Genetic Assignment Tests to Identify the Probable Geographic Origin of a Captive Specimen of Military Macaw (Ara militaris) in Mexico: Implications for Conservation
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Extraction and Genotyping
2.2. Data Analysis
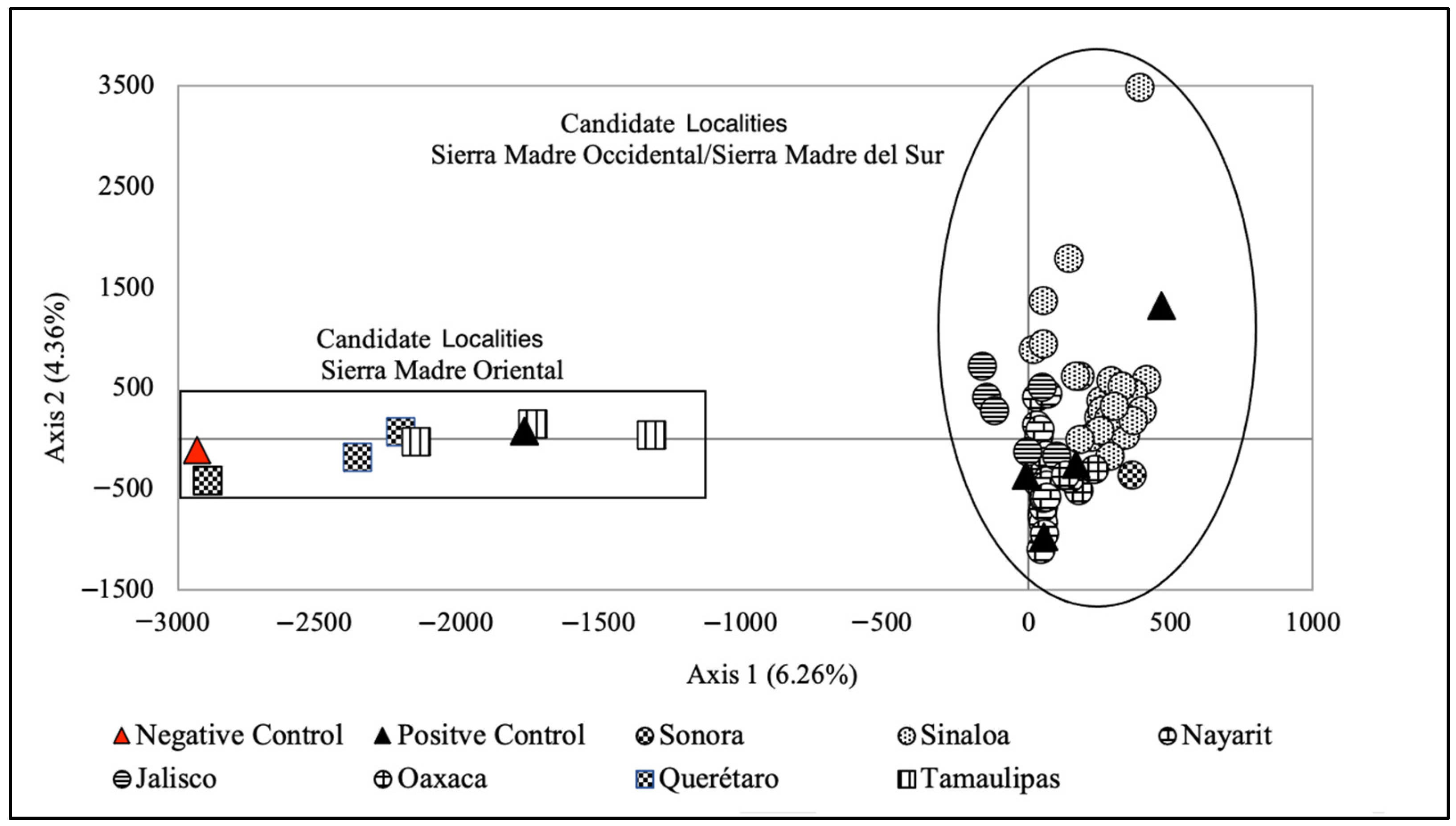
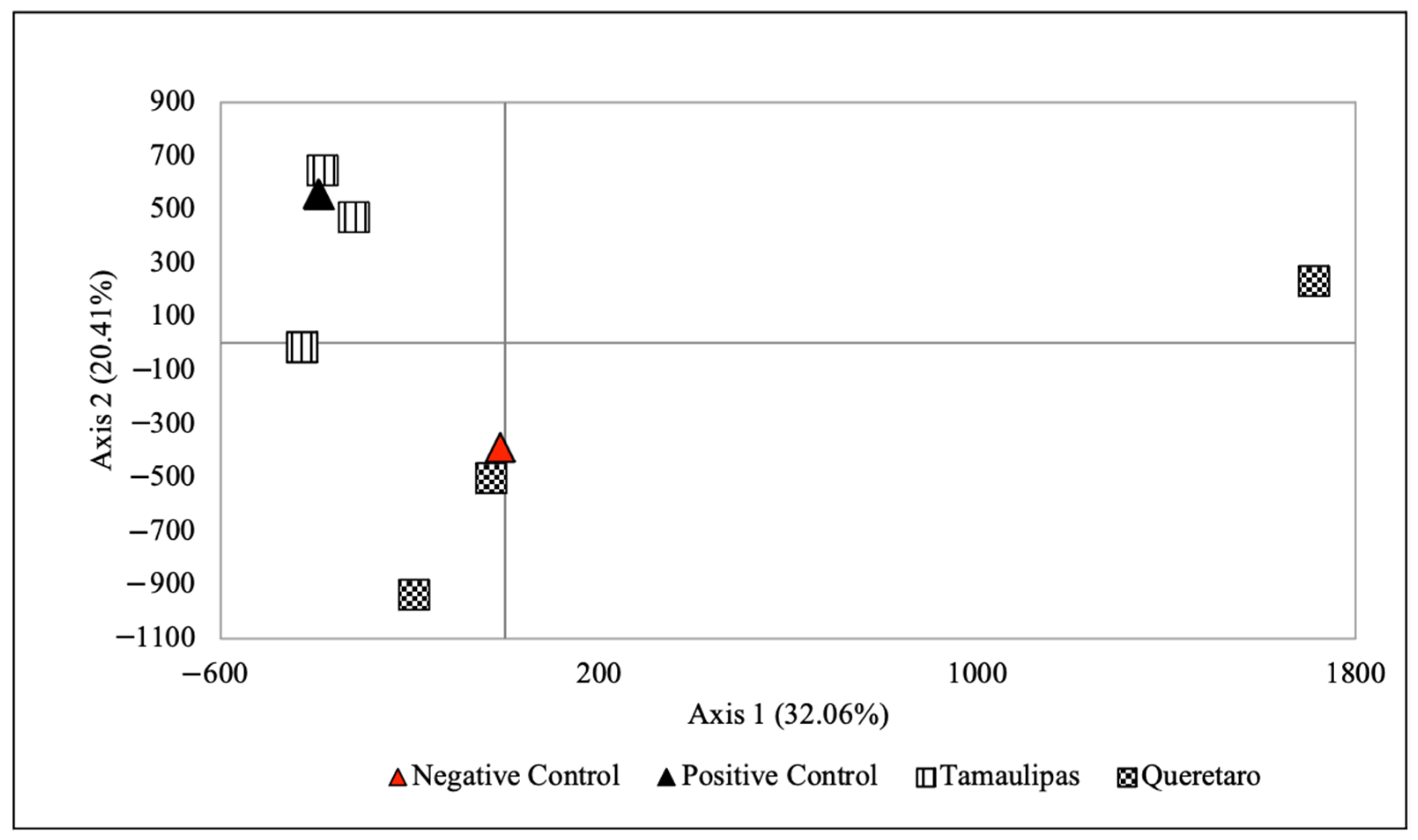
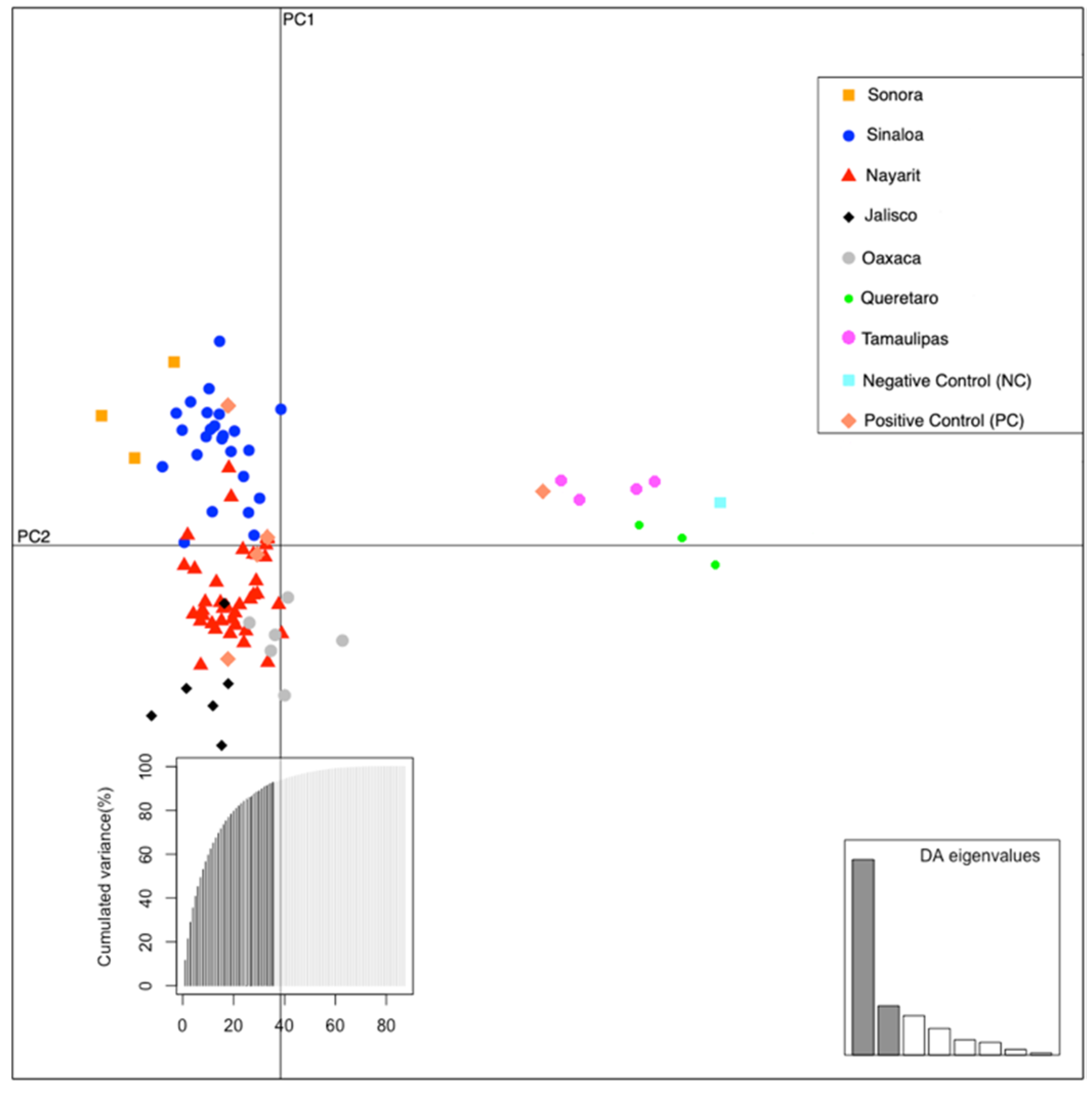
2.3. Factorial Correspondence Analysis (FCA)
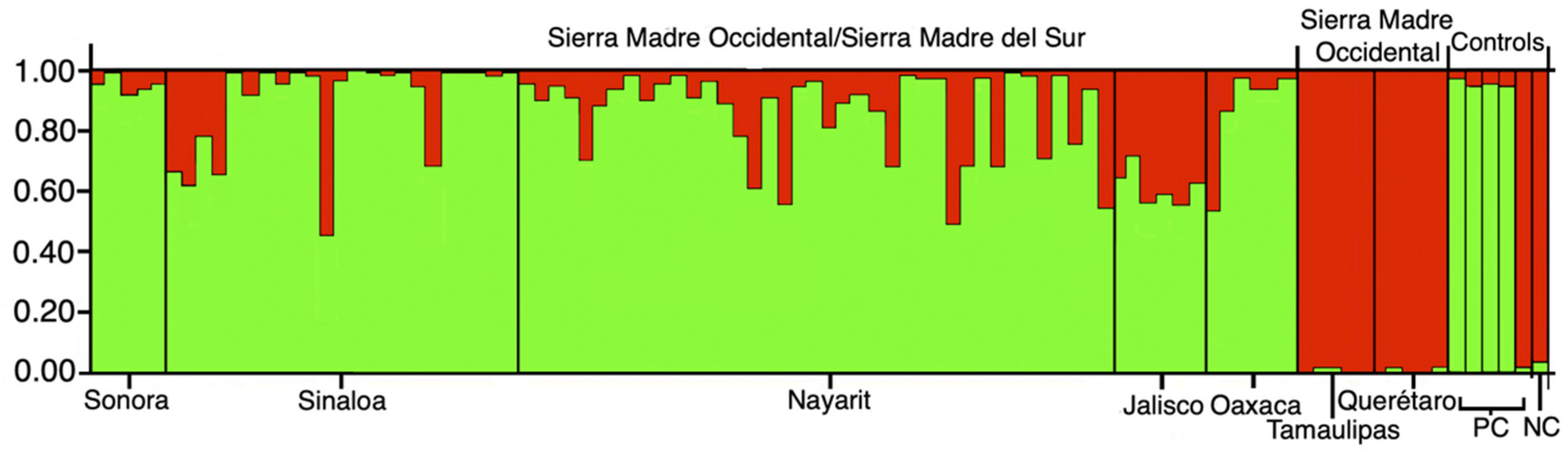
2.4. Genetic Assignment Analysis
3. Results
3.1. FCA
3.2. Genetic Assignment
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Juniper, T.; Parr, M. Parrots: A Guide to the Parrots of the World; Yale University Press: New Haven, CT, USA, 1998. [Google Scholar]
- Collar, N.J. Globally threatened parrots; criteria, characteristics and cures. Int. Zoo. Yearbk. 2000, 37, 21–35. [Google Scholar] [CrossRef]
- Berlanga, H.; Gómez de Silva, H.; Vargas-Canales, V.M.; Rodríguez-Contreras, V.; Sánchez-González, L.A.; Ortega-Álvarez, R.; Claderón-Parra, R. Aves de México: Lista Actualizada de Especies y Nombres Communes; CONABIO Press: Mexico City, Mexico, 2015. [Google Scholar]
- Norma Oficial Mexicana 059 SEMARNAT. Protección ambiental—Especies Nativas de México de Flora y Fauna Silvestres—Categorías de Riesgo y Especificaciones Para su Inclusión, Exclusión o Cambio. Lista de Especies en Riesgo. México. Available online: https://www.gob.mx/profepa/documentos/norma-oficial-mexicana-nom-059-semarnat-2010 (accessed on 13 December 2020).
- IUCN. Red List of Threatened Species. Available online: https://www.iucnredlist.org/ (accessed on 24 April 2020).
- Collar, N.J. Family Psittacidae (parrots). In Handbook of the Birds of the World; del Hoyo, J., Elliott, A., Sargatal, J., Eds.; Bird Life Internationa: Barcelona, Spain, 1997; pp. 280–477. [Google Scholar]
- Macías, C.C.; Iñigo-Elías, E.E.; Enkerlin-Hoeflich, E.C. Proyecto de Recuperación de Especies Prioritarias: Proyecto Nacional para la Conservación, Manejo y Aprovechamiento Sustentable de los Psitácidos de México; SEMARNAT Press: Mexico City, Mexico, 2000. [Google Scholar]
- Iñigo-Elias, E.; Ramos, M. The psittacine trade in Mexico. In Neotropical Wildlife Use and Conservation; Robinson, J.G., Redford, K.H., Eds.; University of Chicago: Chicago, IL, USA, 1991; pp. 380–392. [Google Scholar]
- Collar, N.J.; Juniper, A.T. Dimensions and causes of the parrot conservation crisis. In New World Parrots in Crisis: Solutions from Conservation Biology; Beissinger, S.R., Snyder, N.F.R., Eds.; Smithsonian Institution: Washington, DC, USA, 1992; pp. 1–24. [Google Scholar]
- Ríos-Muñoz, C.A.; Navarro-Sigüenza, A.G. Efectos del cambio de uso de suelo en la disponibilidad hipotética de hábitat para los psitácidos de México. Ornitol. Neotrop. 2009, 20, 491–509. [Google Scholar]
- Monterrubio-Rico, T.C.; de Labra-Hernández, M.A.; Ortega-Rodríguez, J.M.; Cancino-Murillo, R.; Villaseñor-Gómez, F. Distribución actual y potencial de la guacamaya verde en Michoacán, México. Rev. Mex. Biodivers. 2011, 82, 1311–1319. [Google Scholar] [CrossRef]
- Marín-Togo, M.C.; Monterrubio-Rico, T.C.; Renton, K.; Rubio-Rocha, Y.; Macías-Caballero, C.; Ortega-Rodríguez, J.M.; Cancino-Murillo, R. Reduced current distribution of Psittacidae on the Mexican Pacific coast: Potential impacts of habitat loss and capture for trade. Biodivers. Conserv. 2012, 21, 451–473. [Google Scholar] [CrossRef]
- Rivera- Ortíz, F.A.; Oyama, K.; Ríos-Muñoz, C.A.; Solórzano, S.; Navarro-Sigüenza, A.G.; Arizmendi, M.C. Habitat characterization and modeling of the potential distribution of the Military Macaw (Ara militaris) in Mexico. Rev. Mex. Biodivers. 2013, 84, 1200–1215. [Google Scholar] [CrossRef]
- Monterrubio-Rico, T.C.; Charre-Medellín, J.F.; Pacheco-Figueroa, C.; Arriaga-Weiss, S.; Valdez-Leal, J.D.; Cancino-Murillo, R.; Escalona-Segura, G.; Bonilla-Ruiz, C.; Rubio-Rocha, Y. Distribución potencial histórica y contemporánea de la familia Psittacidae en México. Rev. Mex. Biodivers. 2016, 87, 1103–1117. [Google Scholar] [CrossRef]
- Monterrubio-Rico, T.C.; Álvarez-Jara, M.; Téllez-García, L. Hábitat de anidación de Amazona oratrix (Psittaciformes: Psittacidae) en el Pacífico Central, México. Rev. Mex. Biodivers. 2014, 62, 1053–1072. [Google Scholar] [CrossRef]
- Cantú, J.C.; Sánchez, M.E.; Grosselet, M.; Silva, J. Tráfico Ilegal de Pericos en México: Una Evaluación Detallada; Defenders of Wildlife/Teyeliz, Press: Washington, DC, USA, 2007; pp. 3–74. [Google Scholar]
- SEMARNAT. Ley General del Equilibrio Ecológico y la Protección al Ambiente. México. Available online: https://biblioteca.semarnat.gob.mx/janium/Documentos/Ciga/agenda/DOFsr/148.pdf (accessed on 24 April 2020).
- Moya, H.; Peters, E.; Zamorano, P. La importancia de un enfoque regional para la conservación del hábitat natural en la frontera norte de México. In Temas Sobre la Conservación de Vertebrados Silvestres en México; Sánchez, O., Zamorano, P., Peters, E., Moya, H., Eds.; SEMARNAT: Mexico City, Mexico, 2011; pp. 1–67. [Google Scholar]
- Lynch, M. The genetic interpretation of inbreeding depression and outbreeding depression. Evolution 1991, 45, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Edmands, S. Between a rock and a hard place: Evaluating the relatives risks of inbreeding and outbreeding for conservation and management. Mol. Ecol. 2007, 16, 463–475. [Google Scholar] [CrossRef]
- Fernandes, G.A.; Caparroz, R. DNA sequence analysis to guide the release of blue-and-yellow macaws (Ara ararauna, Psittaciformes, Aves) from the illegal trade back into the wild. Mol. Biol. Rep. 2013, 40, 2757–2762. [Google Scholar] [CrossRef]
- Pertoldi, C.; Bijlsma, R.; Loeschke, V. Conservation genetics in a globally changing environment: Present problems, paradoxes and future challenges. Biodivers. Conserv. 2007, 16, 4147–4163. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. Introduction to Conservation Genetics; Cambridge University Press: Cambridge, UK, 2010; pp. 161–181. [Google Scholar]
- Allendorf, F.W.; Luikart, G.H.; Aitken, S.N. Conservation and the Genetics of Populations; Wiley-Blackwell Press: Oxford, UK, 2012; pp. 52–73. [Google Scholar]
- Qiu-Hong, W.; Hua, W.; Tsutomu, F.; Sheng-Guo, F. Which genetic marker for which conservation issue? Electrophoresis 2004, 25, 2165–2176. [Google Scholar] [CrossRef]
- Manel, S.; Berthier, P.; Luikart, G. Detecting Wildlife Poaching: Identifying the Origin of Individuals with Bayesian Assignment Test and Multilocus Genotypes. Conserv. Biol. 2002, 16, 650–659. [Google Scholar] [CrossRef]
- Burns, C.E.; Ciofi, C.; Beheregaray, L.B.; Fritts, T.H.; Gibbs, J.P.; Márquez, C.; Milinkovitch, M.C.; Powell, J.R.; Caccone, A. The origin of captive Galápagos tortoises based on DNA analysis: Implications for the management of natural populations. Anim. Conserv. 2003, 6, 329–337. [Google Scholar] [CrossRef]
- Presti, F.T.; Guedes, N.M.R.; Antas, P.T.Z.; Miyaki, C.Y. Population genetic structure in Hyacinth Macaws (Anodorhynchus hyacinthinus) and identification of the probable origin of confiscated individuals. J. Heredity 2015, 106, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Velo-Antón, G.; Godinho, R.; Ayres, C.; Ferrand, N.; Cordero-Rivera, A. Assignment tests applied to relocate individuals of unknown origin in a threatened species, the European pond turtle (Emys orbicularis). Amphibia-Reptilia 2007, 28, 475–484. [Google Scholar] [CrossRef]
- Iñigo, E.E. Las Guacamayas verde y escarlata en México. Biodiversitas 1999, 25, 7–11. [Google Scholar]
- Howell, S.N.G.; Webb, S. A Guide to the Birds of Mexico and Northern Central America; Oxford University Press: Oxford, UK, 1995; p. 337. [Google Scholar]
- Arizmendi, M.C.; Marquez-Valdelamar, L. Áreas de Importancia para la Conservación de las Aves en México; CIPAMEX AC Press: Mexico City, Mexico, 2000; pp. 7, 23, 57, 59, 76, 79, 127. [Google Scholar]
- CITES. Appendices I, II and II to the Convention on International Trade in Endangered Species of wild Fauna and Flora. USA. Available online: https://www.cites.org/ (accessed on 13 December 2020).
- Rivera-Ortíz, F.A.; Solórzano, S.; Arizmendi, M.C.; Dávila-Aranda, P.; Oyama, K. Genetic diversity and structure of the Military Macaw (Ara militaris) in Mexico: Implications for conservation. Trop. Conserv. Sci. 2017, 10, 1–12. [Google Scholar] [CrossRef]
- Leeton, P.; Christidis, L. Feathers from Museum Bird Skins—A good source of DNA for Phylogenetic Studies. Condor 1993, 95, 465–466. [Google Scholar] [CrossRef]
- Caparroz, R.; Miyaki, C.Y.; Baker, A.J. Characterization of microsatellite loci in Blue-and-Gold Macaw, Ara ararauna (Psittaciforme: Aves). Mol. Ecol. Notes 2003, 3, 441–443. [Google Scholar] [CrossRef]
- Russello, M.A.; Calcagnotto, D.; DeSalle, R.; Amato, G. Characterization of microsatellite loci in the endangered St. Vicent Parrot, Amazona guildingii. Mol. Ecol. Notes 2001, 1, 162–164. [Google Scholar] [CrossRef]
- Russello, M.A.; Lin, K.; Amato, G.; Caccone, A. Additional microsatellite loci for the endangered St. Vicent Parrot, Amazona guildingii. Conserv. Genet. 2005, 6, 643–645. [Google Scholar] [CrossRef]
- Morin, P.A.; Chambers, K.E.; Boesch, C.; Vigilant, L. Quantitative polymerase chain reaction analysis of DNA from noninvasive samples for accurate microsatellite genotyping of wild chimpanzees (Pantroglodytes verus). Mol. Ecol. 2001, 10, 1835–1844. [Google Scholar] [CrossRef]
- Garrido-Garduño, T.; Vázquez-Domínguez, E. Métodos de análisis genéticos, espaciales y de conectividad en genética del paisaje. Rev. Mex. Biodivers. 2013, 84, 1031–1054. [Google Scholar] [CrossRef]
- Banks, M.; Eichert, W.; Olsen, J.B. Which genetic loci have greater population assignment power? Bioinf. Appl. Note 2003, 19, 1436–1438. [Google Scholar] [CrossRef] [PubMed]
- Yue, G.H.; Xia, J.H.; Liu, P.; Liu, F.; Sun, F.; Lin, G. Tracing asian aeabass individuals to single fish farms using microsatellites. PLoS ONE 2012, 7, e52721. [Google Scholar] [CrossRef]
- Belkhir, K.; Borsa, P.; Chikhi, L.; Raufaste, N.; Bonhomme, F. GENETIX 4.05, Logiciel Sous Windows TM Pour la Génetique des Populations. Francia. Available online: https://kimura.univ-montp2.fr/genetix/ (accessed on 20 August 2020).
- Hair, J.F.; Anderson, R.E.; Tatham, R.L.; Black, W.C. Análisis Multivariante; Pearson Prentice Hall Press: Madrid, Spain, 1999; pp. 92–98. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J. Inference of population structure using multilocus genotypes data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Jakobsson, M.; & Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef]
- Rosenberg, N.A. DISTRUCT: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- R Core Team R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria; Available online: https://www.R-project.org/ (accessed on 3 April 2021).
- Piry, S.; Alapetite, A.; Cornuet, J.M.; Paetkau, D.; Baudouin, L.; Estoup, A. GENECLASS2: A Software for Genetic Assignment and First-Generation Migrant Detection. J. Heredity 2004, 95, 536–539. [Google Scholar] [CrossRef] [PubMed]
- Paetkau, D.; Calvert, W.; Stirling, I.; Strobeck, C. Microsatellite analysis of population structure in Canadian polar bears. Mol. Ecol. 1995, 4, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Paetkau, D.; Slade, R.; Burden, M.; Estoup, A. Genetic assignment methods for the direct, real-time estimation of migration rate: A simulation-based exploration o accuracy and power. Mol. Ecol. 2004, 13, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Rannala, B.; Mountain, J. Detecting immigration by using multilocus genotypes. Proc. Natl. Acad. Sci. USA 1997, 94, 9197–9201. [Google Scholar] [CrossRef] [PubMed]
- Cornuet, J.M.; Piry, S.; Luikart, G.; Estoup, A.; Solignac, M. New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics 1999, 153, 1989–2000. [Google Scholar] [CrossRef]
- Nei, M. The theory and estimation of genetic distance. In Genetic Structure of Population; Morton, N.E., Ed.; University Press: Honolulu, HI, USA, 1973; pp. 45–54. [Google Scholar]
- Nei, M.; Tajima, F.; Tateno, Y. Accuracy of estimated phylogenetic trees from molecular data. J. Mol. Evol. 1983, 9, 153–170. [Google Scholar] [CrossRef]
- Ogden, R.; Dawnay, N.; McEwing, R. Wildlife DNA forensics—Bridging the gap between conservation genetics and law enforcement. Endanger. Species Res. 2009, 9, 179–195. [Google Scholar] [CrossRef]
- Eberhard, J.R.; Iñigo-elias, E.; Enkerlin-Hoeflich, E.; Cun, E.P. Phylogeography of the Military Macaw (Ara militaris) and the Great Green Macaw (A. Ambiguus) Based on MTDNA Sequence Data. Wilson J. Ornithol. 2015, 127, 661–669. [Google Scholar] [CrossRef]
- Hauser, L.; Seamons, T.R.; Dauer, M.; Naish, K.A.; Quinn, T.P. An empirical verification of population assignment methods by marking and parentage data: Hatchery and wild steelhead (Oncorhynchus mikiss) in Forks Creek, Washington, USA. Mol. Ecol. 2006, 15, 3157–3173. [Google Scholar] [CrossRef]
- Ogden, R.; Linacre, A. Wildlife forensic science: A review of genetic geographic origin assignment. Forensic Sci. Int. Genet. 2015, 18, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Palomera-García, C.; Santana, E.; Amparán-Salido, R. Patrones de distribución de la avifauna en tres estados del occidente de México. An. Inst. Biol. 1994, 5, 137–175. [Google Scholar]
- Gaucín, R.N. Biología de la Conservación de la Guacamaya Verde (Ara militaris) en el Sótano del Barro, Querétaro. Master’s Thesis, Universidad Autónoma de Querétaro, Santiago de Querétaro, Mexico, March 2000. [Google Scholar]
- Honnen, A.C.; Petersen, B.; Kabler, L.; Elmeros, M.; Ross, A.; Sommer, R.S.; Zachos, F.E. Genetic structure of Eurasian otter (Lutra lutra, Carnivora: Mustelidae) populations from the western Baltic sea region and its implications for the recolonization of north-western Germany. J. Zool. Syst. Evol. Res. 2011, 49, 169–175. [Google Scholar] [CrossRef]
- García-Navas, V.; Ferrer, E.S.; Sanz, J.J.; Ortego, J. The role of immigration and local adaptation on fine-scale genotypic and phenotypic population divergence in a less mobile passerine. J. Evol. Biol. 2014, 27, 1590–1603. [Google Scholar] [CrossRef] [PubMed]
- Mateus, J.C.; Russo-Almeida, P.A. Traceability of 9 Portuguese cattle breed with PDO products in the market using microsatellites. Food Control 2015, 47, 487–492. [Google Scholar] [CrossRef]
- Tonteri, A.; Je, A.; Zubchenko, A.V.; Lumme, J.; Primmer, C.R. Microsatellites reveal clear genetic boundaries among Atlantic salmon (Salmo salar) populations from the Barents and White seas, northwest Russia. Can. J. Fish. Aquat. Sci. 2009, 66, 717–735. [Google Scholar] [CrossRef]
- Wasser, K.S.; Clark, W.J.; Drori, O.; Kisamo, E.S.; Mailand, C.; Mutayoba, B.; Stephens, M. Combating the illegal trade in African Elephant Ivory whit DNA forensics. Conserv. Biol. 2008, 22, 1065–1071. [Google Scholar] [CrossRef]
- Millions, D.G.; Swanson, B.J. An application of Manel’s model: Detecting Bobcat Poaching in Michigan. Wildl. Soc. Bull. 2006, 34, 150–155. [Google Scholar] [CrossRef]
- Gonclaves-Nazareno, A.; Sedrez dos Reis, M. Where did they come from? Genetic diversity and forensic investigation of the threatened palm species Butia eriospatha. Conserv. Gent. 2014, 15, 441–452. [Google Scholar] [CrossRef]
| Locus | Score | Score (%) | A (%) |
|---|---|---|---|
| UnaCT21 | 139.474 | 15.556 | 83% |
| UnaCT32 | 118.573 | 13.225 | |
| UnaCT74 | 116.432 | 12.986 | |
| UnaCT43 | 114.071 | 12.723 | |
| UnaGT55 | 110.096 | 12.279 | |
| AgGT17 | 109.722 | 12.237 | |
| AgGT19 | 98.694 | 11.007 | |
| AgGT32 | 89.51 | 9.983 |
| Run | Genetic Group 1 | Genetic Group 2 |
|---|---|---|
| Assignment Percentages | ||
| 1 | 2.9 | 97.1 |
| 2 | 0.4 | 99.6 |
| 3 | 97.2 | 92.8 |
| 4 | 97.2 | 92.8 |
| 5 | 2.7 | 98.3 |
| 6 | 1.6 | 98.4 |
| 7 | 0.8 | 99.2 |
| 8 | 97.3 | 90.7 |
| 9 | 0.5 | 99.5 |
| 10 | 1.8 | 98.2 |
| Analysis Type | |||||
|---|---|---|---|---|---|
| Genetic Groups | Controls | Frequencies | Bayesian | Nei’s Genetic Distance (1983) | Minimal Nei Distance (1973) |
| Sierra Madre Occidental/Sierra Madre del Sur | Sinaloa | 0.756 * | 0.705 * | 0.771 * | 0.816 * |
| Nayarit (Ind. 1) | 0.745 * | 0.723 * | 0.700 * | 0.801 * | |
| Nayarit (Ind. 2) | 0.742 * | 0.737 * | 0.715 * | 0.810 * | |
| Oaxaca | 0.778 * | 0.756 * | 0.789 * | 0.836 * | |
| Sierra Madre Oriental | Tamaulipas | 0.764 * | 0.758 * | 0.764 * | 0.799 * |
| Unknown individual | 0.712 * | 0.748 * | 0.79 * | 0.824 * | |
| Analysis Type | ||||
|---|---|---|---|---|
| Candidate Populations | Frequencies | Bayesian | Genetic Distance of Nei (1983) | Minimal Distance of Nei (1973) |
| Sonora | 0.00 | 0.00 | 0.00 | 0.00 |
| Sinaloa | 0.00 | 0.00 | 0.00 | 0.00 |
| Nayarit (Ind. 1) | 0.00 | 0.00 | 0.00 | 0.00 |
| Nayarit (Ind. 2) | 0.00 | 0.00 | 0.00 | 0.00 |
| Jalisco | 0.00 | 0.00 | 0.00 | 0.00 |
| Oaxaca | 0.00 | 0.00 | 0.00 | 0.00 |
| Querétaro | 0.303 * | 0.327 * | 0.373 * | 0.461 * |
| Tamaulipas | 0.00 | 0.00 | 0.224 * | 0.418 * |
| Analysis Type | ||||
|---|---|---|---|---|
| Candidate Populations | Positive Controls (• = TAM, + = SIN, ♢ = NAY and ▸ = Oax) | |||
| Frequencies | Bayesian | Genetic Distance of Nei (1983) | Minimal Distance of Nei (1973) | |
| Sonora | 0.00 | 0.00 | 0.00 | 0.00 |
| Sinaloa | 0.306 +* | 0.298 +* | 0.471 +* | 0.501 +* |
| Nayarit (Ind. 1) | 0.349 ♢* | 0.376 ♢* | 0.401 ♢* | 0.550 ♢* |
| Nayarit (Ind. 2) | 0.449 ♢* | 0.376 ♢* | 0.401 ♢* | 0.505 ♢* |
| Jalisco | 0.00 | 0.00 | 0.00 | 0.00 |
| Oaxaca | 0.408 ▸* | 0.378 ▸* | 0.439 ▸* | 0.451 ▸* |
| Querétaro | 0.00 | 0.00 | 0.00 | 0.00 |
| Tamaulipas | 0.35.7 •* | 0.310 •* | 0.309 •* | 0.457 •* |
| The Criterion of the Algorithm | Number of Individuals | Percentage (%) |
|---|---|---|
| Frequencies | 67 | 77.9 |
| Bayesian | 74 | 86.0 |
| Nei’s Genetic distance (1983) | 78 | 90.6 |
| Minimal Nei distance (1973) | 56 | 65.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rivera-Ortíz, F.A.; Juan-Espinosa, J.; Solórzano, S.; Contreras-González, A.M.; Arizmendi, M.d.C. Genetic Assignment Tests to Identify the Probable Geographic Origin of a Captive Specimen of Military Macaw (Ara militaris) in Mexico: Implications for Conservation. Diversity 2021, 13, 245. https://doi.org/10.3390/d13060245
Rivera-Ortíz FA, Juan-Espinosa J, Solórzano S, Contreras-González AM, Arizmendi MdC. Genetic Assignment Tests to Identify the Probable Geographic Origin of a Captive Specimen of Military Macaw (Ara militaris) in Mexico: Implications for Conservation. Diversity. 2021; 13(6):245. https://doi.org/10.3390/d13060245
Chicago/Turabian StyleRivera-Ortíz, Francisco A., Jessica Juan-Espinosa, Sofía Solórzano, Ana M. Contreras-González, and María del C. Arizmendi. 2021. "Genetic Assignment Tests to Identify the Probable Geographic Origin of a Captive Specimen of Military Macaw (Ara militaris) in Mexico: Implications for Conservation" Diversity 13, no. 6: 245. https://doi.org/10.3390/d13060245
APA StyleRivera-Ortíz, F. A., Juan-Espinosa, J., Solórzano, S., Contreras-González, A. M., & Arizmendi, M. d. C. (2021). Genetic Assignment Tests to Identify the Probable Geographic Origin of a Captive Specimen of Military Macaw (Ara militaris) in Mexico: Implications for Conservation. Diversity, 13(6), 245. https://doi.org/10.3390/d13060245






