Abstract
In this study, we used a metagenomic approach to analyze microbial communities, antibiotic resistance gene diversity, and human pathogenic bacterium composition in two typical landfills in China. Results showed that the phyla Proteobacteria, Bacteroidetes, and Actinobacteria were predominant in the two landfills, and archaea and fungi were also detected. The genera Methanoculleus, Lysobacter, and Pseudomonas were predominantly present in all samples. sul2, sul1, tetX, and adeF were the four most abundant antibiotic resistance genes. Sixty-nine bacterial pathogens were identified from the two landfills, with Klebsiella pneumoniae, Bordetella pertussis, Pseudomonas aeruginosa, and Bacillus cereus as the major pathogenic microorganisms, indicating the existence of potential environmental risk in landfills. In addition, KEGG pathway analysis indicated the presence of antibiotic resistance genes typically associated with human antibiotic resistance bacterial strains. These results provide insights into the risk of pathogens in landfills, which is important for controlling the potential secondary transmission of pathogens and reducing workers’ health risk during landfill excavation.
1. Introduction
Municipal landfills have a high diversity of microorganisms and can be considered as bioreactors for producing biogas through biological degradation of municipal waste [1]. Because of their relatively lower operation cost, larger treatment scale, and eco-friendly biogas resource utilization, sanitary landfilling is one of the primary methods of municipal solid waste disposal in China. The output of urban waste in the country has increased over the years, and the amount of domestic waste generated in more than 196 big and medium-sized cities had reached 235.6 million tons by the end of 2019 [2]. Over 50% of total municipal solid waste is treated in nearly 2600 sanitary landfills nationwide [3], and some of them are operating at full capacity; however, building new landfills is troubled by scarce land resources, especially in areas with rapid urbanization. The imbalance between waste production and treatment capacity is intensifying the predicament of garbage siege. On the other hand, due to historical factors, there are many non-sanitary landfills in China, and the irregular waste disposal from these landfills has caused severe pollution threats to soil and groundwater [4,5], as well as the risk on surrounding public health [6].
Under increasing pressure caused by the imbalance between waste production and treatment capacity in urban waste disposal, the government has been making an effort to recycle the old landfills in China. Landfill excavation/mining is a typical way to recycle material from landfilled refuse, which has been implemented in Europe and the USA in the past era [7]. In brief, landfill excavation/mining is to excavate the old landfilled refuse, sieve out the available matter, recycle it properly, such as composting or mulch, and the unusable part of the refuse is usually incinerated [8]. The basic idea is to recover the recyclable material from the landfilled refuse, and reuse the landfill volume [7]. Conceivably, the demolishment of the old landfill has accompanied some controversies, of which the most critical issue by far is the considerable risk of spreading pathogen microbes.
Landfills differ from each other due to the different refuse sources and local environments, which significantly impact the distribution of microbial communities [1]. Municipal solid waste might contain pathogens that may infect people when the landfilled refuse is being stirred and excavated [9]. We cannot ignore the fact that during the exploitation process, potential pathogens may come in contact with the human body, particularly through dermal contact or aerosol inhalation which might pose a potential occupational health and safety risk to the workers [10]. Though landfills’ environmental health risk is an important topic facing human activities, detailed research on landfill pathogens and their risk assessment are minimal.
Suzhou and Wuhan are both new first-tier cities in China. Urban growth brings a rapid substantial increase in waste production and leads to the proximity of expanding residential areas to landfills. Hence, there is a growing call for waste resource utilization by ectopic disposal in these two cities. In this study, two typical landfills were selected. The landfill in Suzhou, like most non-sanitary landfills, can be seen as an open storage ground for surrounding urban waste without environmental pollution control measures. On the contrary, the Wuhan landfill is a standard sanitary landfill, where urban waste is covered by soil and industrial film, covering effective pollution treatment measures such as the liner system, landfill gas conductive emission system, leachate collection, etc. Samples were collected at different depths considering the differences in waste stability. Metagenomic analysis was performed using the high-throughput Illumina HiSeq platform to investigate the microbial community composition, the abundance distribution and diversity of both antibiotic resistance genes (ARGs), and human pathogenic bacteria (HPBs) in landfills. This study aimed to provide a greater understanding of the pathogenic risk related to landfill excavation, which contributes to taking necessary measures to safely treat landfill refuse.
2. Materials and Methods
2.1. Sample Collection
All samples were from Suzhou (East China) and Wuhan (Central China) landfills (Figure 1). The Suzhou landfill (SZ) (31°60″ N 120°50″ E) is an open waste accumulation site located in Wujiang district and has been in use for less than one year. The Wuhan landfill (WH) (31°21″ N 114°12″ E) is a sanitary landfill, covered by high-density polyethylene film, located in Jiangxia district and has been in use for more than three years. Both represent typical landfill types in China. In total, we collected six samples, named SZ1, SZ2, and SZ3 (from SZ), and WH1, WH2, and WH3 (from WH), indicating the waste at vertical depths of 1.0, 2.0, and 3.0 m under the surface layer, respectively. In the SZ landfill, five sampling points were opted for on a relative flat slope of waste body, including a central point and four sub-points located 2.0 m around the central collection point. In the WH landfill, the soil samples were collected from a landfill cell using the same sampling method as that of SZ. For each sample, soils from five sampling points were gathered and mixed; then, we removed large size waste through a 10.0 mm screen, and obtained the final soil sample by the quartering method. Soil samples were collected at each landfill employing an agricultural soil sampler. To avoid cross contaminations, we cleaned both the soil sampler and the screen before excavating a new point. All soil samples were stored at −20 °C.

Figure 1.
The geographical location of two sampling landfills. Suzhou landfill (SZ) and Wuhan landfill (WH) in China.
2.2. Illumina Sequencing
DNA was extracted from a 2.0 g sample using the DNeasy PowerSoil kit (QIAGEN, Hilden, Germany). The DNA purity and degradation degree were evaluated by 1% agarose gel electrophoresis, and the DNA concentration was determined using the Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA). Qualified DNA samples were randomly fragmented to 350 bp by Ultrasonicator (Covaris, Woburn, MA, USA), then the DNA fragments were end-polished, A-tailed, and ligated with the Illumina sequencing adaptor for purification and Polymerase Chain Reaction (PCR) amplification. The whole libraries were initially quantified using the Qubit 2.0, the insert size was analyzed using the Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA) and quantified again by Quantitative Real-Time PCR (effective library concentration >3 nM). The clustering of the indexed samples was performed on a cBot Cluster Generation System according to the manufacturer’s instructions. After cluster generation, the libraries were sequenced on an Illumina HiSeq platform at Novogene Bioinformatics Technology Co., Ltd. (Tianjin, China).
2.3. Data Analysis
Readfq (V8) was used to preprocess the raw data from the sequencing platform to obtain the clean data for subsequent analyses. The clean data were analyzed using MEGAHIT (V1.0.4-beta, parameters used: --presets meta-large --min-count 2 --k-min 27 --k-max 87 --k-step 10) [11]. The obtained scaftigs with a minimum length of 500 bp were subjected to open reading frame (ORF) prediction using MetaGeneMark (V2.10) [12], following which ORFs with less than 100 nucleotides were removed. The CD-HIT software (V4.5.8) was used to obtain a non-redundant gene catalogue from the predicted ORFs (clustered with 95% identity and 90% coverage), and the most extended sequence as the representative sequence was selected [13,14]. Next, the clean sequence data were aligned against the gene catalogue using Bowtie2 (V2.2.4; parameters used [15]: --end-to-end --sensitive -I 200 -X 400). A curated gene catalogue (unigenes) was subsequently produced after discarding genes which were covered by less than two reads in each sample. To generate the gene abundance table, we calculated the relative abundance of each gene in each sample using the formula as described in [16].
The obtained unigenes were aligned with bacterial, fungal, archaeal, and viral sequences, which were extracted from NCBI’s NR database (V20180102) using DIAMOND (V0.9.9.110; parameters used [17]: blastp, 1e-5). Then, the LCA (lowest common ancestor) algorithm was used for taxonomic classification of each gene [18,19,20]. For functional annotation, we used DIAMOND software to blast unigenes to the KEGG database (V20180101; parameters used: blastp, 1e-5), and the best blast hit was selected for subsequent analysis [15,21,22]. To analyze the relative abundance of each ARO (antibiotic resistance ontology), we used the RGI (Resistance Gene Identifier) software to align our unigenes against the CARD database [23,24,25]. To identify the correlation between AROs and MGEs (mobile genetic elements) in our metagenomic data, we used Blastall (parameters used [26]: -m 8 -a 4 -e 1e-30 -b 10) to align our scaftigs, which were annotated as ARO sequences; again, searching in databases of NCBI Plasmids, NCBI NR, and Isfinder. The genomic database of each HPB in our samples was obtained using the HPB database [27], and the HPB information extracted from the databases could be found in NCBI GenBank (V20180102). Sequencing data have been submitted to the NCBI’s Sequence Read Archive (SRA) under accession number PRJNA718480.
3. Results and Discussion
3.1. Sequence Data
In this study, a total of 1,550,555 scaftigs were obtained by Illumina HiSeq sequencing with a total length of 1,660,639,666 bp in six samples, an average length of 1071 bp, a maximum length of 280,023 bp, and an N50 of 1121.83 bp. Table 1 provides detailed information on the assembled data.

Table 1.
Summary of the sequence data for six samples.
3.2. Microbial Taxonomy Composition at the Phylum Level
Refuse sources and local environments have a significant impact on the distribution of microbial communities in landfills. In this study, bacteria, archaea, and fungi were found in all six samples from both SZ and WH, demonstrating the landfills’ microbial diversity (Figure 2a). One hundred and fifty-two phyla were identified in the six samples. The most dominant prokaryotic phyla in SZ were Proteobacteria (67.59%), Actinobacteria (12.06%), Bacteroidetes (5.98%), Euryarchaeota (4.01%), and Firmicutes (3.46%), and the others were less than 1% of the total bacterial community. By contrast, phyla Proteobacteria (27.94%), Bacteroidetes (22.83%), Actinobacteria (16.83%), Euryarchaeota (10.43%), Deinococcus-Thermus (4.49%), and Firmicutes (2.55%) were dominant in WH. Overall, the two landfills were dominated by Proteobacteria, Bacteroidetes, and Actinobacteria. In addition, Ascomycota was identified in all samples. The overall microbial composition at the phylum level was relatively similar for different sampling depths in the same landfill (Figure 2b).
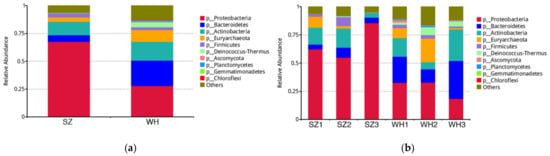
Figure 2.
Relative abundances at the phylum level: (a) Top 10 relative abundances in both landfills at the phylum level. SZ, Suzhou landfill; WH, Wuhan landfill. (b) Top 10 relative abundances in each sample at the phylum level. SZ1, SZ2, SZ3 from the Suzhou landfill; WH1, WH2, WH3 from the Wuhan landfill.
Proteobacteria was the most abundant group in both landfill samples, and was overwhelmingly abundant in the SZ landfill (67.59%) (Figure 2a). A high abundance of Proteobacteria was previously found in landfills [28,29,30], which could play a vital role in organic matter degradation [28]. For instance, Zainun et al. [29] found that Proteobacteria were predominant in both active and closed non-sanitary landfills, and within this phylum, Gammaproteobacteria were the most abundant class detected in the active landfill, indicating their potential role in the decomposition of organic matter. Besides, it was reported that the order Pseudomonadales, from Gammaproteobacteria, are associated with pollutant removal and have been found to be dominant bacteria in aged landfill leachate [31] further suggesting that the prominent function of Pseudomonadales spp. In degrading organic substances.
In this study, the relative abundance of Bacteroidetes was higher in WH (22.83%) than in SZ (5.98%). On the contrary, Proteobacteria was evidently lower in WH (27.94%) than in SZ (67.59%) (Figure 2a). Since samples from the Wuhan landfill had a longer buried age, the difference in the predominant bacterial profiles of the two landfills corresponds to the scenario observed by KöChling et al., in which Bacteroidetes replaced Proteobacteria as the most dominant bacteria in older landfills [32].
There was a low abundance of Deinococcus-Thermus in SZ (0.22%), which was however enriched in WH (4.49%) (Figure 2b). The different richness of Deinococcus-Thermus between SZ and WH is believed to be attributed to the different content of refractory organics in landfills [33].
Euryarchaeota is a strictly anaerobic methanogen that belongs to the archaea. Methanogenesis is the final stage of waste degradation in the landfill stabilization process [34], and all methanogens detected in landfill samples so far belong to Euryarchaeota [1]. Thus, the high abundance of Euryarchaeota (in WH 10.43% and in SZ 4.01%) indicates that methanogenesis exists in both landfills. What should be of concern is that the Wuhan landfill has excellent facilities for landfill gas controlment. In contrast, the Suzhou landfill, a non-sanitary landfill lacking a gas discharge guiding system, which makes it difficult to vent combustible landfill gas within the waste body, poses a potential threat to the safety of the landfill operation environment.
Ascomycota fungi were found in two landfills with relative abundances of 0.08% (SZ) and 0.97% (WH), respectively. It is worth mentioning that few studies have reported the presence of fungal populations in municipal landfills, probably as bacteria and archaea have the ability to metabolize a broader range of organic and inorganic substrates, and possess functional dominance over fungi [1]. Since few studies have detected fungi in landfill samples using culture-dependent techniques [1], and since culture-independent techniques capture all intact DNA regardless of whether it comes from dead, living, or un-degraded DNA, our results need to be further confirmed.
3.3. Microbial Taxonomy Composition at the Genus Level
At the genus level, 2451 genera were observed in all samples. Dominant genera presented in >1% of the total bacterial community in SZ were Brevundimonas (9.33%), Lysobacter (8.45%), Thiobacillus (2.82%), Pseudomonas (2.43%), Psychrobacter (2.36%), Methanofollis (1.63%), Microbacterium (1.38%), and Methanoculleus (1.02%); whereas WH samples were dominated by Methanoculleus (8.97%), Microbacterium (7.00%), Luteimonas (3.57%), Truepera (3.07%), Lysobacter (1.81%), and Pseudomonas (1.13%). From the results, the microbial genus composition differed significantly between the two landfill samples (Figure 3a). Besides, there were some unique microbial compositions from different depths of the same landfill. High richness of Brevundimonas (17.78%) was identified in the shallow layer SZ1, then gradually decreased in the middle SZ2 (5.89%) and deep SZ3 (4.34%); while in WH, relative abundance of Brevundimonas was <1% in all samples, which may be related to the aerobiosis of these microorganisms (Figure 3b).
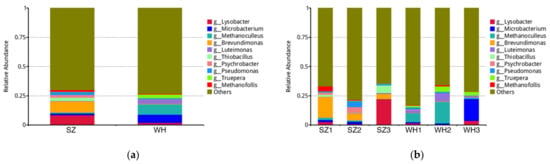
Figure 3.
Relative abundances at the genus level: (a) Top 10 relative abundances in both landfills at the genus level. (b) Top 10 relative abundances in each sample at the genus level.
Dominant genera identified in all the samples were Methanoculleus, Lysobacter, and Pseudomonas. Methanoculleus is a hydrogenotrophic methanogen that reacts with H2 and CO2 to convert acetate to methane [35], described as major players in landfills and other anaerobic digestion systems [34]. Methanoculleus was the richest genus in WH, but low in SZ in comparison. Owing to the fact that methanogenesis is the final activity in landfill waste decomposition [34], we speculate that the Wuhan landfill is in a more stable state overall. It has been reported that Lysobacter was detected in various soils, river sludges, along with old leachate [36,37,38]. Lysobacter has a strong proteolytic ability to lyse various microorganisms, such as Gram-negative bacteria, Gram-positive bacteria (including actinomycetes), cyanobacteria, fungi, green algae, and nematodes [39]. A variety of compounds with potent antibacterial or antifungal activity have been isolated from Lysobacter, suggesting that this bacterial group is a potential source of novel antibiotics [40]. In addition, Pseudomonas had a similar abundance in both landfills, which is known as organic matter degrading bacteria [41] with the potential to accelerate denitrification [42]. Some Pseudomonas cause disease in diverse host organisms, including the opportunistic pathogen, P. aeruginosa, and the plant pathogen, P. syringae. The pathogenicity of these bacteria has received widespread attention [43].
Brevundimonas is a group of aerobic gram-negative bacteria [44] which contains some emerging, global opportunistic pathogens [45]. We observed that Brevundimonas appeared as the most abundant genus in SZ (9.33%) and was enriched in the shallow layer SZ1 (17.78%), but a lower count in WH (0.35%). Considering the potentially invasive infections caused by Brevundimonas, the Suzhou landfill may have a higher risk of exposure to potentially infectious pathogens during the excavation/mining of landfilled refuse, necessitating appropriate protective measures. Thiobacillus was moderately abundant in the SZ samples (2.82%), on the contrary, barely found in WH (0.01%). Thiobacillus is a group of sulfur-oxidizing bacteria that can oxidize sulfides to sulfates [46]. Thiobacillus’ richness in SZ deep samples (SZ3 6.55%) indicates that landfill gas such as H2S is generated within the waste body. Therefore, effective methods that control odor pollution during refuse disposal are needed.
The second most dominant genus in WH was Microbacterium, which was found in high abundance in the deep sample WH3 (18.72%), but low in the shallow WH1 (1.08%) and middle WH2 (1.21%). Microbacterium is reported to be the dominant genus in landfills [47], widely distributed in the environment and has recently been considered a rare pathogen in humans [48]. For instance, Lau et al. reported and described the first culture-documented central venous catheter-related Microbacterium bacteremia [49]. The low number of Microbacterium infections reported at present may be explained by the relatively weak pathogenicity, from which we hypothesize that the environmental risk due to Microbacterium in Wuhan landfills is relatively low.
Some Luteimonas (3.57%) and Truepera (3.07%) were identified in WH, but both with abundance below 0.5% in SZ. The genus Luteimonas from Xanthomonadaceae had been discovered in various environments such as compost generated from food waste [50], hexachlorocyclohexane (HCH)-contaminated dumpsite [51], and wastewater treatment plants [52], which indicates that these bacteria could have a versatile role in the soil ecosystem. Recently, Zhou et al. isolated a Luteimonas sp. as a chitinolytic bacterium from landfill soil [53], suggesting that members of this bacteria may have the ability to degrade insoluble macromolecular organic matter. A previous study showed that Truepera from Deinococcus-Thermus plays an essential role in cellulose degradation, acid production, and methane production [54], and the enrichment of Truepera was related to the high concentrations of refractory organics in the environment [33]. Thus, the difference of Truepera abundance between SZ and WH probably reflect the relative abundance of refractory organics or the different stability, as WH has experienced a more extended period of biodegradation, and the characteristic of landfilled refuse shifts slowly from promptly biodegradable towards refractory.
3.4. Gene Function Prediction of the Microbial Communities
Genes of the same function were categorized into six putative metabolic pathways by the KEGG database. The relative abundance of KEGG level 1 was similar in each sample (Figure 4a). The KEGG pathways in both landfills showed a similar abundance of genes that are involved in metabolism (amino acid metabolism, carbohydrate metabolism, energy metabolism, cofactors and vitamins metabolism, and nucleotide metabolism), genetic information processing (translation, and replication and repair), environmental information processing (membrane transport, and signal transduction), cellular processes (cellular community—prokaryotes), organismal systems (endocrine system, aging, environmental adaptation, nervous system, etc.), and human diseases (antimicrobial resistance, infectious bacterial diseases, cancers, endocrine and metabolic diseases, antineoplastic resistance, neurodegenerative diseases, cardiovascular diseases, infectious parasites, immune diseases, and drug dependence) (Figure 4b).
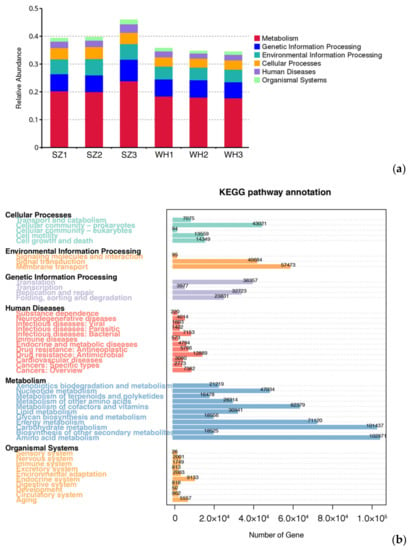
Figure 4.
Gene function prediction by KEGG: (a) Relative abundances of KEGG level 1 in each sample. (b) The number of genes annotated in KEGG level 2.
3.5. ARGs Diversity
Although naturally originated antibiotic resistance is widespread in natural environments, it is considered that the misuse of antibiotics could promote the acquisition and retention of resistance genes in indigenous microbes in human-impacted environments [55]. Therefore, ARGs are currently identified as environmental contaminants [56]. The main sources of ARGs include the natural evolution of environmental microorganisms and horizontal gene transfer between environmental microorganisms and pathogens. Most ARGs in the environments can transfer to HPB via mobile genetic elements (MGEs) such as transposons, integrons, and plasmids, resulting in drug resistance in pathogens [57]. During the exploitation of landfilled refuse, antibiotic-resistant bacteria in the soil usually diffuse into the air as bioaerosols; therefore, ARGs in the refuse can transfer to the environment and lead to a severe threat on human health. Consequently, detailed investigations on the abundance of ARGs in landfill soil can speculate the bioaerosol potential of ARGs, which is necessary for the environmental risk assessment of landfills and waste resource disposal.
A total of 376 ARG subtypes, belonging to 28 drug classes, were detected in the six samples. Among these, macrolide, aminoglycoside, cephalosporin, and tetracycline were the dominant classes in both landfills (Table S1). As shown in Figure 5a, the ARG abundance among the samples from both SZ and WH varied greatly, and the lowest was found in SZ1 and the highest in WH3, respectively. Overall, more ARGs were observed in WH. This result could reflect that WH has a longer landfill age, since the abundance of ARGs may be positively related to landfill age, and the older landfill sites are considered to serve as a reservoir for ARGs [58].
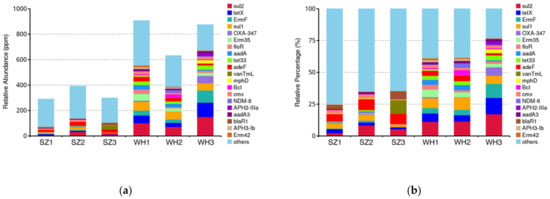
Figure 5.
Distribution of antibiotic resistance genes (ARGs) in each sample: (a) Relative abundance of the top 20 ARGs in each sample. (b) The relative percentage of the top 20 ARGs of the total abundances of ARGs in each sample.
The relative abundances of the top 20 ARG subtypes among all samples are shown in Figure 5b, accounting for 25.06–77.04% of the total ARG abundances. The resistance genes sul2, sul1, tetX, and adeF were the four most dominant subtypes in the six samples, and sul2 was the predominant ARG subtype in all samples, ranging from 2.37 (SZ1) to 17.19% (WH3) (Figure 5b). sul2 affiliates with the sulfonamide resistance genes, and a high abundance of sul2 was previously detected in leachate samples [59]. In the present study, Proteobacteria was the most likely host of ARGs in all samples (Figure 6). Similarly, sul1 is commonly present in leachate as well [60], and we found that sul1 co-occurs with Bacteroidetes in the WH landfill (Table 2). tetX is a class of tetracycline resistance genes, Deinococcus-Thermus and Actinobacteria were the possible main hosts of these genes in WH; while in SZ, they were detected in the phylum Proteobacteria (Table 2). adeF is a fluoroquinolone and tetracycline resistance gene, identified in the Proteobacteria in both landfills (Table 2).
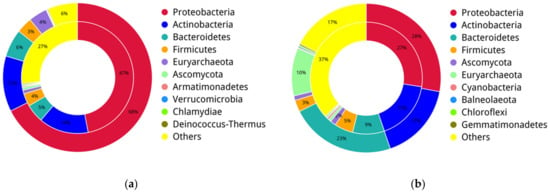
Figure 6.
Relationship between ARGs distribution with taxonomic compositions at the phylum level in SZ (a) and WH (b). The inner circle is the ARG distribution for each phylum. The outer circle is the taxonomic composition at the phylum level.

Table 2.
The co-occurrence between top 20 ARG subtypes and microbial taxa in both landfills.
ErmF, Erm35, and tet33 were the three dominant ARG subtypes in WH (Figure 5b). Both ErmF and Erm35 are multi-drug resistance genes, including macrolide, lincosamide and streptogramin, which were detected in Bacteroidetes, and tet33, a tetracycline resistance gene, mainly found in Actinobacteria (Table 2). In addition, the cephalosporin-penam resistance gene Bcl, was found in the phylum Proteobacteria of WH samples, but absent in that of SZ samples (Table 2). From our results, these detected antibiotic resistance genes were mainly distributed in Proteobacteria, Bacteroidetes, and Actinobacteria (Figure 6). Interestingly, tetX was present in the unique dominant phylum Deinococcus-Thermus in WH, while in SZ, it appeared in Proteobacteria (Table 2).
To further explore the risk of ARG dissemination in the landfills, we investigated the correlation between AGRs and MGEs. A total of 103 plasmids, 53 integrons, and 4 insertion sequences were identified as ARG carriers in the two landfills (Figure S1). In our research, we observed a higher diversity of both plasmids and integrons (plasmids 60; integrons 37) in WH as compared to SZ (plasmids 43; integrons 16). Of the insertion sequences, two (IS5564 and ISBbi1) were identified in SZ, and three (IS5564, IS6100, and IS401) were found in WH. These four different insertion sequences were originated from Bifidobacterium bifidum, Corynebacterium striatum, Mycobacterium fortuitum, and Burkholderia cepacia. Among the antibiotic resistance plasmids identified, the Ralstonia solanacearum CFBP2957 plasmid RCFBPv3_mp was most prevalent in SZ (Figure S2), which was correlated to 3 ARGs (lnuE, adeF and OXA-50). Escherichia coli ACN001 plasmid pACN001-A was most prevalent in WH (Figure S2), with correlation to four ARGs (AAC3-Iic, adeF, Mrx, and mphA). Of the integrons, Pseudomonas aeruginosa VRFPA04 was the most abundant in both SZ and WH (Figure S3), which was related to 18 ARGs.
A previous study has showed that MGEs had the greatest impact on ARG abundance in landfills compared with the concentration of antibiotics and heavy metals, as well as environmental factors [61]. In our study, MGEs conferring antibiotic resistance were more diverse in WH, which suggested that ARGs in this landfill have a higher possibility to be horizontally transferred among microbes, and it may lead to public health risks when they are ultimately transferred into pathogens.
3.6. HPB Species Diversity
Several studies have shown that municipal waste landfills are a source of potentially pathogenic bacteria for humans [10,62]. Understanding pathogens in landfilled refuse can help assess the possible pathogenic risks in the landfill environment and develop appropriate prevention measures [63]. In this study, a total of 69 different species of bacterial pathogens were identified from all samples using the HPB database, of which 64 pathogens were detected in SZ and 39 in WH, respectively. The composition of the top 20 HPB species in the two landfills was characterized in Figure 7a, and Klebsiella pneumoniae, Bordetella pertussis, Pseudomonas aeruginosa, and Bacillus cereus were the main composition of pathogenic bacteria in both landfills, followed by Staphylococcus aureus, Bartonella quintana, Brucella melitensis, Staphylococcus epidermidis, Enterococcus faecalis, Brucella suis, Bordetella bronchiseptica, and Mycobacterium tuberculosis. All these HPBs had a relative abundance of more than 1% of the total bacterial community. To further elucidate the differences among HPB distributions between SZ and WH, we obtained the biomarker species using liner discriminant analysis effect size (LefSe) analysis. The distribution diagram of HPB differences at all levels and the cladogram based on significantly different HPBs obtained by LefSe analysis [64] showed that the abundances of 15 HPBs were significantly enriched in the SZ landfill (liner discriminant analysis (LDA) score > 4). However, no significant bacterial pathogen was identified in the WH samples (Figure 8a). The cladogram indicated HPB species from the families Micrococcaceae, Moraxellaceae, and Pseudomonadaceae, the order Pseudomonadales, as well as the class Gammaproteobacteria were enriched in SZ (Figure 8b).
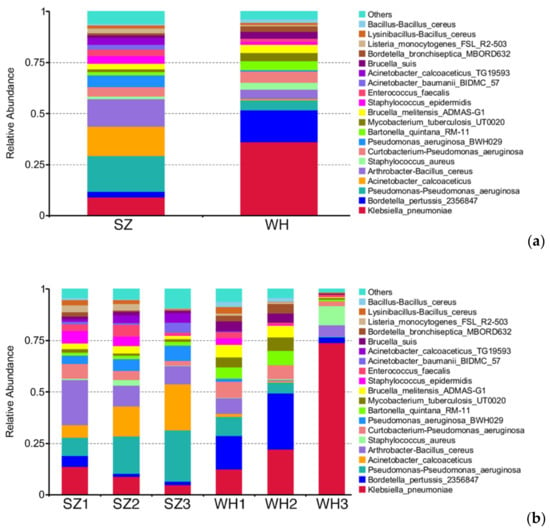
Figure 7.
Relative abundances of human pathogenic bacteria (HPB) at the species level: (a) Top 20 relative abundances of HPB in both landfills at the species level; (b) top 20 relative abundances of HPB in each sample at the species level.
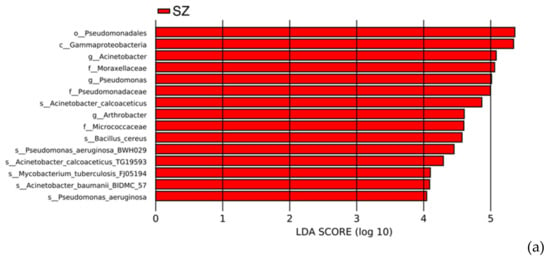
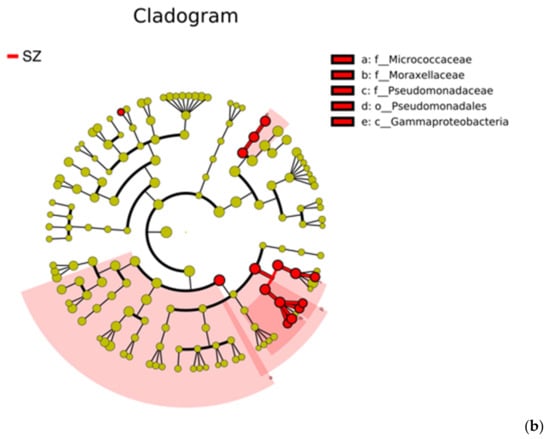
Figure 8.
LDA score diagram and cladograms by LEfSe: (a) The distribution of LDA score for differential HPB species (LDA > 4). Red bars represent the more abundant HPB species in SZ; (b) cladogram for the different HPB species from SZ. Circles radiating from inner to outer represent taxonomic levels from phylum to species. Yellow nodes represent the species without significant differences, and red nodes indicate the microbial taxa that play an essential role in SZ.
An abundant amount of the bacterial pathogens, K. pneumoniae and B. pertussis, were found in both landfills, and their relative abundance was higher in WH (37.02%) compared to that in SZ (9.38%) (Figure 7a). Klebsiella pneumoniae was the predominant pathogen in WH. This species is an opportunistic pathogen as well, which can enter the blood circulation or other tissues through the nasopharynx causing human infection [65]. Most K. pneumoniae are antibiotic-susceptible, but studies have found that a type of carbapenem-resistant hypervirulent K. pneumoniae may cause untreatable severe disease in the healthy population [66]. Given the high abundance of K. pneumonia in WH, hypervirulent strains are more likely to exist, posing potential health risks to workers.
A larger number of B. pertussis was detected in WH (15.49%) than in SZ (2.76%) (Figure 6a), which is known as a highly infectious kind of respiratory pathogen with hosts restricted to humans [67] and may be transmitted directly between hosts via aerosol droplets [68]. Despite the widespread use of effective vaccines, B. pertussis can still be transmitted asymptomatically via vaccinated individuals [69]. Since B. pertussis was the second dominant pathogen in WH (Figure 6a), the risk of infection with B. pertussis cannot be ignored during landfilled refuse excavation, especially its transmission among the relevant workers.
Pseudomonas aeruginosa was similarly dominant in both landfills (SZ 29.56% and WH 11.13%), which is a common opportunistic pathogen and a major cause for nosocomial infections. P. aeruginosa often infects the trachea and urinary tract, causes bloodstream infections, and is considered as a “superbug”, which is very difficult to eradicate due to its high resistance to multiple antibiotics [70].
Two members of the genus Acinetobacter, A. calcoaceticus and A. baumannii, were found in SZ (17.63% and 2.24%, respectively), while the relative abundance of these two pathogens in WH was each less than 1%. Particularly, A. baumannii was barely detected in WH, thus we consider these bacteria to be the characteristic pathogens of SZ. Both the bacteria are opportunistic pathogens that often appear clinically as A. calcoaceticus-A. baumannii complex [71], can cause zoonosis, with a strong environmental adaptability due to their characteristics of multi-drug resistance [72]. Previous studies have shown abundant numbers of zoonotic pathogens in landfills, and interactions between humans, animals, and microorganisms through landfills may be a factor contributing to public health risks [73]. Therefore, their presence in large numbers suggests that there could be more zoonotic pathogens present in the Suzhou landfill environment, prompting further concern for potential human or animal infections.
In SZ, its higher abundance of the opportunistic foodborne pathogen L. monocytogenes than that of WH, poses a severe threat to food safety (Figure 7a). Infection with such pathogens predisposes people to highly lethal listeriosis [66], and there were reports of community outbreaks of listeriosis caused by the consumption of unhygienic food [74].
We are usually more concerned with the risk of pathogenic bacteria in shallow waste, as the excavation of this part of refuse is the first step in landfill excavation/mining. According to the relative abundances of the top 20 HPB species in each sample (Figure 7b), we found two pathogens, B. cereus and S. epidermidis, were more dominant in shallow samples. Bacillus cereus is well adapted for growth in the intestinal flora of animals, which are commonly found in food production environments, and they are one of the main foodborne opportunistic pathogens [75]. This pathogenic bacterium was enriched in shallow layers from both landfills SZ and WH, and its high presence may be related to the undegraded food waste in the landfills. Besides, S. epidermidis was also highly abundant in the shallow layer samples from both landfills. This group of bacteria is commonly found in the human epidermis, mainly enriched in the axillae, head, and nares [76]. Although S. epidermidis infections rarely cause severe diseases, it is currently seen as an important opportunistic pathogen because of its high frequency and great difficulty to eradicate [77]. Therefore, prolonged and direct skin contact during landfill excavation/mining may cause chronic infection by S. epidermidis.
4. Conclusions
The municipal landfill is a particularly effective and prevalent tool for disposal of the urban garbage in China. However, it is not a once-and-for-all waste disposal, especially in some fast-growing cities, where the rapidly increasing output of waste puts severe pressure on local landfills. Nowadays, excavating/mining landfilled refuse is being implemented in China to release landfill volume and to recover recyclable material. It is necessary to evaluate the environmental sanitation risk to control and prevent the hazard of spreading pathogens during the excavating/mining of landfilled refuse. Our study used metagenomic analysis to investigate the microbial compositions, ARG diversity, and HPB risk in two typical municipal landfills in China. Potential human pathogen risk was identified in both landfills, and more types and numbers of pathogens were detected in the non-sanitary landfill. To excavate landfilled refuse hygienically, we suggest that processing the more stable and sanitary landfills first along with disinfecting the refuse before excavation would potentially minimize the potential risk of secondary pathogen transmission to workers. Protective measures for exposed skin are needed to prevent pathogen infection from inhalation or skin contact during excavation/mining of landfilled refuse.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/d13060230/s1, Figure S1: The numbers of MGEs in six samples, Figure S2: The abundance of integrons in six samples, Figure S3: The abundance of plasmids in six samples, Table S1: Numbers of ARG drug classes detected in six samples.
Author Contributions
Conceptualization, S.C., J.W., M.X., and S.W.; methodology, S.W. and J.T.; validation, S.W. and J.T.; investigation, S.W., Y.P., and J.W.; writing—original draft preparation, S.W. and M.X.; writing—review and editing, S.C., J.W., and F.Y.; supervision, S.C., J.W., and F.Y.; funding acquisition, S.C., J.W., and M.X. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key R & D Program of China (2018YFC1901402 and 2018YFC1901404).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Sequence data in this study have been deposited in the NCBI sequence reads archive (SRA) with accession number PRJNA718480.
Acknowledgments
We would like to thank Qiang Wang from Nanjing University, for grammatical editing of this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sekhohola-Dlamini, L.; Tekere, M. Microbiology of municipal solid waste landfills: A review of microbial dynamics and ecological influences in waste bioprocessing. Biodegradation 2020, 31, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Ecology and Environment of the People’s Republic of China. Statical Yearbook of Prevention and Control of Environmental Pollution by Solid Waste in Big and Medium-Sized Cities; Ministry of Ecology and Environment of the People’s Republic of China: Beijing, China, 2020.
- Ministry of Housing and Urban-Rural Development of the People’s Republic of China (MOHURD). China Urban Rural Construction Statistical Yearbook; Ministry of Housing and Urban-Rural Development of the People’s Republic of China (MOHURD): Beijing, China, 2020.
- Baziene, K.; Tetsman, I.; Albrektiene, R. Level of Pollution on Surrounding Environment from Landfill Aftercare. Int. J. Environ. Res. Public Health 2020, 17, 2007. [Google Scholar] [CrossRef] [PubMed]
- Hussein, M.; Yoneda, K.; Mohd-Zaki, Z.; Amir, A.; Othman, N. Heavy metals in leachate, impacted soils and natural soils of different landfills in Malaysia: An alarming threat. Chemosphere 2021, 267, 128874. [Google Scholar] [CrossRef] [PubMed]
- Gujre, N.; Mitra, S.; Soni, A.; Agnihotri, R.; Rangan, L.; Rene, E.R.; Sharma, M.P. Speciation, contamination, ecological and human health risks assessment of heavy metals in soils dumped with municipal solid wastes. Chemosphere 2021, 262, 128013. [Google Scholar] [CrossRef]
- Ziyang, L.; Luochun, W.; Nanwen, Z.; Youcai, Z. Martial recycling from renewable landfill and associated risks: A review. Chemosphere 2015, 131, 91–103. [Google Scholar] [CrossRef]
- Wanka, S.; Münnich, K.; Fricke, K. Landfill Mining—Wet mechanical treatment of fine MSW with a wet jigger. Waste Manag. 2017, 59, 316–323. [Google Scholar] [CrossRef]
- Gerba, C.P.; Tamimi, A.H.; Pettigrew, C.; Weisbrod, A.V.; Rajagopalan, V. Sources of microbial pathogens in municipal solid waste landfills in the United States of America. Waste Manag. Res. 2011, 29, 781–790. [Google Scholar] [CrossRef]
- Nair, A.T. Bioaerosols in the landfill environment: An overview of microbial diversity and potential health hazards. Aerobiologia 2021, 4, 1–19. [Google Scholar]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Nielsen, H.B.; Almeida, M.; Juncker, A.S.; Rasmussen, S.; Li, J.; Sunagawa, S.; Plichta, D.R.; Gautier, L.; Pedersen, A.G.; Le Chatelier, E.; et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat. Biotechnol. 2014, 32, 822–828. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Li, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; Nielsen, T.; et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014, 32, 834–841. [Google Scholar] [CrossRef]
- Li, Y.; Cao, W.; Liang, S.; Yamasaki, S.; Chen, X.; Shi, L.; Ye, L. Metagenomic characterization of bacterial community and antibiotic resistance genes in representative ready-to-eat food in southern China. Sci. Rep. 2020, 10, 15175. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Huson, D.H.; Mitra, S.; Ruscheweyh, H.J.; Weber, N.; Schuster, S.C. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011, 21, 1552–1560. [Google Scholar] [CrossRef]
- Karlsson, F.H.; Tremaroli, V.; Nookaew, I.; Bergström, G.; Behre, C.J.; Fagerberg, B.; Nielsen, J.; Bäckhed, F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013, 498, 99–103. [Google Scholar] [CrossRef]
- Karlsson, F.H.; Fåk, F.; Nookaew, I.; Tremaroli, V.; Fagerberg, B.; Petranovic, D.; Bäckhed, F.; Nielsen, J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun. 2012, 3, 1245. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Hattori, M.; Aoki-Kinoshita, K.F.; Itoh, M.; Kawashima, S.; Katayama, T.; Araki, M.; Hirakawa, M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006, 34, D354–D357. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014, 42, D199–D205. [Google Scholar] [CrossRef]
- Martínez, J.L.; Coque, T.M.; Baquero, F. What is a resistance gene? Ranking risk in resistomes. Nat. Rev. Microbiol. 2015, 13, 116–123. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- McArthur, A.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef]
- Ma, L.; Xia, Y.; Li, B.; Yang, Y.; Li, L.G.; Tiedje, J.M.; Zhang, T. Metagenomic Assembly Reveals Hosts of Antibiotic Resistance Genes and the Shared Resistome in Pig, Chicken, and Human Feces. Environ. Sci. Technol. 2016, 50, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Wang, H.; Cai, L.; Yu, Y. Prevalence of antibiotic resistance genes and bacterial pathogens in long-term manured greenhouse soils as revealed by metagenomic survey. Environ. Sci. Technol. 2015, 49, 1095–1104. [Google Scholar] [CrossRef] [PubMed]
- Zainun, M.Y.; Simarani, K. Metagenomics profiling for assessing microbial diversity in both active and closed landfills. Sci. Total Environ. 2018, 616–617, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Thakur, K.; Chownk, M.; Kumar, V.; Purohit, A.; Vashisht, A.; Kumar, V.; Yadav, S.K. Bioprospecting potential of microbial communities in solid waste landfills for novel enzymes through metagenomic approach. World J. Microbiol. Biotechnol. 2020, 36, 34. [Google Scholar] [CrossRef] [PubMed]
- Morita, A.K.M.; Sakamoto, I.K.; Varesche, M.B.A.; Wendland, E. Microbial structure and diversity in non-sanitary landfills and association with physicochemical parameters. Environ. Sci. Pollut. Res. Int. 2020, 27, 40690–40705. [Google Scholar] [CrossRef]
- Xie, B.; Xiong, S.; Liang, S.; Hu, C.; Zhang, X.; Lu, J. Performance and bacterial compositions of aged refuse reactors treating mature landfill leachate. Bioresour. Technol. 2012, 103, 71–77. [Google Scholar] [CrossRef]
- Köchling, T.; Sanz, J.L.; Gavazza, S.; Florencio, L. Analysis of microbial community structure and composition in leachates from a young landfill by 454 pyrosequencing. Appl. Microbiol. Biotechnol. 2015, 99, 5657–5668. [Google Scholar] [CrossRef]
- Wen, P.; Huang, Y.; Qiu, Z.; Li, Q. Microbial response during treatment of different types of landfill leachate in a semi-aerobic aged refuse biofilter. Chemosphere 2021, 262, 127822. [Google Scholar] [CrossRef]
- Song, L.; Wang, Y.; Tang, W.; Lei, Y. Archaeal community diversity in municipal waste landfill sites. Appl. Microbiol. Biotechnol. 2015, 99, 6125–6137. [Google Scholar] [CrossRef]
- Gaspari, M.; Treu, L.; Zhu, X.; Palù, M.; Angelidaki, I.; Campanaro, S.; Kougias, P.G. Microbial dynamics in biogas digesters treating lipid-rich substrates via genome-centric metagenomics. Sci. Total Environ. 2021, 778, 146296. [Google Scholar] [CrossRef]
- Osman, J.R.; Wang, Y.; Jaubert, C.; Nguyen, T.N.; Fernandes, G.R.; DuBow, M.S. The bacterial communities of surface soils from desert sites in the eastern Utah (USA) portion of the Colorado Plateau. Microbiol. Res. 2021, 244, 126664. [Google Scholar] [CrossRef]
- Iwasaki, Y.; Ichino, T.; Saito, A. Transition of the Bacterial Community and Culturable Chitinolytic Bacteria in Chitin-treated Upland Soil: From Streptomyces to Methionine-auxotrophic Lysobacter and Other Genera. Microbes Environ. 2020, 35, ME19070. [Google Scholar] [CrossRef]
- Mwaikono, K.S.; Maina, S.; Sebastian, A.; Schilling, M.; Kapur, V.; Gwakisa, P. High-throughput sequencing of 16S rRNA Gene Reveals Substantial Bacterial Diversity on the Municipal Dumpsite. BMC Microbiol. 2016, 16, 145. [Google Scholar] [CrossRef]
- Christensen, P.; Cook, F.D. Lysobacter, a new genus of nonfruiting, gliding bacteria with a high base ratio. Int. J. Syst. Bacteriol. 1978, 28, 367–393. [Google Scholar] [CrossRef]
- Panthee, S.; Hamamoto, H.; Paudel, A.; Sekimizu, K. Lysobacter species: A potential source of novel antibiotics. Arch. Microbiol. 2016, 198, 839–845. [Google Scholar] [CrossRef]
- Horel, A.; Mortazavi, B.; Sobecky, P.A. Input of organic matter enhances degradation of weathered diesel fuel in sub-tropical sediments. Sci. Total Environ. 2015, 533, 82–90. [Google Scholar] [CrossRef]
- Pishgar, R.; Dominic, J.A.; Sheng, Z.; Tay, J.H. Denitrification performance and microbial versatility in response to different selection pressures. Bioresour. Technol. 2019, 281, 72–83. [Google Scholar] [CrossRef]
- Silby, M.W.; Winstanley, C.; Godfrey, S.A.; Levy, S.B.; Jackson, R.W. Pseudomonas genomes: Diverse and adaptable. FEMS Microbiol. Rev. 2011, 35, 652–680. [Google Scholar] [CrossRef] [PubMed]
- Segers, P.; Vancanneyt, M.; Pot, B.; Torck, U.; Hoste, B.; Dewettinck, D.; Falsen, E.; Kersters, K.; De Vos, P. Classification of Pseudomonas diminuta Leifson and Hugh 1954 and Pseudomonas vesicularis Büsing, Döll, and Freytag 1953 in Brevundimonas gen. nov. as Brevundimonas diminuta comb. nov. and Brevundimonas vesicularis comb. nov., respectively. Int. J. Syst. Bacteriol. 1994, 44, 499–510. [Google Scholar] [CrossRef]
- Ryan, M.P.; Pembroke, J.T. Brevundimonas spp: Emerging global opportunistic pathogens. Virulence 2018, 9, 480–493. [Google Scholar] [CrossRef] [PubMed]
- Xia, F.F.; Su, Y.; Wei, X.M.; He, Y.H.; Wu, Z.C.; Ghulam, A.; He, R. Diversity and activity of sulphur-oxidizing bacteria and sulphate-reducing bacteria in landfill cover soils. Lett. Appl. Microbiol. 2014, 59, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Heo, Y.; Park, J.; Lim, S.I.; Hur, H.G.; Kim, D.; Park, K. Size-resolved culturable airborne bacteria sampled in rice field, sanitary landfill, and waste incineration sites. J. Environ. Monit. 2010, 12, 1619–1624. [Google Scholar] [CrossRef]
- Amano, J.; Hase, R.; Otsuka, Y.; Tsuchimochi, T.; Noguchi, Y.; Igarashi, S. Catheter-related bloodstream infection by Microbacterium paraoxydans in a pediatric patient with B-cell precursor acute lymphocytic leukemia: A case report and review of literature on Microbacterium bacteremia. J. Infect. Chemother. 2019, 25, 806–810. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.; Woo, P.C.; Woo, G.K.; Yuen, K.Y. Catheter-related Microbacterium bacteremia identified by 16S rRNA gene sequencing. J. Clin. Microbiol. 2002, 40, 2681–2685. [Google Scholar] [CrossRef] [PubMed]
- Young, C.C.; Kämpfer, P.; Chen, W.M.; Yen, W.S.; Arun, A.B.; Lai, W.A.; Shen, F.T.; Rekha, P.D.; Lin, K.Y.; Chou, J.H. Luteimonas composti sp. nov., a moderately thermophilic bacterium isolated from food waste. Int. J. Syst. Evol. Microbiol. 2007, 57, 741–744. [Google Scholar] [CrossRef]
- Rani, P.; Mukherjee, U.; Verma, H.; Kamra, K.; Lal, R. Luteimonas tolerans sp. nov., isolated from hexachlorocyclohexane-contaminated soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 1851–1856. [Google Scholar] [CrossRef]
- Siddiqi, M.Z.; Yeon, J.M.; Choi, H.; Lee, J.H.; Kim, S.Y.; Wee, J.H.; Im, W.T. Luteimonas granuli sp. nov., Isolated from Granules of the Wastewater Treatment Plant. Curr. Microbiol. 2020, 77, 2002–2007. [Google Scholar] [CrossRef]
- Zhou, J.; Chen, J.; Ma, J.; Xu, N.; Xin, F.; Zhang, W.; Zhang, H.; Dong, W.; Jiang, M. Luteimonas wenzhouensis Sp. Nov., A Chitinolytic Bacterium Isolated from a Landfill Soil. Curr. Microbiol. 2021, 78, 383–388. [Google Scholar] [CrossRef]
- Wang, X.; Cao, A.; Zhao, G.; Zhou, C.; Xu, R. Microbial community structure and diversity in a municipal solid waste landfill. Waste Manag. 2017, 66, 79–87. [Google Scholar] [CrossRef]
- Chen, B.; Yang, Y.; Liang, X.; Yu, K.; Zhang, T.; Li, X. Metagenomic profiles of antibiotic resistance genes (ARGs) between human impacted estuary and deep ocean sediments. Environ. Sci. Technol. 2013, 47, 12753–12760. [Google Scholar] [CrossRef]
- Ben Maamar, S.; Glawe, A.J.; Brown, T.K.; Hellgeth, N.; Hu, J.; Wang, J.P.; Huttenhower, C.; Hartmann, E.M. Mobilizable antibiotic resistance genes are present in dust microbial communities. PLoS Pathog. 2020, 16, e1008211. [Google Scholar] [CrossRef]
- Pérez, J.; Contreras-Moreno, F.J.; Marcos-Torres, F.J.; Moraleda-Muñoz, A.; Muñoz-Dorado, J. The antibiotic crisis: How bacterial predators can help. Comput. Struct. Biotechnol. J. 2020, 18, 2547–2555. [Google Scholar] [CrossRef]
- Wu, D.; Huang, X.H.; Sun, J.Z.; Graham, D.W.; Xie, B. Antibiotic Resistance Genes and Associated Microbial Community Conditions in Aging Landfill Systems. Environ. Sci. Technol. 2017, 51, 12859–12861. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, W.; Qiao, J.; Song, L. Occurrence and prevalence of antibiotic resistance in landfill leachate. Environ. Sci. Pollut. Res. Int. 2015, 22, 12525–12533. [Google Scholar] [CrossRef]
- Zhao, R.; Feng, J.; Yin, X.; Liu, J.; Fu, W.; Berendonk, T.U.; Zhang, T.; Li, X.; Li, B. Antibiotic resistome in landfill leachate from different cities of China deciphered by metagenomic analysis. Water Res. 2018, 134, 126–139. [Google Scholar] [CrossRef]
- Yao, L.; Li, Y.; Li, Z.; Shen, D.; Feng, H.; Zhou, H.; Wang, M. Prevalence of fluoroquinolone, macrolide and sulfonamide-related resistance genes in landfills from East China, mainly driven by MGEs. Ecotoxicol. Environ. Saf. 2020, 190, 110131. [Google Scholar] [CrossRef]
- Kalwasińska, A.; Burkowska, A. Municipal landfill sites as sources of microorganisms potentially pathogenic to humans. Environ. Sci. Process. Impacts 2013, 15, 1078–1086. [Google Scholar] [CrossRef]
- De Mandal, S.; Mathipi, V.; Muthukumaran, R.B.; Gurusubramanian, G.; Lalnunmawii, E.; Kumar, N.S. Amplicon sequencing and imputed metagenomic analysis of waste soil and sediment microbiome reveals unique bacterial communities and their functional attributes. Environ. Monit. Assess. 2019, 191, 778. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Wang, G.; Zhao, G.; Chao, X.; Xie, L.; Wang, H. The Characteristic of Virulence, Biofilm and Antibiotic Resistance of Klebsiella pneumoniae. Int. J. Environ. Res. Public Health 2020, 28, 6278. [Google Scholar] [CrossRef]
- Lee, C.R.; Lee, J.H.; Park, K.S.; Jeon, J.H.; Kim, Y.B.; Cha, C.J.; Jeong, B.C.; Lee, S.H. Antimicrobial Resistance of Hypervirulent Klebsiella pneumoniae: Epidemiology, Hypervirulence-Associated Determinants, and Resistance Mechanisms. Front. Cell Infect. Microbiol. 2017, 7, 483. [Google Scholar] [CrossRef]
- Trainor, E.A.; Nicholson, T.L.; Merkel, T.J. Bordetella pertussis transmission. Pathog. Dis. 2015, 73, ftv068. [Google Scholar] [CrossRef]
- Coote, J.G. Environmental sensing mechanisms in Bordetella. Adv. Microb. Physiol. 2001, 44, 141–181. [Google Scholar]
- Zhang, Y.; Ran, Z.; Tian, M.; Zhou, Y.; Yang, J.; Yin, J.; Lu, D.; Li, R.; Zhong, J. Commensal Microbes Affect Host Humoral Immunity to Bordetella pertussis Infection. Infect. Immun. 2019, 87, e00421-19. [Google Scholar] [CrossRef]
- Breidenstein, E.B.; de la Fuente-Núñez, C.; Hancock, R.E. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef]
- Kuzi, S.; Blum, S.E.; Kahane, N.; Adler, A.; Hussein, O.; Segev, G.; Aroch, I. Multi-drug-resistant Acinetobacter calcoaceticus-Acinetobacter baumannii complex infection outbreak in dogs and cats in a veterinary hospital. J. Small Anim. Pract. 2016, 57, 617–625. [Google Scholar] [CrossRef]
- Nocera, F.P.; Attili, A.R.; De Martino, L. Acinetobacter baumannii: Its Clinical Significance in Human and Veterinary Medicine. Pathogens 2021, 10, 127. [Google Scholar] [CrossRef]
- Matereke, L.T.; Okoh, A.I. Listeria monocytogenes Virulence, Antimicrobial Resistance and Environmental Persistence: A Review. Pathogens 2020, 9, 528. [Google Scholar] [CrossRef] [PubMed]
- Lüth, S.; Halbedel, S.; Rosner, B.; Wilking, H.; Holzer, A.; Roedel, A.; Dieckmann, R.; Vincze, S.; Prager, R.; Flieger, A.; et al. Backtracking and forward checking of human listeriosis clusters identified a multiclonal outbreak linked to Listeria monocytogenes in meat products of a single producer. Emerg. Microbes Infect. 2020, 9, 1600–1608. [Google Scholar] [CrossRef] [PubMed]
- Bottone, E.J. Bacillus cereus, a volatile human pathogen. Clin. Microbiol. Rev. 2010, 23, 382–398. [Google Scholar] [CrossRef] [PubMed]
- Kloos, W.E.; Musselwhite, M.S. Distribution and persistence of Staphylococcus and Micrococcus species and other aerobic bacteria on human skin. Appl. Microbiol. 1975, 30, 381–385. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus epidermidis—The ‘accidental’ pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).