Assessing Temporal Patterns and Species Composition of Glass Eel (Anguilla spp.) Cohorts in Sumatra and Java Using DNA Barcodes
Abstract
1. Introduction
2. Materials and Methods
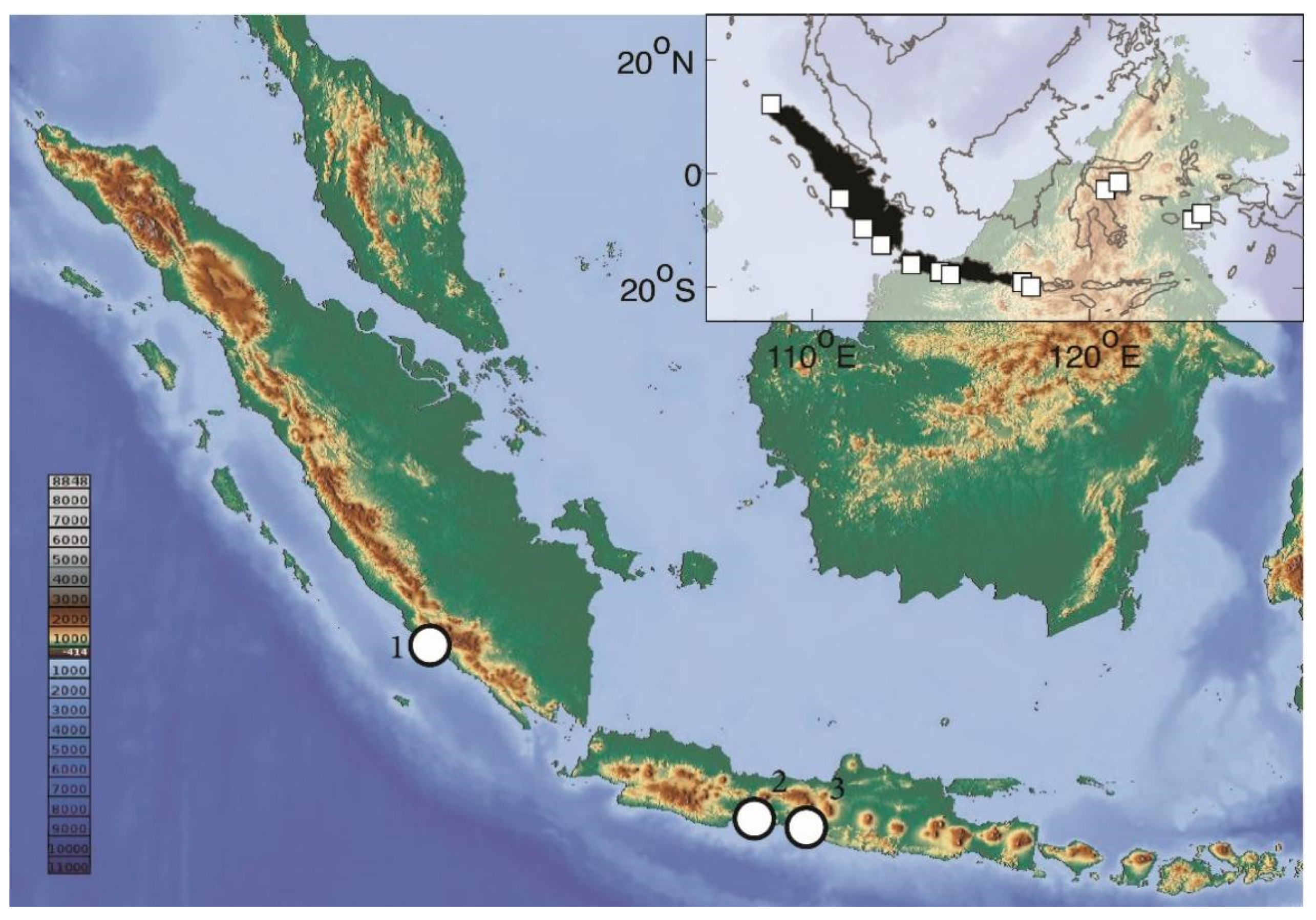
2.1. Sampling
2.2. DNA Extraction, Amplification and Sequencing
2.3. Data Analysis
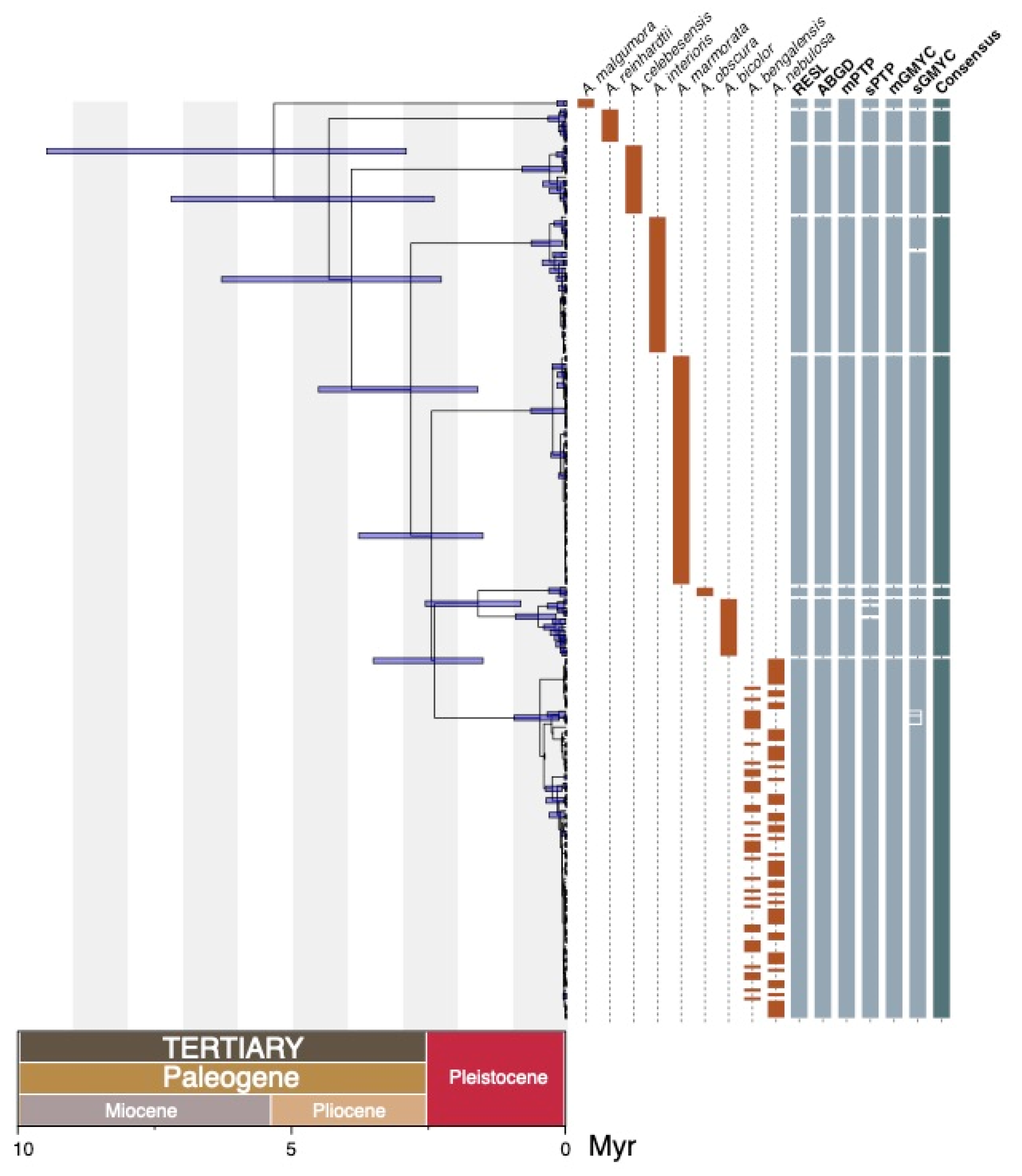
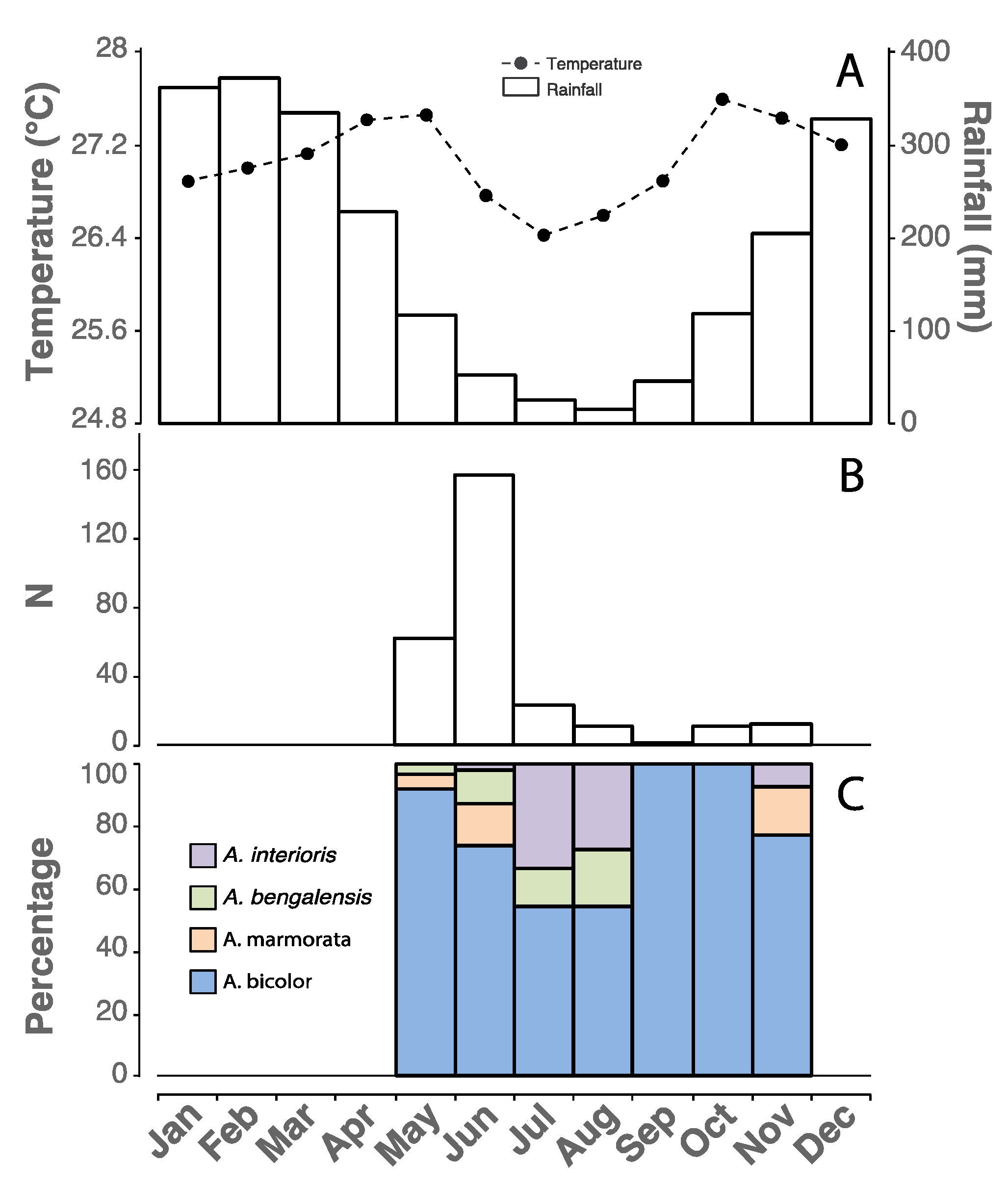
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Froese, R.; Pauly, D. Fishbase. Available online: http://www.fishbase.org (accessed on 15 December 2020).
- Kuroki, M.; Miller, M.J.; Tsukamoto, K. Diversity of early life-history traits in freshwater eels and the evolution of their oceanic migrations. Can. J. Zool. 2014, 92, 749–770. [Google Scholar] [CrossRef]
- Tesch, F.W.; Rohlf, N. Migration from continental waters to the spawning grounds. In Eel Biology; Springer: Berlin, Germany, 2003; pp. 223–234. [Google Scholar]
- Elliott, J.M.; Tesch, F.-W. The Eel: Biology and Management of Anguillid Eels. J. Anim. Ecol. 1978, 47, 1033. [Google Scholar] [CrossRef]
- Watanabe, S. Taxonomy of the Freshwater Eels, Genus Anguilla Schrank, 1798. In Eel Biology; Springer: Tokyo, Japan, 2003; pp. 3–18. [Google Scholar]
- Jacoby, D.M.; Casselman, J.M.; Crook, V.; DeLucia, M.-B.; Ahn, H.; Kaifu, K.; Kurwie, T.; Sasal, P.; Silfvergrip, A.M.; Smith, K.G.; et al. Synergistic patterns of threat and the challenges facing global anguillid eel conservation. Glob. Ecol. Conserv. 2015, 4, 321–333. [Google Scholar] [CrossRef]
- Sugeha, H.Y.; Suharti, S.R.; Wouthuyzen, S.; Sumadhiharga, K. Biodiversity, distribution and abundance of the tropical anguillid eels in the Indonesian waters. Mar. Res. Indones. 2008, 33, 129–138. [Google Scholar] [CrossRef]
- Ringuet, S.; Muto, F.; Raymakers, C. Eels: Their harvest and trade in Europe and Asia. Traffic Bull. Int. 2002, 19, 80–106. [Google Scholar]
- FAO. Des Pêches Et De L’aquaculture; FAO: Rome, Italy, 2018; ISBN 9789251306925. [Google Scholar]
- Tsukamoto, K.; Kuroki, M. Eels and Humans; Springer: Tokyo, Japan, 2014; ISBN 4431545298. [Google Scholar]
- Shiraishi, H.; Crook, V. Eel Market Dynamics: An Analysis of Anguilla Production; TRAFFIC: Tokyo, Japan, 2015. [Google Scholar]
- BKIPM. Eels Trade in Indonesia; BKIPM: Jakarta, Indonesia, 2018.
- Tanaka, H.; Kagawa, H.; Ohta, H. Production of leptocephali of Japanese eel (Anguilla japonica) in captivity. Aquaculture 2001, 201, 51–60. [Google Scholar] [CrossRef]
- Tanaka, H.; Kagawa, H.; Ohta, H.; Unuma, T.; Nomura, K. The first production of glass eel in captivity: Fish reproductive physiology facilitates great progress in aquaculture. Fish Physiol. Biochem. 2003, 28, 493–497. [Google Scholar] [CrossRef]
- Friedland, K.D.; Miller, M.J.; Knights, B. Oceanic changes in the Sargasso Sea and declines in recruitment of the European eel. ICES J. Mar. Sci. 2007, 64, 519–530. [Google Scholar] [CrossRef]
- Wirth, T.; Bernatchez, L. Decline of North Atlantic eels: A fatal synergy? Proc. R. Soc. B Boil. Sci. 2003, 270, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Dekker, W. Management of the eel is slipping through our hands! Distribute control and orchestrate national protection. ICES J. Mar. Sci. 2016, 73, 2442–2452. [Google Scholar] [CrossRef]
- Righton, D.; Walker, A.M. Anguillids: Conserving a global fishery. J. Fish Biol. 2013, 83, 754–765. [Google Scholar] [CrossRef] [PubMed]
- Bonhommeau, S.; Chassot, E.; Rivot, E. Fluctuations in European eel (Anguilla anguilla) recruitment resulting from environmental changes in the Sargasso Sea. Fish. Oceanogr. 2008, 17, 32–44. [Google Scholar] [CrossRef]
- Arai, T.; Kadir, S.R.A. Opportunistic spawning of tropical anguillid eels Anguilla bicolor bicolor and A. bengalensis bengalensis. Sci. Rep. 2017, 7, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Arai, T.; Limbong, D.; Otake, T.; Tsukamoto, K. Recruitment mechanisms of tropical eels Anguilla spp. and implications for the evolution of oceanic migration in the genus Anguilla. Mar. Ecol. Prog. Ser. 2001, 216, 253–264. [Google Scholar] [CrossRef]
- Fahmi, M.R.; Solihin, D.D.; Soewardi, K.; Pouyaud, L.; Berrebi, P. Molecular phylogeny and genetic diversity of freshwater Anguilla eels in indonesian waters based on mitochondrial sequences. Vie Milieu-Life Environ. 2015, 65, 139–150. [Google Scholar]
- Eschmeyer, W.N.; Fricke, R.; van der Laan, R. Catalog of Fishes Electronic Version. Available online: https://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.aspg (accessed on 15 December 2020).
- Aoyama, J. Life History and Evolution of Migration in Catadromous Eels (Genus: Anguilla). Aqua-BioScience Monogr. 2009, 2, 1–42. [Google Scholar] [CrossRef]
- Aoyama, J.; Wouthuyzen, S.; Miller, M.J.; Minegishi, Y.; Kuroki, M.; Suharti, S.R.; Kawakami, T.; Sumardiharga, K.O.; Tsukamoto, K. Distribution of leptocephali of the freshwater eels, genus Anguilla, in the waters off west Sumatra in the Indian Ocean. Environ. Boil. Fishes 2007, 80, 445–452. [Google Scholar] [CrossRef]
- Ndobe, S.; Serdiati, N.; Moore, A. Species Composition Of Glass Eels Recruiting To The Palu River. J. Agroecol. 2015, 1, 12–23. [Google Scholar]
- Shirotori, F.; Ishikawa, T.; Tanaka, C.; Aoyama, J.; Shinoda, A.; Yambot, A.V.; Yoshinaga, T. Species composition of anguillid glass eels recruited at southern Mindanao Island, the Philippines. Fish. Sci. 2016, 82, 915–922. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; Dewaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B Boil. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.; Ratnasingham, S.; De Waard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B Boil. Sci. 2003, 270, S96–S99. [Google Scholar] [CrossRef] [PubMed]
- Dahruddin, H.; Hutama, A.; Busson, F.; Sauri, S.; Hanner, R.; Keith, P.; Hadi, D.; Hubert, N. Revisiting the ichthyodiversity of Java and Bali through DNA barcodes: Taxonomic coverage, identification accuracy, cryptic diversity and identification of exotic species. Mol. Ecol. Resour. 2017, 17, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Hutama, A.; Dahruddin, H.; Busson, F.; Sauri, S.; Keith, P.; Hadiaty, R.K.; Hanner, R.; Suryobroto, B.; Hubert, N. Identifying spatially concordant evolutionary significant units across multiple species through DNA barcodes: Application to the conservation genetics of the freshwater fishes of Java and Bali. Glob. Ecol. Conserv. 2017, 12, 170–187. [Google Scholar] [CrossRef]
- Hubert, N.; Lumbantobing, D.; Sholihah, A.; Dahruddin, H.; Delrieu-Trottin, E.; Busson, F.; Sauri, S.; Hadiaty, R.; Keith, P. Revisiting species boundaries and distribution ranges of Nemacheilus spp. (Cypriniformes: Nemacheilidae) and Rasbora spp. (Cypriniformes: Cyprinidae) in Java, Bali and Lombok through DNA barcodes: Implications for conservation in a biodiversity hotspot. Conserv. Genet. 2019, 20, 517–529. [Google Scholar] [CrossRef]
- Sholihah, A.; Delrieu-Trottin, E.; Sukmono, T.; Dahruddin, H.; Risdawati, R.; Elvyra, R.; Wibowo, A.; Kustiati, K.; Busson, F.; Sauri, S.; et al. Disentangling the taxonomy of the subfamily Rasborinae (Cypriniformes, Danionidae) in Sundaland using DNA barcodes. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.; Abidin, M.Z.; Pulungan, C.P.; De Bruyn, M.; Nor, S.A.M. DNA Barcoding Reveals High Cryptic Diversity of the Freshwater Halfbeak Genus Hemirhamphodon from Sundaland. PLoS ONE 2016, 11, e0163596. [Google Scholar] [CrossRef] [PubMed]
- Farhana, S.N.; Muchlisin, Z.A.; Duong, T.Y.; Tanyaros, S.; Page, L.M.; Zhao, Y.; Adamson, E.A.S.; Khaironizam, Z.; De Bruyn, M.; Azizah, M.N.S. Exploring hidden diversity in Southeast Asia’s Dermogenys spp. (Beloniformes: Zenarchopteridae) through DNA barcoding. Sci. Rep. 2018, 8, 10787. [Google Scholar] [CrossRef]
- Hubert, N.; Hadiaty, R.K.; Paradis, E.; Pouyaud, L. Cryptic Diversity in Indo-Australian Rainbowfishes Revealed by DNA Barcoding: Implications for Conservation in a Biodiversity Hotspot Candidate. PLoS ONE 2012, 7, e40627. [Google Scholar] [CrossRef]
- Wibowo, A.; Sloterdijk, H.; Ulrich, S.P. Identifying Sumatran Peat Swamp Fish Larvae through DNA Barcoding, Evidence of Complete Life History Pattern. Procedia Chem. 2015, 14, 76–84. [Google Scholar] [CrossRef]
- Wibowo, A.; Wahlberg, N.; Vasemägi, A. DNA barcoding of fish larvae reveals uncharacterised biodiversity in tropical peat swamps of New Guinea, Indonesia. Mar. Freshw. Res. 2017, 68, 1079–1087. [Google Scholar] [CrossRef]
- Kottelat, M.; Whitten, A.J.; Kartikasari, N.; Wirjoatmodjo, S. Freshwater Fishes of Western Indonesia and Sulawesi; PERIPLUS: Jakarta, Indonesia, 1993. [Google Scholar]
- Keith, P.; Marquet, G.; Lord, C.; Kalfatak, D.; Vigneux, E. Poissons et Crustacés d’Eau Douce du Vanuatu; Société Française d’Ichtyologie: Paris, France, 2010. [Google Scholar]
- Keith, P.; Marquet, G.; Gerbeaux, P.; Vigneux, E.; Lord, C. Poissons et Crustacés d’Eau Douce de Polynésie; Société Française d’Ichthyologie: Paris, France, 2013. [Google Scholar]
- Hubert, N.; Hanner, R.; Holm, E.; Mandrak, N.E.; Taylor, E.; Burridge, M.; Watkinson, D.; Dumont, P.; Curry, A.; Bentzen, P.; et al. Identifying Canadian Freshwater Fishes through DNA Barcodes. PLoS ONE 2008, 3, e2490. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, N.V.; Zemlak, T.S.; Hanner, R.H.; Hebert, P.D.N. Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Notes 2007, 7, 544–548. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System (www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Avise, J.C. Molecular Markers, Natural History and Evolution; Hall C.& Ed.: New York, NY, USA, 1989. [Google Scholar]
- Moritz, C. Defining “Evolutionary Significant Units” for conservation. Trends Ecol. Evol. 1994, 9, 373–375. [Google Scholar] [CrossRef]
- Vogler, A.P.; DeSalle, R. Diagnosing Units of Conservation Management. Conserv. Biol. 1994, 8, 354–363. [Google Scholar] [CrossRef]
- Kekkonen, M.; Mutanen, M.; Kaila, L.; Nieminen, M.; Hebert, P.D.N. Delineating Species with DNA Barcodes: A Case of Taxon Dependent Method Performance in Moths. PLoS ONE 2015, 10, e0122481. [Google Scholar] [CrossRef] [PubMed]
- Limmon, G.; Delrieu-Trottin, E.; Patikawa, J.; Rijoly, F.; Dahruddin, H.; Busson, F.; Steinke, D.; Hubert, N. Assessing species diversity of Coral Triangle artisanal fisheries: A DNA barcode reference library for the shore fishes retailed at Ambon harbor (Indonesia). Ecol. Evol. 2020, 10, 3356–3366. [Google Scholar] [CrossRef] [PubMed]
- Delrieu-Trottin, E.; Durand, J.; Limmon, G.; Sukmono, T.; Sugeha, H.Y.; Chen, W.; Busson, F.; Borsa, P.; Dahruddin, H.; Sauri, S. Biodiversity inventory of the grey mullets (Actinopterygii: Mugilidae) of the Indo-Australian Archipelago through the iterative use of DNA-based species delimitation and specimen assignment methods. Evol. Appl. 2020, 13, 1451–1467. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Hubert, N.; Huang, Y.; Wang, X.; Gan, X.; Peng, Z.; He, S. DNA barcoding the ichthyofauna of the Yangtze River: Insights from the molecular inventory of a mega-diverse temperate fauna. Mol. Ecol. Resour. 2019, 19, 1278–1291. [Google Scholar] [CrossRef] [PubMed]
- Ratnasingham, S.; Hebert, P.D.N. A DNA-Based Registry for All Animal Species: The Barcode Index Number (BIN) System. PLoS ONE 2013, 8, e66213. [Google Scholar] [CrossRef] [PubMed]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2011, 21, 1864–1877. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef] [PubMed]
- Kapli, P.; Lutteropp, S.; Zhang, J.; Kobert, K.; Pavlidis, P.; Stamatakis, A.; Flouri, T. Multi-rate Poisson Tree Processes for single-locus species delimitation under Maximum Likelihood and Markov Chain Monte Carlo. Bioinformatics 2017, 33, 1630–1638. [Google Scholar] [CrossRef]
- Fujisawa, T.; Barraclough, T.G. Delimiting Species Using Single-Locus Data and the Generalized Mixed Yule Coalescent Approach: A Revised Method and Evaluation on Simulated Data Sets. Syst. Biol. 2013, 62, 707–724. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.-H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [PubMed]
- Bermingham, E.; McCafferty, S.S.; Martin, A.P. Fish Biogeography and Molecular Clocks: Perspectives from the Panamanian Isthmus. In Molecular Systematics of Fishes; Academic Press: San Diego, CA, USA, 1997; pp. 113–126. [Google Scholar]
- Ogilvie, H.A.; Bouckaert, R.R.; Drummond, A.J. StarBEAST2 Brings Faster Species Tree Inference and Accurate Estimates of Substitution Rates. Mol. Biol. Evol. 2017, 34, 2101–2114. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.Y.W.; Larson, G. Molecular clocks: When timesare a-changin’. Trends Genet. 2006, 22, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.-B.; Hao, M.-D.; Yang, C.-Q.; Shi, Z.-Y. BarcodingR: An integrated r package for species identification using DNA barcodes. Methods Ecol. Evol. 2016, 8, 627–634. [Google Scholar] [CrossRef]
- Zhang, A.-B.; Muster, C.; Liang, H.-B.; Zhu, C.-D.; Crozier, R.; Wan, P.; Feng, J.; Ward, R.D. A fuzzy-set-theory-based approach to analyse species membership in DNA barcoding. Mol. Ecol. 2011, 21, 1848–1863. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Han, H.; Hu, X.; Li, X.; Zhu, C.; Ho, S.Y.W.; Ward, R.D.; Zhang, A. Quantifying species diversity with a DNA barcoding-based method: Tibetan moth species (Noctuidae) on the Qinghai-Tibetan Plateau. PLoS ONE 2013, 8, e64428. [Google Scholar] [CrossRef] [PubMed]
- Paradis, E. pegas: An R package for population genetics with an integrated-modular approach. Bioinformatics 2010, 26, 419–420. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Tajima, F. DNA Polymorphism Detectable by Restriction Endonucleases. Genetics 1981, 97, 145–163. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Watterson, G. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 1975, 7, 256–276. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Paradis, E.; Schliep, K. ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2018, 35, 526–528. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.D.J.; Collins, R.A.; Boyer, S.; Lefort, C.; Malumbres-Olarte, J.; Vink, C.J.; Cruickshank, R.H. Spider: An R package for the analysis of species identity and evolution, with particular reference to DNA barcoding. Mol. Ecol. Resour. 2012, 12, 562–565. [Google Scholar] [CrossRef]
- Muchlisin, Z.A.; Batubara, A.S.; Fadli, N.; Muhammadar, A.A.; Utami, A.I.; Farhana, N.; Siti-Azizah, M.N. Assessing the species composition of tropical eels (Anguillidae) in Aceh Waters, Indonesia, with DNA barcoding gene cox1. F1000Research 2017, 6, 258. [Google Scholar] [CrossRef] [PubMed]
- Hanzen, C.; Lucas, M.C.; O’Brien, G.; Downs, C.T.; Willows-Munro, S. African freshwater eel species (Anguilla spp.) identification through DNA barcoding. Mar. Freshw. Res. 2020, 71, 1543. [Google Scholar] [CrossRef]
- Stein, F.M.; Wong, J.C.Y.; Sheng, V.; Law, C.S.W.; Schröder, B.; Baker, D.M. First genetic evidence of illegal trade in endangered European eel (Anguilla anguilla) from Europe to Asia. Conserv. Genet. Resour. 2016, 8, 533–537. [Google Scholar] [CrossRef]
- Arai, T.; Wong, L.L. Validation of the occurrence of the tropical eels, Anguilla bengalensis bengalensis and A. bicolor bicolor at Langkawi Island in Peninsular Malaysia, Malaysia. Trop. Ecol. 2016, 57, 23–31. [Google Scholar]
- Rosenberg, A.; Norborg, M. Genealogical trees, coalescent theory and the analysis of genetic polymorphisms. Nat. Rev. Genet. 2002, 3, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Hubert, N.; Hanner, R. DNA Barcoding, species delineation and taxonomy: A historical perspective. DNA Barcodes 2015, 3, 44–58. [Google Scholar] [CrossRef]
- Arai, T.; Aoyama, J.; Limbong, D.; Tsukamoto, K. Species composition and inshore migration of the tropical eels Anguilla spp. recruiting to the estuary of the Poigar River, Sulawesi Island. Mar. Ecol. Prog. Ser. 1999, 188, 299–303. [Google Scholar] [CrossRef]
- Sugeha, H.Y.; Arai, T.; Miller, M.J.; Limbong, D.; Tsukamoto, K. Inshore migration of the tropical eels Anguilla spp. recruiting to the Poigar River estuary on north Sulawesi Island. Mar. Ecol. Prog. Ser. 2001, 221, 233–243. [Google Scholar] [CrossRef]
- Fahmi, M.R. Phylogeography of Tropical Eels (Anguilla spp.). In Indonesian Waters; Bogor Agricultural University: Bogor, Indonesia, 2013. [Google Scholar]
- Hubert, N.; Kadarusman; Wibowo, A.; Busson, F.; Caruso, D.; Sulandari, S.; Nafiqoh, N.; Rüber, L.; Pouyaud, L.; Avarre, J.C.; et al. DNA barcoding Indonesian freshwater fishes: Challenges and prospects. DNA Barcodes 2015, 3, 144–169. [Google Scholar] [CrossRef]
- Hubert, N.; Espiau, B.; Meyer, C.; Planes, S. Identifying the ichthyoplankton of a coral reef using DNA barcodes. Mol. Ecol. Resour. 2015, 15, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Collet, A.; Durand, J.-D.; Desmarais, E.; Cerqueira, F.; Cantinelli, T.; Valade, P.; Ponton, D. DNA barcoding post-larvae can improve the knowledge about fish biodiversity: An example from La Reunion, SW Indian Ocean. Mitochondrial DNA Part A 2018, 29, 905–918. [Google Scholar] [CrossRef] [PubMed]
- Steinke, D.; Connell, A.D.; Hebert, P.D. Linking adults and immatures of South African marine fishes. Genome 2016, 59, 959–967. [Google Scholar] [CrossRef]
- Pianka, E.R. On r and K selection. Am. Nat. 1970, 104, 592–597. [Google Scholar] [CrossRef]
- Houde, E.D. Recruitment variability. Fish Reprod. Biol. 2016, 98–187. [Google Scholar]
- UNEP-WCMC Preliminary Overview of the Genus Anguilla. 2015. Available online: https://ec.europa.eu/environment/cites/pdf/reports/Preliminary%20overview%20of%20the%20genus%20Anguilla.pdf (accessed on 15 December 2020).
- Nikolic, N.; Liu, S.; Jacobsen, M.W.; Jónsson, B.; Bernatchez, L.; Gagnaire, P.; Hansen, M.M. Speciation history of European (Anguilla anguilla) and American eel (A. rostrata), analysed using genomic data. Mol. Ecol. 2020, 29, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-L.K.; Feunteun, E.; Miyazawa, Y.; Tsukamoto, K. New clues on the Atlantic eels spawning behavior and area: The Mid-Atlantic Ridge hypothesis. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Arai, T. Ecology and evolution of migration in the freshwater eels of the genus Anguilla Schrank, 1798. Heliyon 2020, 6, 05176. [Google Scholar] [CrossRef] [PubMed]
- Enbody, E.D.; Pettersson, M.E.; Sprehn, C.G.; Palm, S.; Wickström, H.; Andersson, L. Ecological adaptation in European eels is based on phenotypic plasticity. Proc. Natl. Acad. Sci. USA 2021, 118, e2022620118. [Google Scholar] [CrossRef] [PubMed]
- Gilpin, E.; Soulé, M. Minimum viable populations: Processes of species extinction. In Conservation Biology: The Science of Scarcity and Diversity; Soulé, M.E., Ed.; Sinauer: Sunderland, UK, 1986; pp. 19–34. [Google Scholar]
- Fagan, W.F.; Holmes, E.E. Quantifying the extinction vortex. Ecol. Lett. 2005, 9, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Spracklen, D.V.; Reddington, C.L.; A Gaveau, D.L. Industrial concessions, fires and air pollution in Equatorial Asia. Environ. Res. Lett. 2015, 10, 91001. [Google Scholar] [CrossRef]
- Breckwoldt, A.; Dsikowitzky, L.; Baum, G.; Ferse, S.C.; Van Der Wulp, S.; Kusumanti, I.; Ramadhan, A.; Adrianto, L. A review of stressors, uses and management perspectives for the larger Jakarta Bay Area, Indonesia. Mar. Pollut. Bull. 2016, 110, 790–794. [Google Scholar] [CrossRef]
- Hayati, A.; Tiantono, N.; Mirza, M.F.; Putra, I.D.S.; Abdizen, M.M.; Seta, A.R.; Solikha, B.M.; Fu’Adil, M.H.; Putranto, T.W.C.; Affandi, M.; et al. Water quality and fish diversity in the Brantas River, East Java, Indonesia. J. Biol. Res. 2017, 22, 43–49. [Google Scholar] [CrossRef]
- Garg, T.; Hamilton, S.E.; Hochard, J.P.; Kresch, E.P.; Talbot, J. (Not so) gently down the stream: River pollution and health in Indonesia. J. Environ. Econ. Manag. 2018, 92, 35–53. [Google Scholar] [CrossRef]

| K2P Genetic Distance | Assignment Probability | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| BIN | Max. Intraspecific | Min. Interspecific | Species | Fuzzy Average (Min–Max) | Fuzzy Identif. | Bayesian Average (Min–Max) | Bayesian Identif. | BP Average (Min–Max) | BP Identif. |
| BOLDAAD2092 | 0.004 | 0.031 | A. marmorata | 0.753 (0.636–0.758) | A. marmorata | 1 | A. marmorata | 0.949 (0.945–0.949) | A. marmorata |
| BOLDAAD9080 | 0.002 | 0.060 | A. reinhardtii | ||||||
| BOLDAAE4923 | 0.029 | 0.044 | A. bicolor | 0.735 (0.572–1) | A. bicolor | 1 | A. bicolor | 0.885 (0.853–0.963) | A. bicolor |
| BOLDAAJ2664 | 0.015 | 0.038 | A. bengalensis | 0.989 (0.969–1) | A. bengalensis | 1 | A. bengalensis | 0.948 (0.947–0.948) | A. bengalensis |
| A. nebulosa | A. nebulosa | A. nebulosa | A. nebulosa | ||||||
| BOLDADC7451 | 0.008 | 0.038 | A. interioris | 0.789 (0.595–0.919) | A. interioris | 1 | A. interioris | 0.946 (0.936–0.952) | A. interioris |
| BOLDADC7453 | 0 | 0.087 | A. malgumora | ||||||
| BOLDADC7455 | 0.006 | 0.060 | A. celebesensis | ||||||
| BOLDADC8019 | 0.002 | 0.035 | A. obscura | ||||||
| Species | Stage/Scale | N | h | Hd | π | θw | D | p-Value |
|---|---|---|---|---|---|---|---|---|
| A. interioris | Yellow-silver eels/Regional | 35 | 9 | 0.718 | 0.004 | 2.428 | −0.341 | 0.7330 |
| Glass eels/Local | 15 | 2 | 0.514 | 0.004 | 1.230 | 2.155 | 0.0312 | |
| A. marmorata | Yellow-silver eels/Regional | 58 | 6 | 0.539 | 0.003 | 5.381 | −2.158 | 0.0308 |
| Glass eels/Local | 26 | 2 | 0.077 | 0.001 | 0.262 | −1.156 | 0.2479 | |
| A. nebulosa/A. bengalensis | Yellow-silver eels/Regional | 91 | 16 | 0.667 | 0.002 | 4.132 | −2.243 | 0.0249 * |
| Glass eels/Local | 24 | 2 | 0.489 | 0.001 | 0.268 | 1.391 | 0.1642 | |
| A. bicolor | Yellow-silver eels/Regional | 15 | 11 | 0.952 | 0.013 | 6.766 | 0.050 | 0.9603 |
| Glass eels/Local | 213 | 7 | 0.476 | 0.003 | 1.179 | 0.997 | 0.3189 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wibowo, A.; Hubert, N.; Dahruddin, H.; Steinke, D.; Suhaimi, R.A.; Samuel; Atminarso, D.; Anggraeni, D.P.; Trismawanti, I.; Baumgartner, L.J.; et al. Assessing Temporal Patterns and Species Composition of Glass Eel (Anguilla spp.) Cohorts in Sumatra and Java Using DNA Barcodes. Diversity 2021, 13, 193. https://doi.org/10.3390/d13050193
Wibowo A, Hubert N, Dahruddin H, Steinke D, Suhaimi RA, Samuel, Atminarso D, Anggraeni DP, Trismawanti I, Baumgartner LJ, et al. Assessing Temporal Patterns and Species Composition of Glass Eel (Anguilla spp.) Cohorts in Sumatra and Java Using DNA Barcodes. Diversity. 2021; 13(5):193. https://doi.org/10.3390/d13050193
Chicago/Turabian StyleWibowo, Arif, Nicolas Hubert, Hadi Dahruddin, Dirk Steinke, Rezki Antoni Suhaimi, Samuel, Dwi Atminarso, Dian Pamularsih Anggraeni, Ike Trismawanti, Lee J. Baumgartner, and et al. 2021. "Assessing Temporal Patterns and Species Composition of Glass Eel (Anguilla spp.) Cohorts in Sumatra and Java Using DNA Barcodes" Diversity 13, no. 5: 193. https://doi.org/10.3390/d13050193
APA StyleWibowo, A., Hubert, N., Dahruddin, H., Steinke, D., Suhaimi, R. A., Samuel, Atminarso, D., Anggraeni, D. P., Trismawanti, I., Baumgartner, L. J., & Ning, N. (2021). Assessing Temporal Patterns and Species Composition of Glass Eel (Anguilla spp.) Cohorts in Sumatra and Java Using DNA Barcodes. Diversity, 13(5), 193. https://doi.org/10.3390/d13050193







