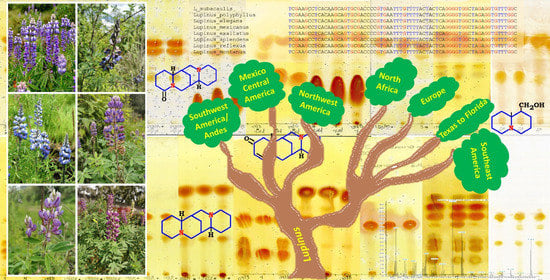
Molecular and Chemical Markers to Illustrate the Complex Diversity of the Genus Lupinus (Fabaceae)
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Sequences and Treatment
2.2. Quinolizine Alkaloids (QA) Database
3. Results
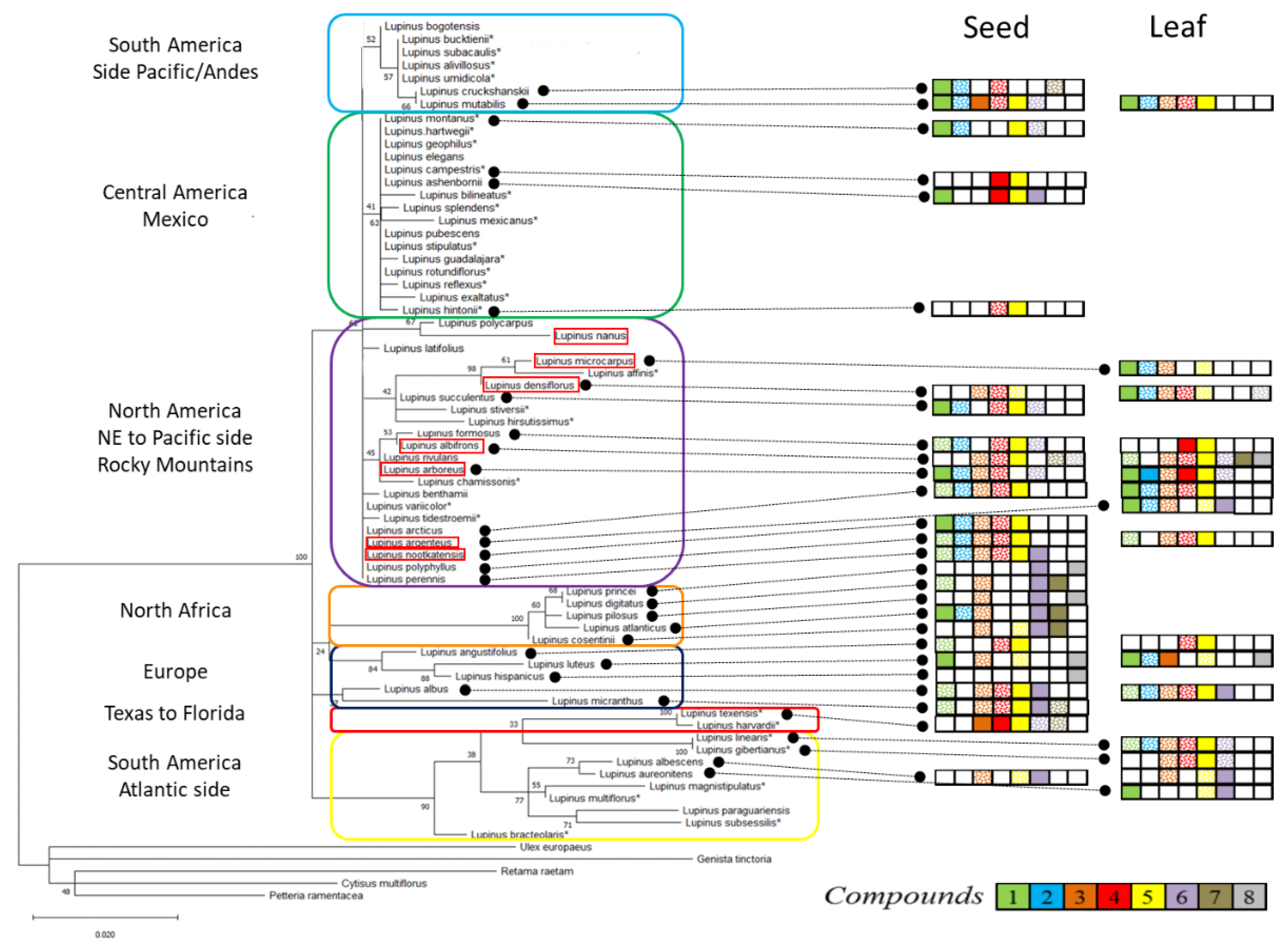
3.1. Phylogenetic Relationships Based on ITS
3.2. Alkaloid Profiles and Relationships
3.3. QA-Based Clustering of Lupinus
4. Discussion
4.1. Phylogenetic Relationships Based on ITS
4.2. QAs and Chemotaxonomy
4.3. Implications for Biogeographic Scenario
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dunn, D.B. Cytotaxonomy and distribution of the New World lupin species. In Proceedings of the Third International Lupin Conference, La Rochelle, France, 4–8 June 1984; pp. 67–85. [Google Scholar]
- Kasprzak, A.; Šafář, J.; Janda, J.; Doležel, J.; Wolko, B.; Naganowska, B. The bacterial artificial chromosome (BAC) library of the narrow-leafed lupin (Lupinus angustifolius L.). Cell. Mol. Biol. Lett. 2006, 11, 396–407. [Google Scholar] [CrossRef] [PubMed]
- Eastwood, R.J.; Drummond, C.S.; Schifino-Wittmann, M.T.; Hughes, C. Diversity and evolutionary history of Lupins–Insights from new phylogenies. In Lupins for Health and Wealth, Proceedings of the 12th International Lupin Conference, Fremantle, Australia, 14–18 September 2008; Palta, J.A., Berger, J.B., Eds.; International Lupin Association: Canterbury, New Zealand, 2008; pp. 346–354. [Google Scholar]
- Bermudez-Torres, K.; Herrera, J.M.; Figueroa Brito, R.; Wink, M.; Legal, L. Activity of quinolizidine alkaloids from three Mexican Lupinus against the lepidopteran crop pest Spodoptera frugiperda. Biocontrol 2009, 54, 459–466. [Google Scholar] [CrossRef]
- Bermúdez-Torres, K.; Legal, L.; Ferval, M. High Altitude Ecosystems in the Era of Climate Change. Lupinus species in Central Mexico in the Era of Climate Change: Adaptation, migration or extinction? In Climate Change Impacts on High-Altitude Ecosystems; Ozturk, M., Hakeem, K.R., Faridah-Hanum, I., Efe, R., Eds.; Springer: Heidelberg, Germany, 2015; Chapter 8; pp. 215–228. [Google Scholar]
- Käss, E.; Wink, M. Molecular phylogeny and phylogeography of Lupinus (Leguminosae) inferred from nucleotide sequences of the rbcL gene ITS 1 + 2 regions of rDNA. Plant Syst. Evol. 1997, 208, 139–167. [Google Scholar] [CrossRef]
- Hughes, C.; Eastwood, R.J. Island radiation on a continental scale: Exceptional rates of plant diversification after uplift of the Andes. Proc. Natl. Acad. Sci. USA 2006, 103, 10334–10339. [Google Scholar] [CrossRef] [PubMed]
- Drummond, C.S. Diversification of Lupinus (Leguminosae) in the western New World: Derived evolution of perennial life history and colonization of montane habitats. Mol. Phylogenet. Evol. 2008, 48, 408–421. [Google Scholar] [CrossRef]
- Drummond, C.S.; Eastwood, R.J.; Miotto, S.T.S.; Hughes, C. Multiple continental radiations and correlates of diversification in Lupinus (Leguminosae): Testing for key innovations with incomplete taxon sampling. Syst. Biol. 2012, 61, 443–460. [Google Scholar] [CrossRef]
- Wink, M.; Botschen, F.; Gosmann, C.; Schafer, H.; Waterman, G. Chemotaxonomy seen from a phylogenetic perspective and evolution of secondary metabolism. Annu. Plant Rev. 2009, 40, 364–433. [Google Scholar]
- Ferval, M.; Legal, L.; Gers, C.; Pélissier, C.; Winterton, P.; Sánchez López, J.A.; Corona-Rangel, M.L.; Bermúdez-Torres, K. When island-like populations at high elevation show genetic divergence despite no morphological variability. The case of Lupinus montanus in Central Mexico. Turk. J. Bot. 2013, 37, 789–801. [Google Scholar] [CrossRef]
- Ferval, M.; Bermúdez-Torres, K.; Gers, C.; Legal, L. Genomic fingerprinting versus nuclear gene sequences: A comparative approach for studying the Lupinus montanus (Fabaceae) species complex. S. Afr. J. Bot. 2013, 89, 106–110. [Google Scholar] [CrossRef][Green Version]
- Aïnouche, A.; Bayer, R.J. Phylogenetic relationships in Lupinus (Fabaceae: Papilionoideae) based on Internal Transcribed Spacer Sequences (ITS) of nuclear ribosomal DNA. Am. J. Bot. 1999, 86, 590–607. [Google Scholar] [CrossRef] [PubMed]
- Aïnouche, A.; Bayer, R.J.; Misset, M.T. Molecular phylogeny, diversification and character evolution in Lupinus (Fabaceae) with special attention to Mediterranean and African lupins. Plant Syst. Evol. 2004, 246, 211–222. [Google Scholar] [CrossRef]
- Huang, D.I.; Friar, E.A. Relationships in the Lupinus albifrons species complex (Fabaceae) based on two highly variable chloroplast regions. Syst. Bot. 2011, 36, 362–370. [Google Scholar] [CrossRef]
- Weitemer, K.A. Phylogeographic Patterns and Intervarietal Relationships within Lupinus lepidus: Morphological Differences, Genetic Similarities. Master Thesis, Portland State University, Portland, OR, USA, 2010. [Google Scholar]
- Drummond, C.S.; Hamilton, M.B. Hierarchical components of genetic variation at a species boundary: Population structure in two sympatric varieties of Lupinus microcarpus (Leguminosae). Mol. Ecol. 2007, 16, 753–769. [Google Scholar] [CrossRef]
- Barrera, L.L.; Trujillo, M.E.; Goodfellow, M.; García, F.J.; Hernández-Lucas, I.; Dávila, G.; Van Berkum, P.; Martínez-Romero, E. Biodiversity of Bradyrhizobia nodulating Lupinus spp. Int. J. Syst. Bacteriol. 1997, 47, 1086–1091. [Google Scholar] [CrossRef]
- Del Moral, R.; Rozzell, L.R. Long-term effects of Lupinus lepidus on vegetation dynamics at Mount St. Helens. Plant Ecol. 2005, 181, 203–215. [Google Scholar] [CrossRef]
- Johnson, N.D.; Liu, B.; Bentley, B.L. The effects of nitrogen fixation, soil nitrate, and defoliation on the growth, alkaloids, and nitrogen levels of Lupinus succulentus (Fabaceae). Oecologia 1987, 74, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Wink, M. Plant breeding: Importance of plant secondary metabolites for protection against pathogens and herbivores. Theor. Appl. Genet. 1988, 75, 225–233. [Google Scholar] [CrossRef]
- Wink, M.; Meißner, C.; Witte, L. Patterns of quinolizidine alkaloids in 56 species of the genus Lupinus. Phytochemistry 1995, 38, 139–153. [Google Scholar] [CrossRef]
- Roberts, M.F.; Wink, M. Alkaloids: Biochemistry, Ecology and Medicinal Applications; Plenum Press: New York, NY, USA; London, UK, 1998. [Google Scholar]
- Bunsupa, S.; Yamazaki, M.; Saito, K. Quinolizidine alkaloids biosynthesis: Recent advances and future prospects. Front. Plant Sci. 2013, 3, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, L.; Desbuquois, C.; Papineau, J.; Charrier, M. Influence of the quinolizidine alkaloid content of Lupinus albus (Fabaceae) on the feeding choices of Helix aspersa (Gastropoda: Pulmonata). J. Molluscan Stud. 1999, 66, 61–68. [Google Scholar] [CrossRef]
- Wink, M. Quinolizidine alkaloids: Biochemistry, metabolism and function in plants and cell suspension cultures. Planta Med. 1987, 53, 509–514. [Google Scholar] [CrossRef]
- De la Vega, R.; Gutierrez, M.P.; Sanz, C.; Calvo, R.; Robredo, L.M.; de la Cuadra, C.; Muzquiz, M. Bactericide-like effect of Lupinus alkaloids. Ind. Crop Prod. 1996, 5, 141–148. [Google Scholar] [CrossRef]
- Chludil, H.D.; Vilariño, M.; Franco, M.L.; Leicach, S.R. Changes in Lupinus albus and Lupinus angustifolius alkaloid profiles in response to mechanical damage. J. Agric. Food Chem. 2009, 57, 6107–6113. [Google Scholar] [CrossRef] [PubMed]
- Chludil, H.D.; Leicach, S.R.; Corbino, G.B.; Barriga, L.G.; Vilariño, M. Genistin and quinolizidine induction in L. angustifolius aerial parts in response to mechanical damage. J. Plant Interact. 2013, 8, 117–124. [Google Scholar] [CrossRef]
- Wink, M.; Waterman, P.G. Chemotaxonomy in relation to molecular phylogeny of plants. Annu. Plant Rev. 1999, 2, 300–341. [Google Scholar]
- Wink, M. Evolution of secondary metabolites in legumes (Fabaceae). S. Afr. J. Bot. 2013, 89, 164–175. [Google Scholar] [CrossRef]
- Van Wyk, B.E. The value of chemosystematics in clarifying relationships in the genistoid tribes of papilionoid legumes. Biochem. Syst. Ecol. 2003, 31, 875–884. [Google Scholar] [CrossRef]
- Zidorn, C.; Stuppner, H. Chemosystematics of taxa from the Leondoton section Oporinia. Biochem. Syst. Ecol. 2001, 29, 827–837. [Google Scholar] [CrossRef]
- Máximo, P.; Lourenço, A.; Tei, A.; Wink, M. Chemotaxonomy of Portuguese Ulex: Quinolizidine alkaloids as taxonomical markers. Phytochemistry 2006, 67, 1943–1949. [Google Scholar] [CrossRef]
- Larsson, S. The “new” chemosystematics: Phylogeny and phytochemistry. Phytochemistry 2007, 68, 2903–2907. [Google Scholar] [CrossRef] [PubMed]
- Roux, O.; Gers, C.; Legal, L. When, among variations during ontogeny, cuticular compounds of Calliphoridae may be used as phylogenetic markers. Biochem. Syst. Ecol. 2006, 34, 406–416. [Google Scholar] [CrossRef]
- Rønsted, N.; Franzyk, H.; Mølgaard, P.; Jaroszewski, J.W.; Jensen, S.R. Chemotaxonomy and evolution of Plantago L. Plant Syst. Evol. 2003, 242, 63–82. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using Maximum Likelihood, evolutionary distance, and Maximum Parsimony methods. Mol. Biol. Evol. 2005, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.L. PAUP Phylogenetic Analysis Using Parsimony; Version 4.0b10; Sinauer: Sunderland, MA, USA, 2001. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Koichiro Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogeny: An approach using the bootstrap. Evolution 1985, 39, 783–789. [Google Scholar] [CrossRef] [PubMed]
- Ihaka, R.; Gentleman, R. R: A language for data analysis and graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar]
- García-Mateos, R.; Soto-Hernandez, M.; Kelly, D. Alkaloids from six Erythrina species endemic to Mexico. Biochem. Syst. Ecol. 1998, 26, 545–551. [Google Scholar] [CrossRef]
- Święcicki, W.; Czepiel, K.; Wilczura, P.; Barzyk, P.; Kaczmarek, Z.; Kroc, M. Chromatographic Fingerprinting of the Old World Lupins Seed Alkaloids: A Supplemental Toll in Species Discrimination. Plants 2019, 8, 548. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, S.I.; Moates, R.F. Alkaloids of Lupinus diffusus Nutt. Phytochemistry 1967, 6, 137–140. [Google Scholar] [CrossRef]
- Adler, L.S.; Kittelson, P.M. Variation in Lupinus arboreus alkaloid profiles and relationships with multiple herbivores. Biochem. Syst. Ecol. 2004, 32, 371–390. [Google Scholar] [CrossRef]
- Nevado, B.; Atchinson, G.W.; Hughes, C.E.; Filatov, D.A. Widespread adaptative evolution during repeated evolutionary radiations in New World lupins. Nat. Commun. 2016, 7, 12384. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, T. The evolution of chemosystematics. Phytochemistry 2007, 68, 2887–2895. [Google Scholar] [CrossRef]
- Tiffney, B.H.; Manchester, S.R. The use of geological and paleontological evidence in evaluating plant phylogeographic hypotheses in the northern hemisphere Tertiary. Int. J. Plant Sci. 2001, 162, S3–S17. [Google Scholar] [CrossRef]
- Denk, T.; Grímsson, F.; Zetter, R. Episodic migration of oaks to Iceland: Evidence for a North Atlantic “land bridge” in the latest Miocene. Am. J. Bot. 2010, 97, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Milne, R.I. Northern hemisphere plant disjunctions: A window on Tertiary land bridges and climate change? Ann. Bot. 2006, 98, 465–472. [Google Scholar] [CrossRef]
- Grímmson, G.; Denk, T.; Símonarson, A. Middle Miocene floras of Iceland–The early colonization of an island? Rev. Paleobot. Palynol. 2007, 144, 181–219. [Google Scholar] [CrossRef]
- Denk, T.; Grimm, G.W. The oaks of western Eurasia: Traditional classifications and evidence from two nuclear markers. Taxon 2010, 59, 351–366. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bermúdez-Torres, K.; Ferval, M.; Hernández-Sánchez, A.M.; Tei, A.; Gers, C.; Wink, M.; Legal, L. Molecular and Chemical Markers to Illustrate the Complex Diversity of the Genus Lupinus (Fabaceae). Diversity 2021, 13, 263. https://doi.org/10.3390/d13060263
Bermúdez-Torres K, Ferval M, Hernández-Sánchez AM, Tei A, Gers C, Wink M, Legal L. Molecular and Chemical Markers to Illustrate the Complex Diversity of the Genus Lupinus (Fabaceae). Diversity. 2021; 13(6):263. https://doi.org/10.3390/d13060263
Chicago/Turabian StyleBermúdez-Torres, Kalina, Maxime Ferval, Arianna Michelle Hernández-Sánchez, Andreas Tei, Charles Gers, Michael Wink, and Luc Legal. 2021. "Molecular and Chemical Markers to Illustrate the Complex Diversity of the Genus Lupinus (Fabaceae)" Diversity 13, no. 6: 263. https://doi.org/10.3390/d13060263
APA StyleBermúdez-Torres, K., Ferval, M., Hernández-Sánchez, A. M., Tei, A., Gers, C., Wink, M., & Legal, L. (2021). Molecular and Chemical Markers to Illustrate the Complex Diversity of the Genus Lupinus (Fabaceae). Diversity, 13(6), 263. https://doi.org/10.3390/d13060263