Molecular Identification of an Invasive Sarotherodon Species from the Atchakpa Freshwater Reservoir (Ouémé River Basin, Benin) and Comparison within S. melanotheron Using COI Markers
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. Molecular Identification of the Specimens
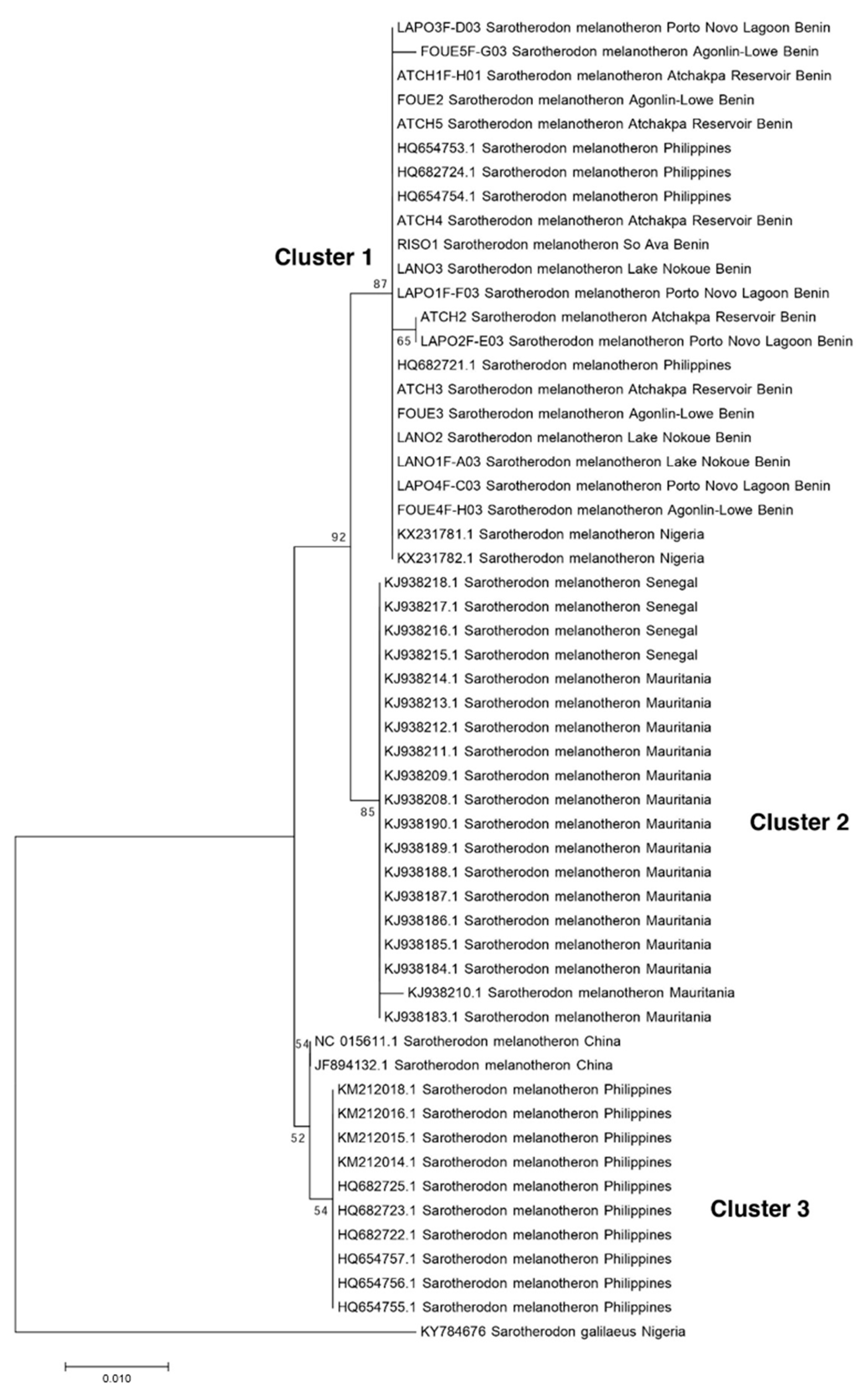
3.2. Comparison with Available Sequences of S. melanotheron
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Trewavas, E.; Teugels, G.G. Sarotherodon. In Check-List of the Freshwater Fishes of Africa (CLOFFA); Daget, J., Gosse, J.-P., Teugels, G.G., Van Den Audenaerde, D.F.E.T., Eds.; Royal Museum for Central Africa: Tervuren, Belgium, 1991; pp. 425–437. [Google Scholar]
- Pauly, D. The biology, fishery and potential for aquaculture of Tilapia melanotheron in a small West African lagoon. Aquaculture 1976, 7, 33–49. [Google Scholar] [CrossRef]
- Adepo-Gourene, B.; Gourene, G. Différentiation morphologique des populations naturelles d’une sous espèce de tilapia Sarotherodon melanotheron melanotheron Rüppell, 1852 (Teleostei; Cichlidae) de Côte d’Ivoire. Sci. Nat. 2008, 5, 15–27. [Google Scholar]
- Laleye, P.; Chikou, A.; Philippart, J.-C.; Teugels, G.; Vandewalle, P. Etude de la diversité ichtyologique du bassin du fleuve Ouémé au Bénin (Afrique de l’ouest). Cybium 2004, 228, 329–339. [Google Scholar]
- Montchowui, E.; Niyonkuru, C.; Ahouansou Montcho, S.; Chikou, A.; Laleye, P. L’ichtyofaune de la rivière Hlan au Bénin (Afrique de l’Ouest). Cybium 2007, 31, 173–176. [Google Scholar]
- Montchowui, E.; Chikou, A.; Kogbeto, M.-J.; Laleye, P. Biodiversité et structure des communautés de poissons du lac Hlan au Bénin. Int. J. Biol. Chem. Sci. 2008, 2, 196–206. [Google Scholar] [CrossRef]
- Fagnon, S.M.; Chikou, A.; Youssao, I.; Laleye, P. Caractérisation morphologique des populations de Sarotherodon melanotheron (Pisces, Cichlidae) en eaux douces et saumâtres au Sud Bénin. Int. J. Biol. Chem. Sci. 2013, 7, 619–630. [Google Scholar] [CrossRef] [Green Version]
- Pèlèbè, E.O.R.; Dohounkpan, G.A.; Agbohessi, T.P.; Imorou Toko, I.; Hounhoedo, A.O.; Lederoun, D. Inventaire partiel des espèces de poisson et évaluation du degré de fraîcheur de Sarotherodon melanotheron vendu au marché des produits de pêche de la commune de Savè (Bénin). Rev. Togol. Sci. 2016, 10, 187–198. [Google Scholar]
- Ordoñez, J.; Asis, A.; Catacutan, B.; Pena, J.D.; Santos, M. First report on the occurrence of invasive black-chin tilapia Sarotherodon melanotheron (Ruppell, 1852) in Manila Bay and of Mayan cichlid Cichlasoma urophthalmus (Gunther, 1862) in the Philippines. Bioinvasions Rec. 2015, 4, 115–124. [Google Scholar] [CrossRef]
- Cassemiro, F.A.S.; Bailly, D.; Da Graça, W.J.; Agostinho, A.A. The invasive potential of tilapias (Osteichthyes, Cichlidae) in the Americas. Hydrobiologia 2017, 817, 133–154. [Google Scholar] [CrossRef]
- Nico, L. Discovery of the invasive Mayan Cichlid fish “Cichlasoma” urophthalmus (Günther 1862) in Thailand, with comments on other introductions and potential impacts. Aquat. Invasions 2007, 2, 197–214. [Google Scholar] [CrossRef]
- Miller, R.R.; Minckley, W.L.; Norris, S.M. Freshwater Fishes of Mexico; University of Chicago Press: Chicago, IL, USA, 2005. [Google Scholar]
- Trivedi, S.; Aloufi, A.A.; Ansari, A.A.; Ghosh, S.K. Role of DNA barcoding in marine biodiversity assessment and conservation: An update. Saudi J. Biol. Sci. 2016, 23, 161–171. [Google Scholar] [CrossRef] [Green Version]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological identifications through DNA barcodes. Proc. Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hebert, P.D.; Ratnasingham, S.; De Waard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B Boil. Sci. 2003, 270, S96–S99. [Google Scholar] [CrossRef] [Green Version]
- Falk, T.M.; Teugels, G.G.; Abban, E.K. Revision and biogeographical analysis of the black-chinned tilapia Sarotherodon melanotheron (Teleostei, Cichlidae): Results of morphometric, allozyme, globin chain and mtDNA studies. J. Nat. Hist. 2003, 37, 2191–2212. [Google Scholar] [CrossRef]
- Falk, T.M.; Teugels, G.G.; Abban, E.K.; Villwock, W.; Renwrantz, L. Morphometric and allozyme variation in the black-chinned tilapiaSarotherodon melanotheron (Perciformes, Cichlidae), with a revision of the subspecies complex. J. Nat. Hist. 2000, 34, 1849–1863. [Google Scholar] [CrossRef]
- Aquilino, S.V.L.; Tango, J.M.; Fontanilla, I.K.C.; Pagulayan, R.C.; Basiao, Z.U.; Ong, P.S.; Quilang, J.P. DNA barcoding of the ichthyofauna of Taal Lake, Philippines. Mol. Ecol. Resour. 2011, 11, 612–619. [Google Scholar] [CrossRef]
- Wohlfarth, G.W.; Hulata, G. Applied Genetics of Tilapias, 2nd ed.; International Center for Living Aquatic Resources Management (ICLARM): Manila, Philippines, 1983. [Google Scholar]
- Randall, J.E. Introductions of marine fishes to the Hawaiian Islands. Bull. Mar. Sci. 1987, 41, 490–502. [Google Scholar]
- Porter, T.M.; Hajibabaei, M. Over 2.5 million COI sequences in GenBank and growing. PLoS ONE 2018, 13, e0200177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paugy, D.; Leveque, C.; Teugels, G.G. The Fresh and Brackish Water Fishes of West Africa; Royal Museum for Central Africa: Tervuren, Belgium, 2003. [Google Scholar]
- Ivanova, N.V.; Zemlak, T.S.; Hanner, R.H.; Hebert, P. Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Notes 2007, 7, 544–548. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Pereira, L.H.G.; Hanner, R.; Foresti, F.; Oliveira, C. Can DNA barcoding accurately discriminate megadiverse Neotropical freshwater fish fauna? BMC Genet. 2013, 14, 20. [Google Scholar] [CrossRef] [Green Version]
- Ward, R.D. DNA barcode divergence among species and genera of birds and fishes. Mol. Ecol. Resour. 2009, 9, 1077–1085. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Aquino, L.M.G.; Tango, J.M.; Canoy, R.J.C.; Fontanilla, I.K.C.; Basiao, Z.U.; Ong, P.S.; Quilang, J.P. DNA barcoding of fishes of Laguna de Bay, Philippines. Mitochondrial DNA 2011, 22, 143–153. [Google Scholar] [CrossRef]
- Falade, M.O.; Opene, A.J.; Benson, O. DNA barcoding of Clarias gariepinus, Coptodon zillii and Sarotherodon melanotheron from Southwestern Nigeria. F1000Research 2016, 5, 1268. [Google Scholar] [CrossRef] [Green Version]
- Kide, N.G.; Dunz, A.; Agnèse, J.-F.; Dilyte, J.; Pariselle, A.; Carneiro, C.; Correia, E.; Brito, J.; Yarba, L.O.; Kone, Y.; et al. Cichlids of the Banc d’Arguin National Park, Mauritania: Insight into the diversity of the genus Coptodon. J. Fish Biol. 2016, 88, 1369–1393. [Google Scholar] [CrossRef] [PubMed]
- He, A.Y.; Tang, S.J.; Jiang, Y.T.; Li, S.F.; Wang, C.H. Complete mitochondrial genome of blackchin tilapiaSarotherodon melanotheron (Perciformes, Cichlidae). Mitochondrial DNA 2011, 22, 171–173. [Google Scholar] [CrossRef] [PubMed]
- Falk, T.M.; Teugels, G.G.; Abban, E.K.; Villwock, W.; Renwrantz, L. Phylogeographic patterns in populations of the black-chinned tilapia complex (Teleostei, Cichlidae) from coastal areas in West Africa: Support for the refuge zone theory. Mol. Phylogenet. Evol. 2003, 27, 81–92. [Google Scholar] [CrossRef]
- Trewavas, E. Tilapias: Taxonomy and Speciation. In Proceedings of the International Conference on the Biology and Culture of Tilapias, Bellagio, Italy, 2–5 September 1980; Pullin, R.S.V., Lowe-McConnell, R.H., Eds.; International Center for Living Aquatic Resources Management (ICLARM): Manila, Philippines, 1982; pp. 3–13. [Google Scholar]
- Englund, R.A. The loss of native biodiversity and continuing nonindigenous species introductions in freshwater, estuarine, and wetland communities of Pearl Harbor, Oahu, Hawaiian Islands. Estuaries 2002, 25, 418–430. [Google Scholar] [CrossRef]
- Li, S.F.; Yan, B.; Cai, W.Q.; Li, T.Y.; Jia, J.H.; Zhang, Y.H. Evaluation of growth, salt tolerance and parents’ heterosis contribution in reciprocal hybrids F2 between Oreochromis niloticus and Sarotherodon melanotheron. J. Fish. China 2008, 32, 335–341. [Google Scholar]



| Aquatic Ecosystems | Number of COI Sequences | Accessions Numbers GenBank | Codes for Our Samples | Country | Status | Reference |
|---|---|---|---|---|---|---|
| Atchakpa reservoir | 05 | MT180099 (1) MT180100 (1) MT180101 (1) MT180102 (2) | ATCH2 ATCH3 ATCH4 ATCH5 and ATCHF-H01 | Benin | This study | |
| Agonlin-Lowé (Ouémé River) | 04 | MT180103 (1) MT180106 (3) | FOUE3 FOUE2, FOUE4F-H03 and FOUE5F-G03 | Benin | Native | This study |
| Sô-Ava (Sô River) | 01 | MT180104 (1) | RISO1 | Benin | Native | This study |
| Porto-Novo Lagoon | 04 | MT180102 (4) | LAPO2F-E03, LAPO1F-F03, LAPO3F-D03 and LAPO4F-C03 | Benin | Native | This study |
| Lake Nokoué | 03 | MT180107 (1) MT180108 (2) | LANO2 LANO3 and LANO1F-A03 | Benin | Native | This study |
| Manila Bay Lake | 04 | KM212014-KM212016; KM212018 | - | Philippines | Feral | [9] |
| Taal Lake | 05 | HQ654753-HQ654757 | - | Philippines | Feral | [18] |
| Laguna de Bay Lake | 05 | HQ682721-HQ682725 | - | Philippines | Feral | [29] |
| Not specified | 01 | MT666031 | - | Hawaii | Feral | Unpublished |
| Odooba River | 02 | KX231781-KX231782 | - | Nigeria | Native | [30] |
| Banc d’Arguin National Park (PNBA) | 16 | KJ938183-KJ938191; KJ938208- KJ938214 | - | Mauritania | Native | [31] |
| Guiers Lake | 04 | KJ938215-KJ938218 | - | Senegal | Native | [31] |
| State Zhongji Tilapia Farm, Hebei Province | 02 | NC_015611 JF894132 | - | China | Farmed | [32] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pèlèbè, R.O.E.; Imorou Toko, I.; Verheyen, E.; Van Steenberge, M. Molecular Identification of an Invasive Sarotherodon Species from the Atchakpa Freshwater Reservoir (Ouémé River Basin, Benin) and Comparison within S. melanotheron Using COI Markers. Diversity 2021, 13, 297. https://doi.org/10.3390/d13070297
Pèlèbè ROE, Imorou Toko I, Verheyen E, Van Steenberge M. Molecular Identification of an Invasive Sarotherodon Species from the Atchakpa Freshwater Reservoir (Ouémé River Basin, Benin) and Comparison within S. melanotheron Using COI Markers. Diversity. 2021; 13(7):297. https://doi.org/10.3390/d13070297
Chicago/Turabian StylePèlèbè, Rodrigue Orobiyi Edéya, Ibrahim Imorou Toko, Erik Verheyen, and Maarten Van Steenberge. 2021. "Molecular Identification of an Invasive Sarotherodon Species from the Atchakpa Freshwater Reservoir (Ouémé River Basin, Benin) and Comparison within S. melanotheron Using COI Markers" Diversity 13, no. 7: 297. https://doi.org/10.3390/d13070297
APA StylePèlèbè, R. O. E., Imorou Toko, I., Verheyen, E., & Van Steenberge, M. (2021). Molecular Identification of an Invasive Sarotherodon Species from the Atchakpa Freshwater Reservoir (Ouémé River Basin, Benin) and Comparison within S. melanotheron Using COI Markers. Diversity, 13(7), 297. https://doi.org/10.3390/d13070297






