Iztaccihuatl–Popocatepetl National Park between April and September 2009. Geographical coordinates are indicated in Table 1. Herbarium voucher material was collected from three flowering individuals per population and deposited at the MEXU herbarium. Seeds were stored in paper bags inside foil bags with silica gel to control humidity, at 4 °C, until use.

Table 1.
Populations for seed collection.
2.2. Disinfestation and Scarification of Seeds
Seeds were disinfested following protocol [37], with slight modifications. Seeds were washed in detergent solution (0.5% w/v) for 5 min, then transferred to 70% (v/v) ethanol for 5 min, immersed in 1.0% (v/v) sodium hypochlorite for 20 min, and finally placed in a solution of benzylpenicillin and nystatin (0.5% w/v, each) for 60 min. Seeds were dried on filter paper under sterile conditions. All disinfestation steps were carried out using continuous mechanical agitation; three washes with sterile distilled water were conducted in between each disinfestation step, except after the treatment with antibiotics. Disinfested seeds were mechanically scarified using a cylinder made of no. 60 wood sandpaper; seeds were placed inside the cylinder and shaken manually in a rotatory and longitudinal manner for 40 min [38]. Scarified seeds were immediately germinated.
2.3. Seed Germination and Plant Growth
Scarified seeds were placed onto wet germination strips (Sartorius grade 190) contained in aluminum trays closed with self-adhesive plastic. Seeds were germinated in a growth chamber at 20 °C, 60% relative humidity (RH), photoperiod of 16 h light/8 h darkness, and monitored daily. Germinated seeds with a radicle of at least 5 mm in length were transplanted to trays of 98 pots containing 20 mL of perlite in each pot and returned to the growth chamber for growth at 20 °C, 60% RH, and photoperiod of 16 h light/8 h darkness. Plantlets at different stages of development, i.e., germination (5 mm radicle); elongation of hypocotyl; emergence of first up to sixth leaf (in some cases), were harvested and dried at 30–34 °C for 24 h. Dry samples were reduced to powder using mortar and pestle and stored in a desiccator at room temperature until subjected to QA extraction.
2.4. Quinolizidine Alkaloids Extraction
QA extraction was performed according to [33]; 300 mg of dried and grinded plant material were resuspended in 20 mL of 1 M HCl and incubated at room temperature with continuous agitation for 24 h. Extraction mixture was centrifuged at 8500 rpm for 10 min and the supernatant recovered and alkalized (pH 12) with 3 M NH4OH. Supernatant was loaded to an Isolute® HM-N column (IST, Biotage, Uppsala, Sweden); alkaloids were eluted with 30 mL of CH2Cl2 (3×) and collected in a round flask. Eluate was concentrated to dryness in a rotary evaporator (40 °C, without vacuum). Alkaloids were resuspended in 1 mL of methanol and stored in an amber vial at 4 °C in darkness until gas liquid chromatography–mass spectrometry (GLC–MS, Agilent, Santa Clara, CA, US) analysis. All materials used for QA extraction were pre-washed with CH2Cl2.
2.5. Identification and Quantification of Quinolizidine Alkaloids by GLC–MS
Separation and identification of QA was performed by GLC–MS following the protocol reported by [33], with some modifications. A gas chromatograph (Agilent 7890A GC) with HP 5 MS column (30 m, 250 μm internal diameter, and 0.25 μm film thickness) (Agilent, Palo Alto, CA, US) coupled to a mass spectrometer (Agilent 5975C MSD, Santa Clara, CA, US) with electronic impact detector (EI) was used. The capillary column had a length of 30 m, an internal diameter of 0.25 mm, and a film thickness of 0.25 µm. Hydrogen was used as carrier gas at a flow rate of 1.5 mL/min (SPLIT mode) and an injector temperature of 280 °C was used. The initial temperature was maintained at 120 °C for 2 min in isothermal flow, followed by a rise in linear isothermal temperature to 300 °C, at a rate of 8 °C/min, and finally maintained for 10 min in isothermal mode at 300 °C. Samples (1 μL) were injected automatically. Each peak in the chromatogram was integrated and identified using the NIST (access on: 17 May 2018) spectrum library [39] and literature data [2]. Kovats retention index was calculated using the retention time of each QA compared to that of the respective alkane (obtained from 1 mg/mL of alkanes). Abundance of each QA was expressed in mg/mL, using the percentage of the area of 1 mg/mL of sparteine as reference. Content of the total and each QA present in the plant material was calculated and expressed in mg/g dried weight (DW). Three biological samples were evaluated for each point and a descriptive and inferential statistical analysis, and an analysis of variance (ANOVA) were carried out.
3. Results
3.1. QA Patterns of Lupinus aschenbornii Seeds and Plantlets
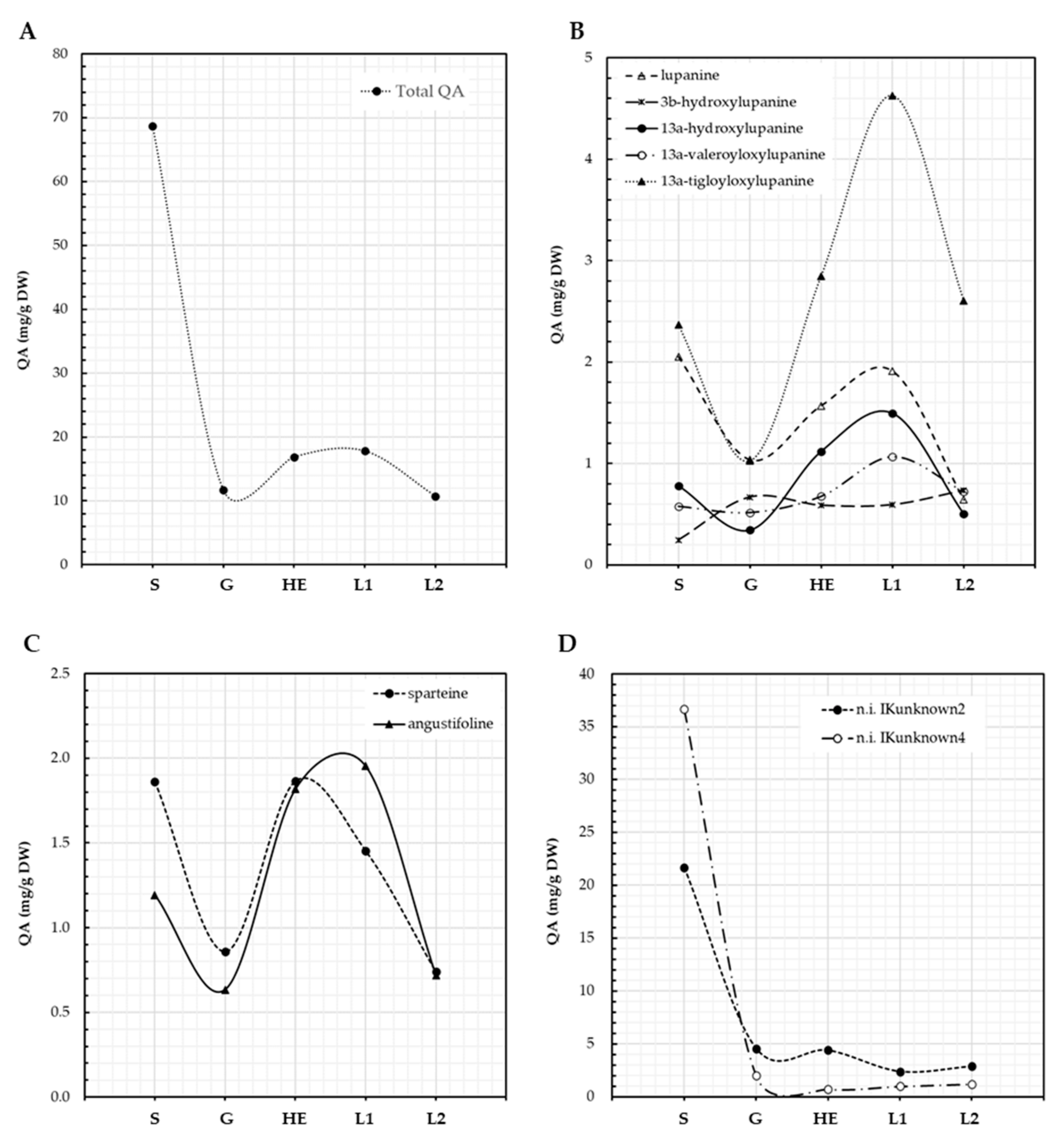
Seeds of L. aschenbornii had a total QA content of 68.7 mg/g DW; this content decreased to 11.7 mg/g DW during germination, as plantlet development progressed an increase in QA content was observed, reaching 16.9 mg/g DW at the hypocotyl elongation stage, suggesting de novo QA biosynthesis (Figure 3A). Total QA content then increased to 17.8 and decreased to 10.7 mg/g DW at the emergence of the first and second leaf, respectively (Figure 3A). The main QA present in L. aschenbornii seeds were 13α-tigloyloxylupanine (3.4%), lupanine (3.0%), sparteine (2.7%), angustifoline (1.7%), 13α-hydroxylupanine (1.1%), 13α-valeroyloxylupanine (0.8%), 3β-hydroxylupanine (0.3%), and two unidentified QA esters, n.i. IKunknown2 (31.6%) and n.i. IKunknown4 (53.4%) (Table 2). Tetrahydrorhombifoline, 11,12-seco-12,13-didehydromultiflorine, and multiflorine were found as traces in L. aschenbornii seeds; however, these QAs were not considered in further analyses since they were not synthesized by the plantlets. Nascent plantlets quickly metabolized all of the main QAs during germination, suggesting their use as a source of nitrogen (Figure 3B–D), and the synthesis of all of them, except 3β-hydroxylupanine, n.i. IKunknown2, and n.i. IKunknown4, was initiated after germination and before hypocotyl elongation (Figure 3B–D). Maximum content of sparteine was observed at the hypocotyl elongation stage, while that of angustifoline, lupanine (and derivatives), n.i. IKunknown2 and n.i. IKunknown4 at the emergence of the first leaf. Moreover, 13α-tigloyloxylupanine (4.6 mg/g DW at the first leaf emergence stage) was the main QA produced by L. aschenbornii plantlets (Figure 3B,D).
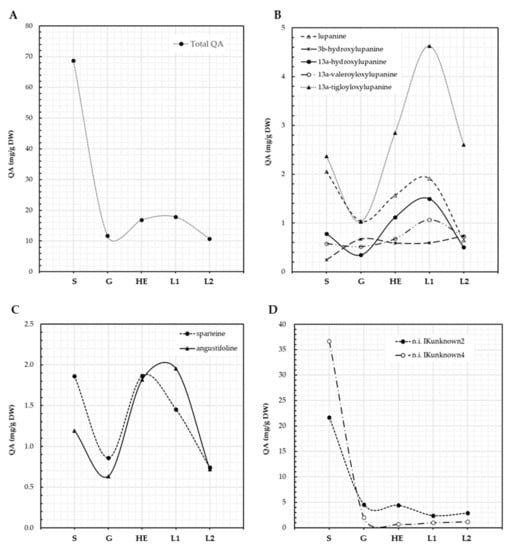
Figure 3.
QA content in L. aschenbornii seeds and plantlets. Total QA (A), lupanine and its derivatives (B), other tetracyclic QA (C), unidentified QA (D). S: seeds, G: germination, HE: hypocotyl elongation, L1: emergence of first leaf, L2: emergence of second leaf.

Table 2.
Alkaloids identified in: Lupinus aschenbornii (A), Lupinus bilineatus (B), and Lupinus montanus (C).
3.2. QA Patterns of Lupinus bilineatus Seeds and Plantlets
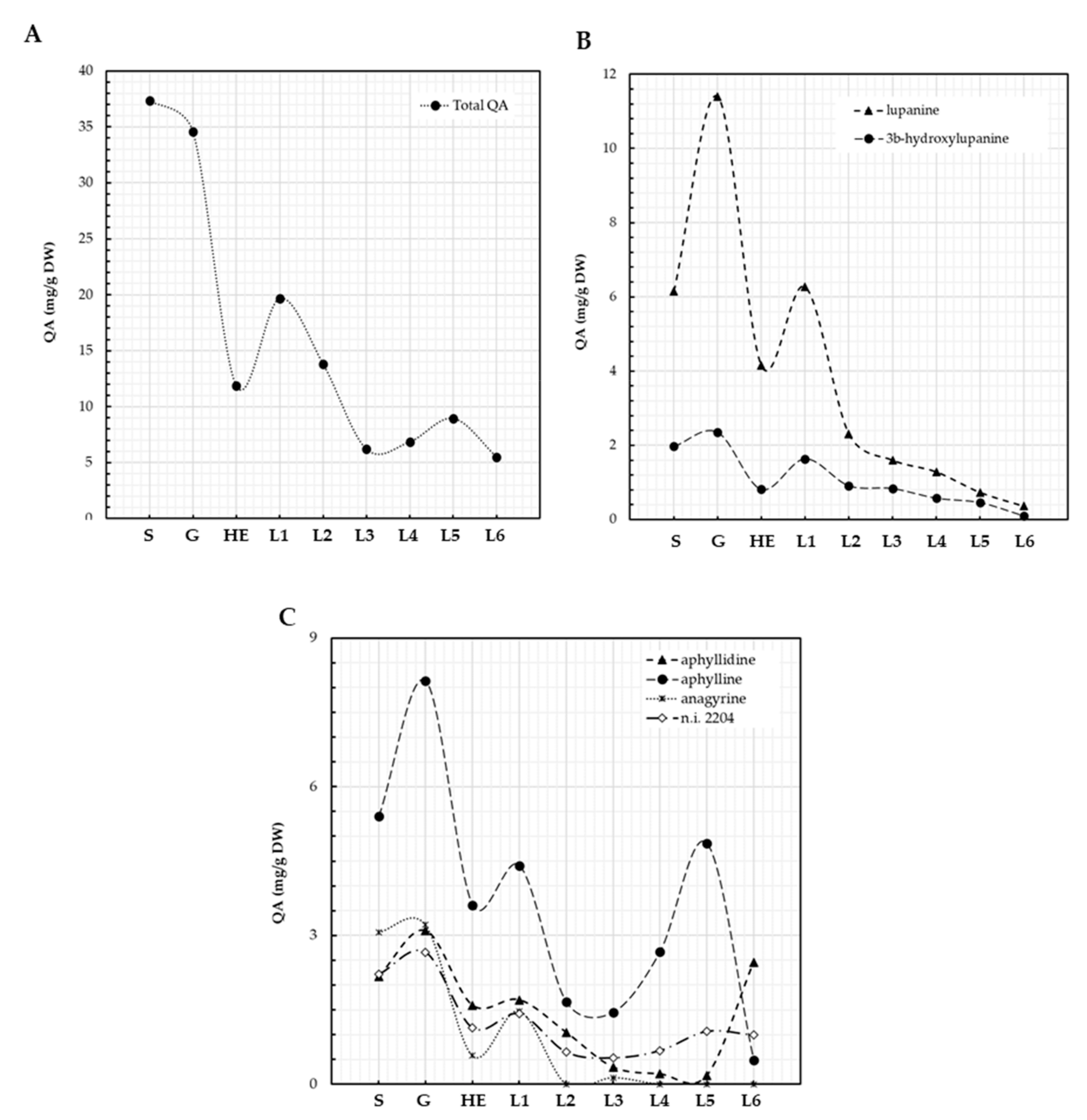
QA content characterization revealed cycles of metabolization and de novo synthesis during L. bilineatus germination and plantlet development (Figure 4A–C). Total content of QAs in L. bilineatus seeds was 37.3 mg/g DW, which slightly decreased to 34.6 mg/g DW during germination. QA content further decreased to 11.9 mg/g DW at the plantlet hypocotyl elongation stage indicating that plantlets were metabolizing these secondary metabolites, it then increased to 19.7 mg/g DW as the first leaf emerged suggesting de novo biosynthesis (Figure 4A). This biosynthesis was not sustained as revealed by the decrease in QA content as plantlet development continued, reaching a minimum of 6.2 mg/g DW at the emergence of third leaf. An increase in QA content was then observed as fourth and fifth leaves emerged, caused mainly by the increase in aphylline, QA content then dropped again to 5.5 mg/g DW in plantlets where the sixth leaf was emerging. The main QAs present in seeds of L. bilineatus were lupanine (16.5%), aphylline (14.5%), anagyrine (8.2%), aphyllidine (5.8%), 3β-hydroxylupanine (5.3%), sparteine (4.0%), and four unidentified QAs, n.i. KIunknown5 (31.3%). n.i. 2204 (5.9%), n.i. 2281 (4.2%), and n.i. 2441 (1.2%); however, only. n.i. 2204 was actively synthesized by the plantlets. Traces of other unidentified QAs (n.i. 1848 and n.i. 1863) were detected in the seeds. Lupanine, 3β-hydroxylupanine, and anagyrine showed similar patterns of metabolization and biosynthesis (Figure 3B), in contrast to aphylline and aphyllidine (Figure 4C).
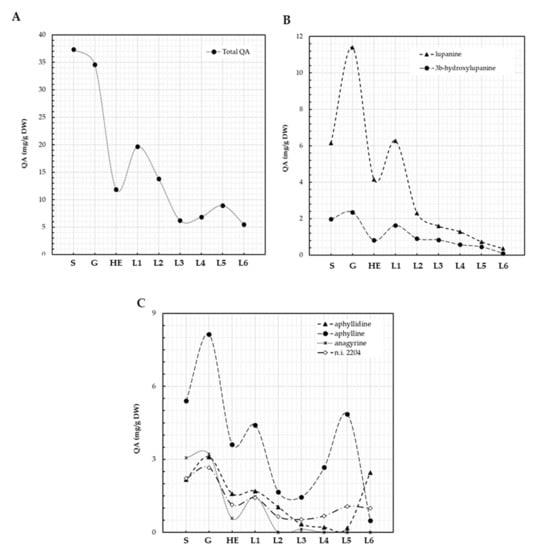
Figure 4.
QA patterns of L. bilineatus seeds and plantlets. Total QA (A), lupanine and its derivatives (B), aphyllidine, and aphylline (C). S: seeds, G: germination, HE: hypocotyl elongation, L1: emergence of first leaf, L2: emergence of second leaf, L3: emergence of third leaf, L4: emergence of fourth leaf, L5: emergence of fifth leaf, L6: emergence of sixth leaf.
3.3. QA Patterns of Lupinus montanus Seeds and Plantlets
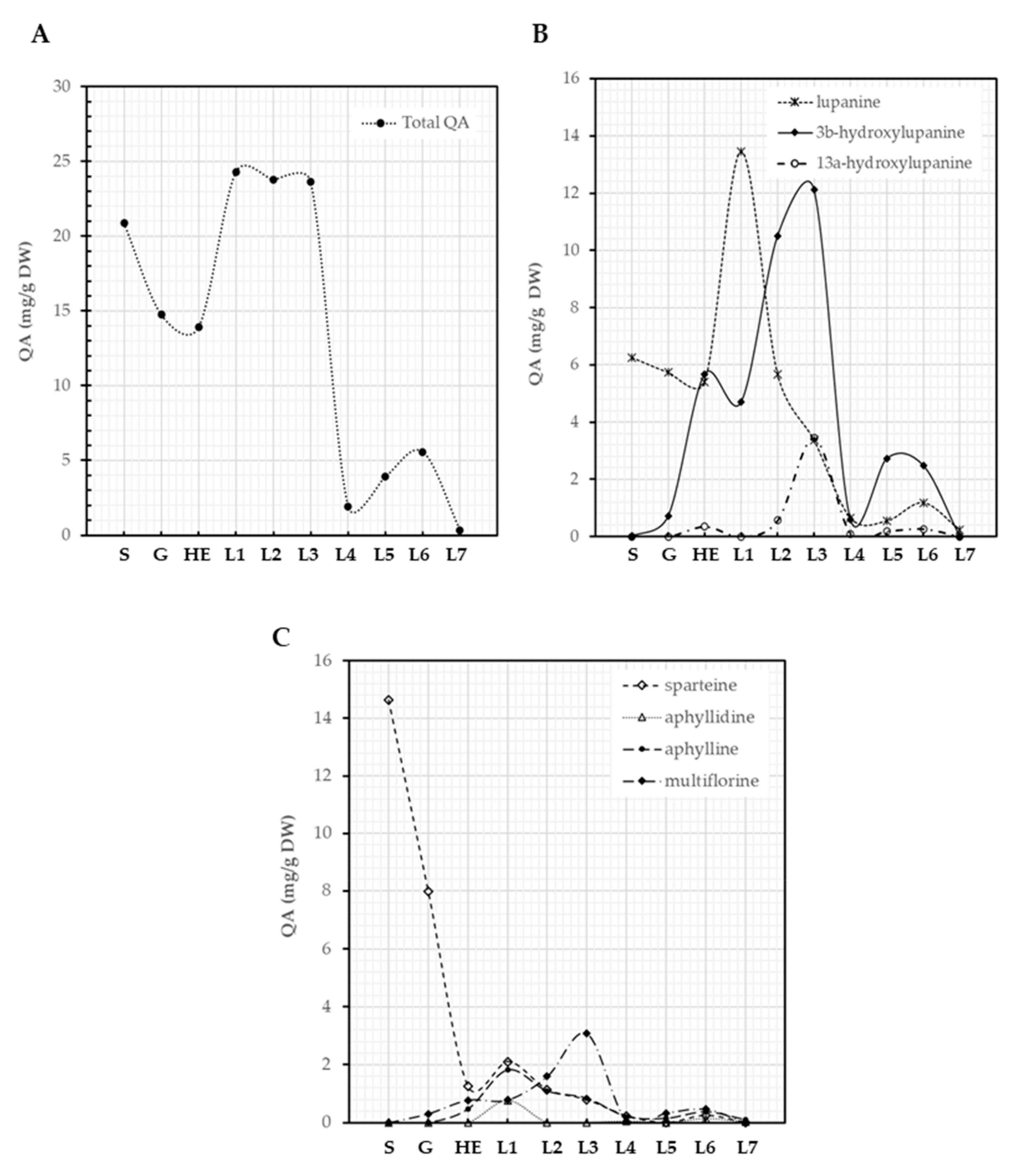
Seeds of L. montanus contained 20.9 mg/g DW of QAs, a reduction of QA content was observed during germination (14.7 mg/g DW) and elongation of the hypocotyl (14.0 mg/g DW), a likely consequence of the use of QAs as a source of nitrogen (Figure 5A). The content of QAs in plantlets increased as their development progressed and was maintained until the emergence of the third leaf, where QAs reached a maximum of 23.6 mg/g DW (Figure 4A), suggesting de novo QA biosynthesis. A steep decrease in QA content (2.0 mg/g DW) was observed at the emergence of the fourth leaf and then a slight increase was detected again at the emergence of the sixth leaf (5.6 mg/g DW), suggesting again the biosynthesis of these compounds. QA content then decreased during the transition to the emergence of the seventh leaf (0.3 mg/g DW) (Figure 5A). Lupinus montanus seeds contained sparteine (70%) and lupanine (29.9%) as main alkaloids with traces of α-isosparteine, 17-oxosparteine, 5,6-dehydrolupanine, 3β-hydroxylupanine, multiflorine, and 17-oxolupanine. Lupanine, 3β-hydroxylupanine, 13α-hydroxylupanine, sparteine, aphyllidine, aphylline, and multiflorine were the main alkaloids produced by L. montanus plantlets (Figure 5B,C). Not all of these QAs followed a similar pattern of metabolization and synthesis. Sparteine was the main QA in seeds and was actively metabolized during germination and the elongation of hypocotyl showing a 91.5% of reduction in content (from 14.6 to 1.3 mg/g DW, Figure 5C). Sparteine and lupanine showed a similar trend of synthesis along the developmental stages characterized (Figure 5B,C). Moreover, 3β-hydroxylupanine, 13α-hydroxylupanine, and multiflorine displayed similar dynamics from germination to the emergence of the seventh leaf (Figure 5B,C). Lupanine (13.5 mg/g DW at the emergence of first leaf) and 3β-hydroxylupanine (12.2 mg/g DW at the emergence of third leaf) were the most abundant QAs in L. montanus plantlets (Figure 5B). De novo biosynthesis of the various QAs was initiated at different developmental stages (Figure 5B,C).
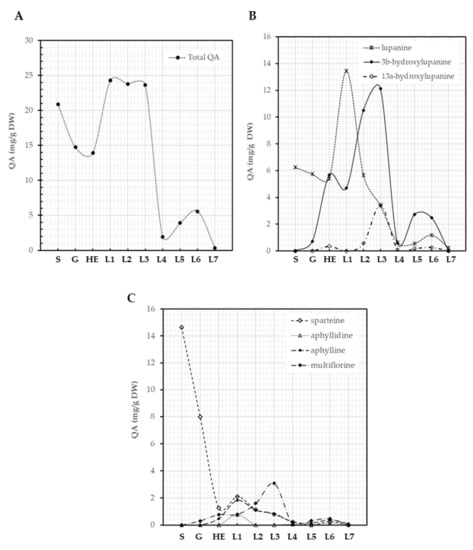
Figure 5.
QA patterns of L. montanus seeds and plantlets. Total QAs (A), lupanine and its derivatives (B), other tetracyclic QA (C). S: seeds, G: germination, HE: hypocotyl elongation, L1: emergence of first leaf, L2: emergence of second leaf, L3: emergence of third leaf, L4: emergence of fourth leaf, L5: emergence of fifth leaf, L6: emergence of sixth leaf, L7: emergence of the seventh leaf.
4. Discussion
QAs are mostly stored in seeds; they protect the seed from potential eaters and serve as source of nitrogen [2,5]. Lupinus aschenbornii, L. bilineatus, and L. montanus seeds contained 68.7, 37.5, and 20.9 mg/g DW of QA, respectively. The authors of [33] noted that seeds of L. aschenbornii contained 3.3 mg QA/g DW, which is about 20 times less than the content (68.7 mg/g DW) determined in this study; however, both reports are within the percentage of QA content (up to 8.0%) detected in lupin seeds [40]. Differences in seed QA content within the same lupin species have been commonly reported, which can be explained by the fact that QA biosynthesis is affected by environmental conditions [17]. Two unidentified QA esters were the main QAs in the L. aschenbornii seeds, IKunknown2 (31.6%) and n.i. IKunknown4 (53.4%). This is in line with previous studies indicating that seeds of L. aschenbornii are rich in QA esters [31,35]; however, it contrasts the reports of N-formylangustifoline or 13α-hydroxylupanine as the most abundant QAs [33,35]. These discrepancies in QA patterns were also observed for L. montanus. In this work, L. montanus seeds had sparteine (70.0%) and lupanine (29.9%) as the main alkaloids, whereas sparteine (89.0%), an unidentified QA n.i. 1940 (6.5%), aphylline (2.4%), and lupanine (1.3%) were the most abundant QAs reported by [31]. We hypothesize that the observed differences in the QA patterns of L. aschenbornii and L. montanus seeds may be due to (1) the result of the effect of environmental factors; (2) differing chemotypes; or (3) the recent origin of the Mexican species, which would imply that its chemical characters might still be established.
QA content decreased during germination of L. aschenbornii, L. montanus, and L. bilineatus seeds; for the latter, the decrease extended to the hypocotyl elongation stage. This phenomenon may be explained by the fact that QAs are used as sources of nitrogen by the nascent plant [22,23]. QAs are an example of the promiscuous use of secondary metabolites by plants for their survival, which maximize the gains from producing molecules of high energetic cost. Metabolization of QAs was faster in L. aschenbornii than in L. montanus and L. bilineatus since a reduction of 85.0% was observed during germination in the former compared to 29.5% and 7.6% in L. montanus and L. bilineatus, respectively. It is logical to think that, the more diverse enzyme machinery for QA catalysis is present in the seeds, the better it is to maximize the use of QAs during these early stages of development. This was clearly observed as L. aschenbornii degraded all of the main QAs present in its seeds, i.e., n.i. IKunknown4, n.i. IKunknown2, 13α-tigloyloxylupanine, lupanine, sparteine, angustifoline, and 13α-hydroxylupanine. Interestingly, a much higher degradation rate was observed for n.i. IKunknown4, the most abundant QA. A similar phenomenon was documented in L. montanus seeds, which degraded sparteine and lupanine during germination, with a higher degradation for sparteine, the most abundant QA, similar behavior was reported by [22] for L. polyphyllus. The behavior of the metabolization of QAs during germination of L. bilineatus was different to the observed in L. aschenbornii and L. montanus and similar to the metabolization of total QAs by L. albus and L. angustifolius reported by [22]. Unexpectedly, an increase of the main QAs identified in the seeds of this species (lupanine, aphylline, aphyllidine, 3β-hydroxylupanine, and sparteine) was observed during germination, even though a decrease in the total QA content was detected during this developmental stage due to the degradation of unidentified QAs. Were the main QAs the products of this degradation? Or were they newly synthesized? We hypothesized that they may be the products of the degradation of other QAs since QAs are synthesized in photosynthetic tissues, which are obviously absent at germinating seeds. On the other hand, active degradation of these main QAs was observed during the hypocotyl elongation stage, in which a steep decrease (68.2%) in QA content was determined and similarly to L. aschenbornii and L. montanus, the most abundant QA, lupanine in this case, was more rapidly degraded.
The developmental stage at which de novo biosynthesis of QAs occurred was species specific, as was reported for other Lupinus species [22]. Lupinus aschenbornii initiated de novo biosynthesis after germination and before hypocotyl elongation. As mentioned before, this species metabolized more rapidly the QAs present in its seeds compared to L. montanus and L. bilineatus, which interestingly started to biosynthesize QAs (mostly) after the elongation of hypocotyl. The onset of QA biosynthesis in the three species was triggered after the decrease of QA. The content of QA was on the range of 11 to 14 mg/g DW before biosynthesis started in all species, raising questions of whether QA concentration is a regulatory mechanism in their de novo biosynthesis.
Clear differences in the biosynthetic capacity of QAs in L. aschenbornii, L. bilineatus, and L. montanus were evident as revealed by their QA patterns. Lupinus bilineatus and L. montanus (mostly) synthesize QA molecules derived from the modification of the rings A and B of a tetracyclic precursor (Figure 1), being these modifications mainly hydroxylation, oxidation, and dehydrogenation. These species do not accumulate esters derived from 13α-hydroxylupanine in contrast to L. aschenbornii, which is rich in these compounds and whose most abundant QAs are the product of tailoring reactions on ring D (Figure 1), i.e., hydroxylation, esterification, and ring cleavage. Are these differences in QA patterns the result of the transcriptional turn on and turn off status of the genes coding for the enzymes that synthesize 13α-hydroxylupanine esters in each species? In this regard, it has been reported that in L. angustifolius the transcriptional regulation of HMT/HLT seems to be under a separate genetic control than the genes LDC and CAO [17]. Lupinus aschenbornii plantlets seemed to have similarities in their QA biosynthetic machinery, with the leaves of L. angustifolius [18].
Similar patterns of metabolization were observed for structurally related QA within each species, suggesting a common regulation in their synthesis. For instance, lupanine and its derivatives (except 3β-hydroxylupanine) and angustifoline showed identical trends from germination to the emergence of the second leaf in L. aschenbornii, whereas lupanine, 3β-hydroxylupanine, and anagyrine did in L. bilineatus from germination to the emergence of the sixth leaf. The dynamics of metabolization and synthesis of aphylline and aphyllidine in L. bilineatus were also alike; however, a clear increase in aphyllidine content was observed after the emergence of the fifth leaf in the plantlets, which coincides with a decrease in the content of aphylline. Interestingly, in L. montanus plantlets, lupanine, sparteine, and aphylline displayed similar trends along the developmental stages characterized, so did the hydroxylated forms of lupanine and multiflorine. Regarding similarities among the three species, an increased peak in the content of most QAs was observed at the emergence of the first leaf, suggesting a high biosynthetic activity with the development of the principal photosynthetic tissues.
Lupinus bilineatus did not accumulate sparteine, the simplest structure of tetracyclic QAs, at least during the developmental stages characterized, which suggests that sparteine is not the precursor of more complex QAs. This is in line with the consensus that tetracyclic QAs are most likely synthesized independently from a common precursor, the diiminium cation (Figure 1) [41]. Aphylline is a QA found in some American Lupinus species, and it is highly accumulated in L. bilineatus and present in L. montanus. When did this biosynthetic capacity first appear? Is it a novel enzyme activity or was it already present in a common ancestor of the lupin American species? The presence of aphylline in Sophora alopecuroides L. [42] suggests that the enzyme(s) involved in the synthesis of aphylline was already present in a common ancestor of the Lupinus and Sophora genus, implying a reversion of the turn off of the aphylline biosynthetic genes in some American lupins as postulated for other QAs [43].
5. Conclusions
The main results of the present research are: (1) QAs were metabolized during germination—the main QAs were the most rapidly catabolized; (2) de novo QA biosynthesis starts once QAs are metabolized, for each evaluated species at a different developmental stage, suggesting that it is regulated in a species-specific manner; (3) structurally related QAs showed similar patterns of metabolization and biosynthesis, suggesting a common regulation; (4) there is no common QA pattern from which the diversity of QAs produced in these species is generated. Lupanine and 3β-lupanine were found in the three species, while sparteine was not present in L. bilineatus, excluding it as the precursor of more complex QAs;. (5) Lupinus montanus showed the most diverse QA pattern since it synthesized four different QA types: sparteine skeleton, lupanine and derivatives, aphylline and derivatives and multiflorine skeleton while Lupinus aschenbornii showed the most complex pattern of lupanine derivatives, what these changes in QA biosynthetic specificities mean in an evolutionary scenario is intriguing.
Author Contributions
Conceptualization, K.B.-T. and A.M.H.-S.; methodology, A.R.-B., E.V.-Z. and N.R.; formal analysis, A.M.H.-S.; investigation, A.R.-B. and G.S.-M.; resources, K.B.-T.; data curation, A.M.H.-S.; writing—original draft preparation, A.M.H.-S.; writing—review and editing, K.B.-T. and A.M.H.-S.; visualization, A.R.-B., A.M.H.-S., G.S.-M., E.V.-Z., N.R., M.W. and K.B.-T.; supervision, A.M.H.-S., G.S.-M., E.V.-Z., N.R., M.W. and K.B.-T.; project administration, K.B.-T.; funding acquisition, K.B.-T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Consejo Nacional de Ciencia y Tecnología (CONACyT A1-S-34633) and the Secretaría de Investigación y Posgrado del Instituto Politécnico Nacional (SIP 20172240, SIP 20180777, and SIP20196663).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available upon request from a corresponding author.
Acknowledgments
A.R.-B. and A.M.H.-S. acknowledge the scholarships granted by CoNaCyT for postgraduate (787734) and postdoctoral studies (740989), respectively.
Conflicts of Interest
M.W. is chief editor of this journal.
References
- Hughes, C.; Eastwood, R. Island radiation on a continental scale: Exceptional rates of plant diversification after uplift of the Andes. Proc. Natl. Acad. Sci. USA 2006, 103, 10334–10339. [Google Scholar] [CrossRef]
- Wink, M.; Meißner, C.; Witte, L. Patterns of quinolizidine alkaloids in 56 species of the genus Lupinus. Phytochemistry 1995, 38, 139–153. [Google Scholar] [CrossRef]
- Bermúdez-Torres, K.; Ferval, M.; Hernández-Sánchez, A.M.; Tei, A.; Gers, C.; Wink, M.; Legal, L. Molecular and Chemical Markers to Illustrate the Complex Diversity of the Genus Lupinus (Fabaceae). Diversity 2021, 13, 263. [Google Scholar] [CrossRef]
- Feitoza, R.B.B.; Lima, H.R.P. Chemosystematic and evolutionary trends of the genistoid clade sensu stricto (Papilionoideae, Fabaceae). Phytochemistry 2021, 183, 112616. [Google Scholar] [CrossRef] [PubMed]
- Wink, M. The role of Quinolizidine Alkaloids in Plant-Insect Interactions. In Plant Insect Interactions; Bernays, E.A., Ed.; CRC Press: Boca Raton, FL, USA, 1992; pp. 131–166. [Google Scholar]
- Hoppe, D.; Hintze, F.; Tebben, P.; Paetow, M.; Ahrens, H.; Schwerdtfeger, J.; Sommerfeld, P.; Haller, J.; Guarnierp, W.; Kolczewsk, S.; et al. Enantioselective synthesis via sparteine induced asymmetric deprotonation. Pure Appl. Chem. 1994, 66, 1479–1486. [Google Scholar] [CrossRef]
- Schmeller, T.; Sauerwein, M.; Sporer, F.; Wink, M.; Müller, W.E. Binding of quinolizidine alkaloids to nicotinic and muscarinic acetylcholine receptors. J. Nat. Prod. 1994, 57, 1316–1319. [Google Scholar] [CrossRef]
- O′Brien, P. Basic instinct: Design, synthesis and evaluation of (+)-sparteine surrogates for asymmetric synthesis. Chem. Commun. 2008, 1, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Quintana, R.; Palma, A.; Rebolledo, R.; Aguilera, A. Effect of an infusion of canelo and bitter lupin on Aegorhinus superciliosus adults. Cienc. Investig. Agrar. 2011, 38, 397–403. [Google Scholar] [CrossRef]
- Villalpando-Vargas, F.; Medina-Ceja, L. Sparteine as an anticonvulsant drug: Evidence and possible mechanism of action. Seizure 2016, 39, 49–55. [Google Scholar] [CrossRef]
- Dubois, O.; Allanic, C.; Charvet, C.L.; Guégnard, F.; Février, H.; Théry-Koné, I.; Cortet, J.; Koch, C.; Bouvier, F.; Fassier, T.; et al. Lupin (Lupinus spp.) seeds exert anthelmintic activity associated with their alkaloid content. Sci. Rep. 2019, 9, 9070. [Google Scholar] [CrossRef] [PubMed]
- Li, J.C.; Dai, W.F.; Liu, D.; Zhang, Z.J.; Jiang, M.Y.; Rao, K.R.; Li, R.T.; Li, H.M. Quinolizidine alkaloids from Sophora alopecuroides with anti-inflammatory and anti-tumor properties. Bioorg. Chem. 2021, 110, 104781. [Google Scholar] [CrossRef]
- Li, Y.; Wang, G.; Liu, J.; Ouyang, L. Quinolizidine alkaloids derivatives from Sophora alopecuroides Linn: Bioactivities, structure-activity relationships and preliminary molecular mechanisms. Eur. J. Med. Chem. 2020, 188, 111972. [Google Scholar] [CrossRef]
- Zou, J.; Zhao, L.; Yi, P.; An, Q.; He, L.; Li, Y.; Lou, H.; Yuan, C.; Gu, W.; Huang, L.; et al. Quinolizidine Alkaloids with Antiviral and Insecticidal Activities from the Seeds of Sophora tonkinensis Gagnep. J. Agric. Food Chem. 2020, 68, 15015–15026. [Google Scholar] [CrossRef]
- Aly, S.H.; Elissawy, A.M.; Allam, A.E.; Farag, S.M.; Eldahshan, O.A.; Elshanawany, M.A.; Singab, A.N.B. New quinolizidine alkaloid and insecticidal activity of Sophora secundiflora and Sophora tomentosa against Culex pipiens (Diptera: Culicidae). Nat. Prod. Res. 2021, 1–13. [Google Scholar] [CrossRef]
- Wink, M. Wounding-Induced Increase of Quinolizidine Alkaloid Accumulation in Lupin Leaves. Z. Naturforsch. Sect. C J. Biosci. 1983, 38, 905–909. [Google Scholar] [CrossRef]
- Frick, K.M.; Foley, R.C.; Kamphuis, L.G.; Siddique, K.H.M.; Garg, G.; Singh, K.B. Characterization of the genetic factors affecting quinolizidine alkaloid biosynthesis and its response to abiotic stress in narrow-leafed lupin (Lupinus angustifolius L.). Plant Cell Environ. 2018, 41, 2155–2168. [Google Scholar] [CrossRef]
- Otterbach, S.L.; Yang, T.; Kato, L.; Janfelt, C.; Geu-Flores, F.; Hancock, R. Quinolizidine alkaloids are transported to seeds of bitter narrow-leafed lupin. J. Exp. Bot. 2019, 70, 5799–5808. [Google Scholar] [CrossRef] [PubMed]
- Wink, M.; Hartmann, T. Localization of the Enzymes of Quinolizidine Alkaloid Biosynthesis in Leaf Chloroplasts of Lupinus polyphyllus. Plant Physiol. 1982, 70, 74–77. [Google Scholar] [CrossRef]
- Wink, M.; Hartmann, T. Enzymatic synthesis of quinolizidine alkaloid esters: A tigloyl-CoA: 13-hydroxylupanine O-tigloyltransferase from Lupinus albus L. Planta 1982, 156, 560–565. [Google Scholar] [CrossRef] [PubMed]
- Czepiel, K.; Krajewski, P.; Wilczura, P.; Bielecka, P.; Święcicki, W.; Kroc, M. Expression profiles of alkaloid-related genes across the organs of narrow-leafed lupin (Lupinus angustifolius L.) and in response to anthracnose infection. Int. J. Mol. Sci. 2021, 22, 2676. [Google Scholar] [CrossRef]
- Wink, M.; Witte, L. Quinolizidine Alkaloids as Nitrogen Source for Lupin Seedlings and Cell Cultures. Verl. Z. Naturforsch. 1985, 40, 767–775. [Google Scholar] [CrossRef]
- Altares, P.; Pedrosa, M.M.; Burbano, C.; Cuadrado, C.; Goyoaga, C.; Muzquiz, M.; Jim, C.; Gloria, D. Alkaloid variation during germination in different lupin species. Food Chem. 2005, 90, 347–355. [Google Scholar] [CrossRef]
- Bunsupa, S.; Katayama, K.; Ikeura, E.; Oikawa, A.; Toyooka, K.; Saito, K.; Yamazakia, M. Lysine decarboxylase catalyzes the first step of quinolizidine alkaloid biosynthesis and coevolved with alkaloid production in leguminosae. Plant Cell 2012, 24, 1202–1216. [Google Scholar] [CrossRef]
- Yang, T.; Nagy, I.; Mancinotti, D.; Otterbach, S.L.; Andersen, T.B.; Motawia, M.S.; Asp, T.; Geu-Flores, F. Transcript profiling of a bitter variety of narrow-leafed lupin to discover alkaloid biosynthetic genes. J. Exp. Bot. 2017, 68, 5527–5537. [Google Scholar] [CrossRef]
- Okada, T.; Hirai, M.Y.; Suzuki, H.; Yamazaki, M.; Saito, K. Molecular characterization of a novel quinolizidine alkaloid O-tigloyltransferase: cDNA cloning, catalytic activity of recombinant protein and expression analysis in Lupinus plants. Plant Cell Physiol. 2005, 46, 233–244. [Google Scholar] [CrossRef]
- Kroc, M.; Koczyk, G.; Kamel, K.A.; Czepiel, K.; Fedorowicz-Strońska, O.; Krajewski, P.; Kosińska, J.; Podkowiński, J.; Wilczura, P.; Święcicki, W. Transcriptome-derived investigation of biosynthesis of quinolizidine alkaloids in narrow-leafed lupin (Lupinus angustifolius L.) highlights candidate genes linked to iucundus locus. Sci. Rep. 2019, 9, 2231. [Google Scholar] [CrossRef] [PubMed]
- Drummond, C.S. Diversification of Lupinus (Leguminosae) in the western New World: Derived evolution of perennial life history and colonization of montane habitats. Mol. Phylogenet. Evol. 2008, 48, 408–421. [Google Scholar] [CrossRef] [PubMed]
- Sousa, S.M.; Delgado, A.S. Leguminosas mexicanas: Fitogeografía, endemismo y orígenes. In Diversidad biológica de México: Orígenes y Distribución; Ramamoorthy, T.P., Bye, R., Lot, A., Fa, J., Eds.; Instituto de Biología, Universidad Nacional Autónoma de México: Ciudad de México, México, 1998; pp. 449–500. [Google Scholar]
- Dunn, D.B.; Lupinus, L. Flora Fanerogámica del Valle de México; Calderón de Rzedowski, G., Rzedowski, J., Eds.; Instituto de Ecología: Ciudad de México, Mexico, 2005; pp. 290–300. ISBN 9788578110796. [Google Scholar]
- Bermúdez-Torres, K.; Martínez Herrera, J.; Figueroa Brito, R.; Wink, M.; Legal, L. Activity of quinolizidine alkaloids from three Mexican Lupinus against the lepidopteran crop pest Spodoptera frugiperda. BioControl 2009, 54, 459–466. [Google Scholar] [CrossRef]
- Ruiz López, M.; García López, P.; Rodríguez Macías, R.; Zamora Natera, J.; Isaac Virgen, M.; Múzquiz, M. Mexican wild lupines as a source of quinolizidine alkaloids of economic potential. Polibotánica 2010, 29, 159–164. [Google Scholar]
- Montes-Hernández, E.; Corona-Rangel, M.L.; Encarnación-Corona, A.; Cantor del Angel, J.A.; Sánchez-López, J.A.; Sporer, F.; Wink, M.; Bermúdez-Torres, K. Quinolizidine alkaloid composition in different organs of Lupinus aschenbornii. Rev. Bras. Farmacogn. 2011, 21, 824–828. [Google Scholar] [CrossRef]
- Bermúdez-Torres, K.; Robledo-Quintos, N.; Barrera-Necha, L.L.; Wink, M. Alkaloid Profile of Leaves and Seeds of Lupinus hintonii C.P. Smith. Z. Naturforsch 2002, 57, 243–247. [Google Scholar] [CrossRef]
- Bermúdez-Torres, K.; Robledo-Quintos, N.; Martínez-Herrera, J.; Tei, A.; Wink, M. Patrón de acumulación de alcaloides en hojas y semillas de Lupinus aschenbornii crecidos en México. Rev. Latinoam. Química 2000, 27, 101–105. [Google Scholar]
- Ferval, M.; Legal, L.; Gers, C.; Pelissier, C.; Winterton, P. When island-like populations at high elevation show genetic divergence despite no morphological variability: The case of Lupinus montanus in Central Mexico. Turk. J. Bot. 2013, 789–801. [Google Scholar] [CrossRef]
- Hernández-Ferretis, E.; Rivera-Meléndez, R.K.; Ramos-Herrera, O.J.; Salinas-Pérez, F.C.; Rodríguez-Monroy, M.; Bermúdez-Torres, K. Effect of scarification treatments on germination of Lupinus montanus HBK seeds. In Lupinus for Health and Wealth. Proceedings of the 12th International Lupin Conference, Fremantle, Australia, 14–18 September 2008; Palta, J.A., Berger, J.B., Eds.; International Lupin Association: Canterbury, New Zealand, 2008; pp. 405–409. [Google Scholar]
- Figueroa-Rodríguez, M.I. Efecto de Especies Nativas y Silvestres de Lupinus sobre la Fertilidad del suelo Agrícola. Master Thesis, Instituto Politécnico Nacional, Ciudad de México, Mexico, 2017. [Google Scholar]
- NIST/EPA/NIH. Mass spectral library. In Mass Spectral Library with Search Program (Data Version: NIST05, Software Version 2.0); U.S. Department of Commerce, Technology Administration, National Institute of Standards and Technology, Standard Reference Data Program: Gaithersburg, MD, USA, 2002. [Google Scholar]
- Gresta, F.; Wink, M.; Prins, U.; Abberton, M.; Capraro, J.; Scarafoni, A.; Hill, G. Lupins in European cropping systems. Legum. Crop. Syst. 2017, 88–108. [Google Scholar] [CrossRef]
- Fraser, A.M.; Robins, D.J. Incorporation of chiral [1-2H]cadaverines into the quinolizidine alkaloids sparteine, lupanine, and angustifoline. J. Chem. Soc. Chem. Commun. 1984, 1477–1479. [Google Scholar] [CrossRef]
- Rizvi, S.A.H.; Ling, S.; Tian, F.; Liu, J.; Zeng, X. Interference mechanism of Sophora alopecuroides L. alkaloids extract on host finding and selection of the Asian citrus psyllid Diaphorina citri Kuwayama (Hemiptera: Psyllidae). Environ. Sci. Pollut. Res. 2019, 26, 1548–1557. [Google Scholar] [CrossRef] [PubMed]
- Wink, M. Evolution of secondary metabolites in legumes (Fabaceae). S. Afr. J. Bot. 2013, 89, 164–175. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).