Genetic Divergence and Polyphyly in the Octocoral Genus Swiftia [Cnidaria: Octocorallia], Including a Species Impacted by the DWH Oil Spill
Abstract
1. Introduction
2. Materials and Methods
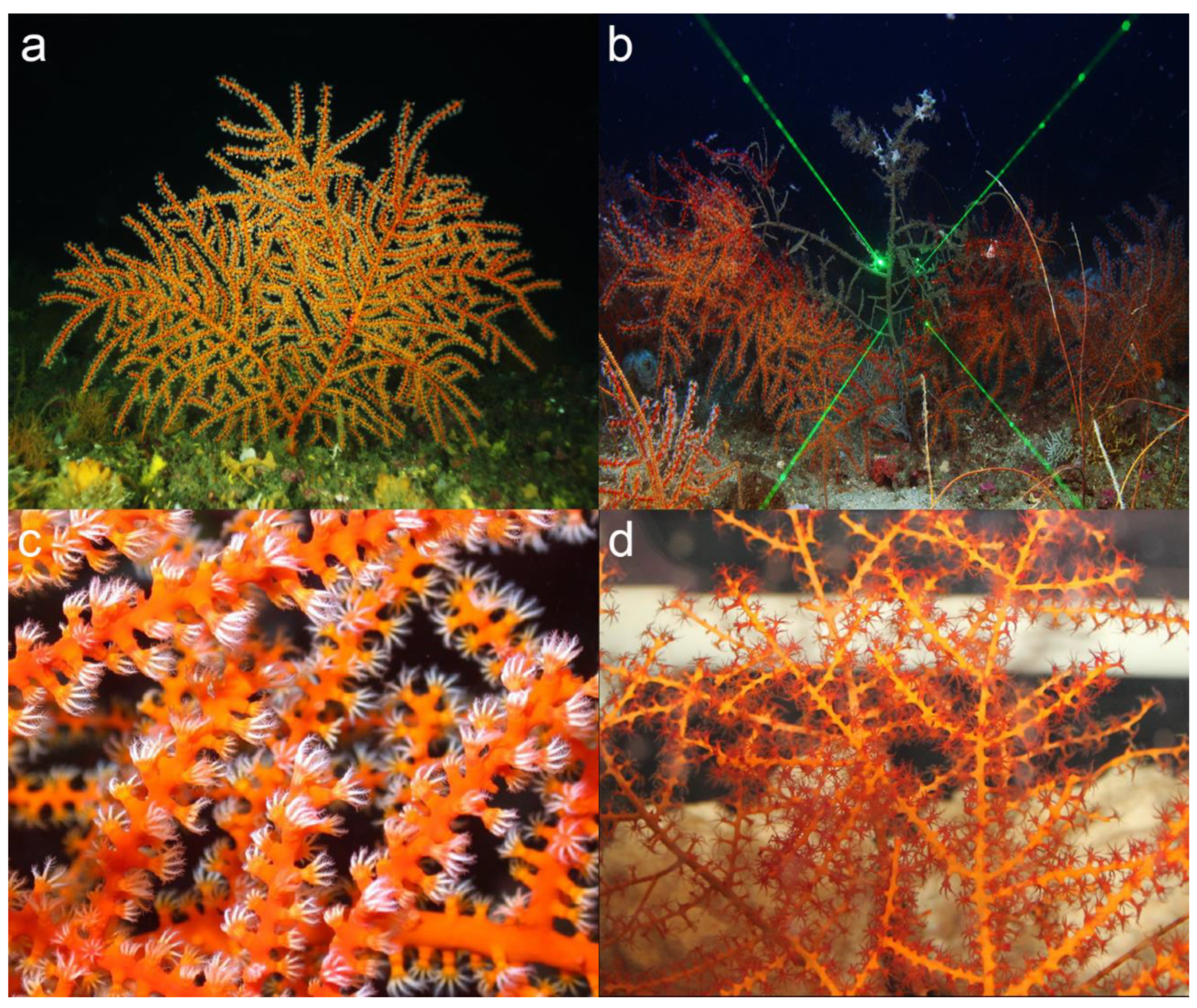
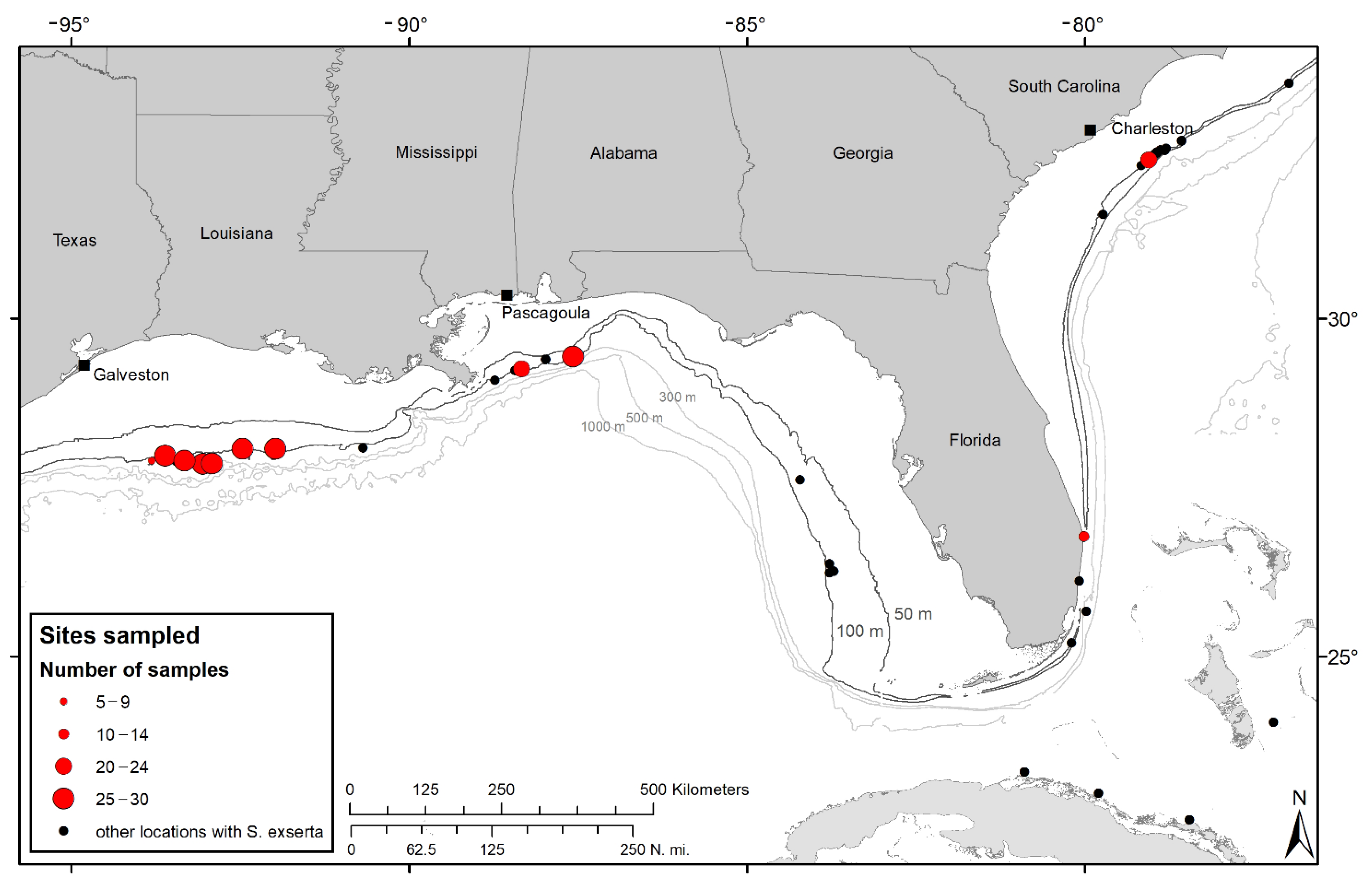
2.1. Sampling and Region of Study
2.2. Molecular Procedures
2.3. Phylogenetic Inference
3. Results
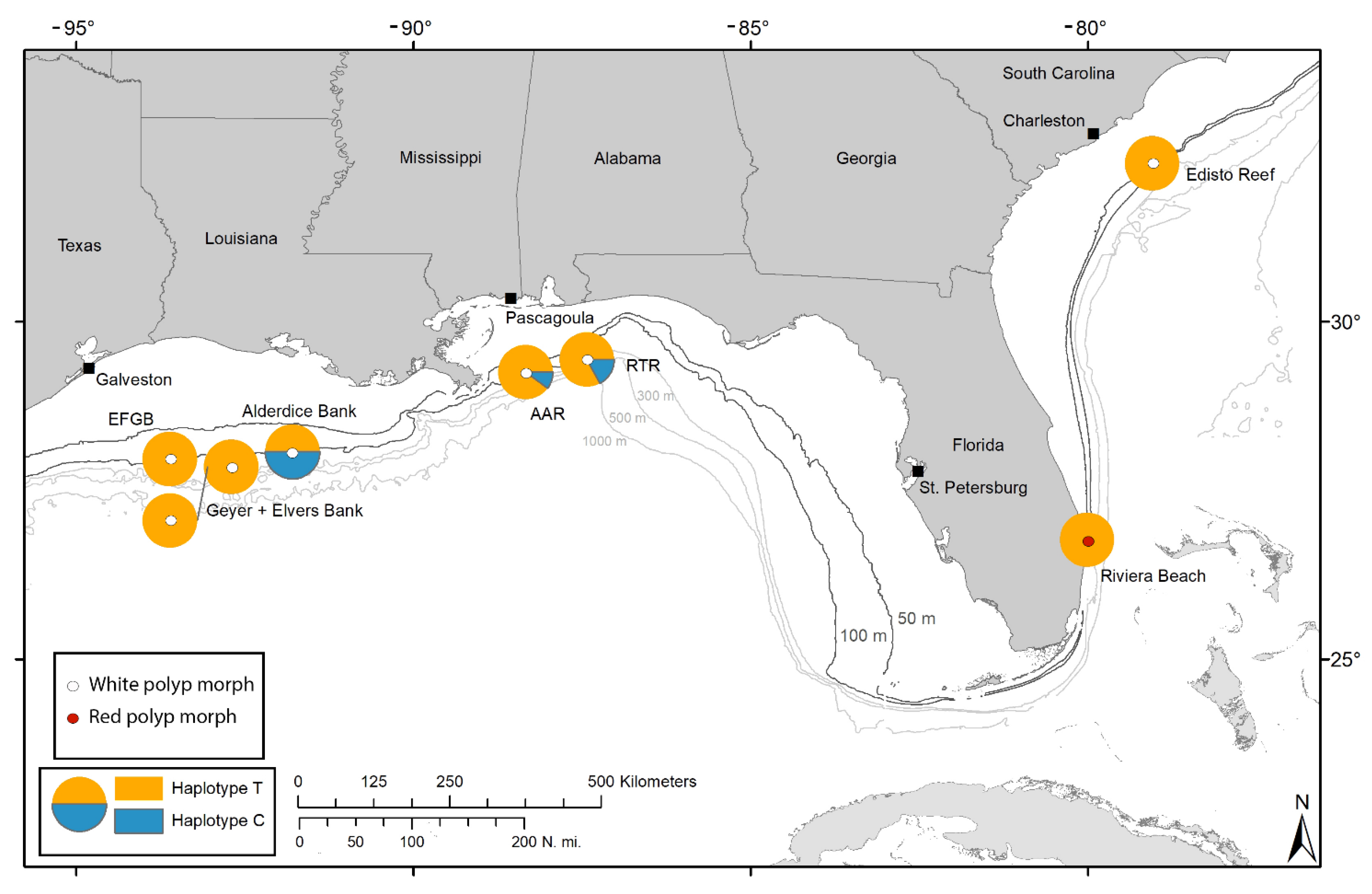
3.1. Haplotype Variation in Swiftia exserta
3.2. Haplotype Variation in Swiftia casta
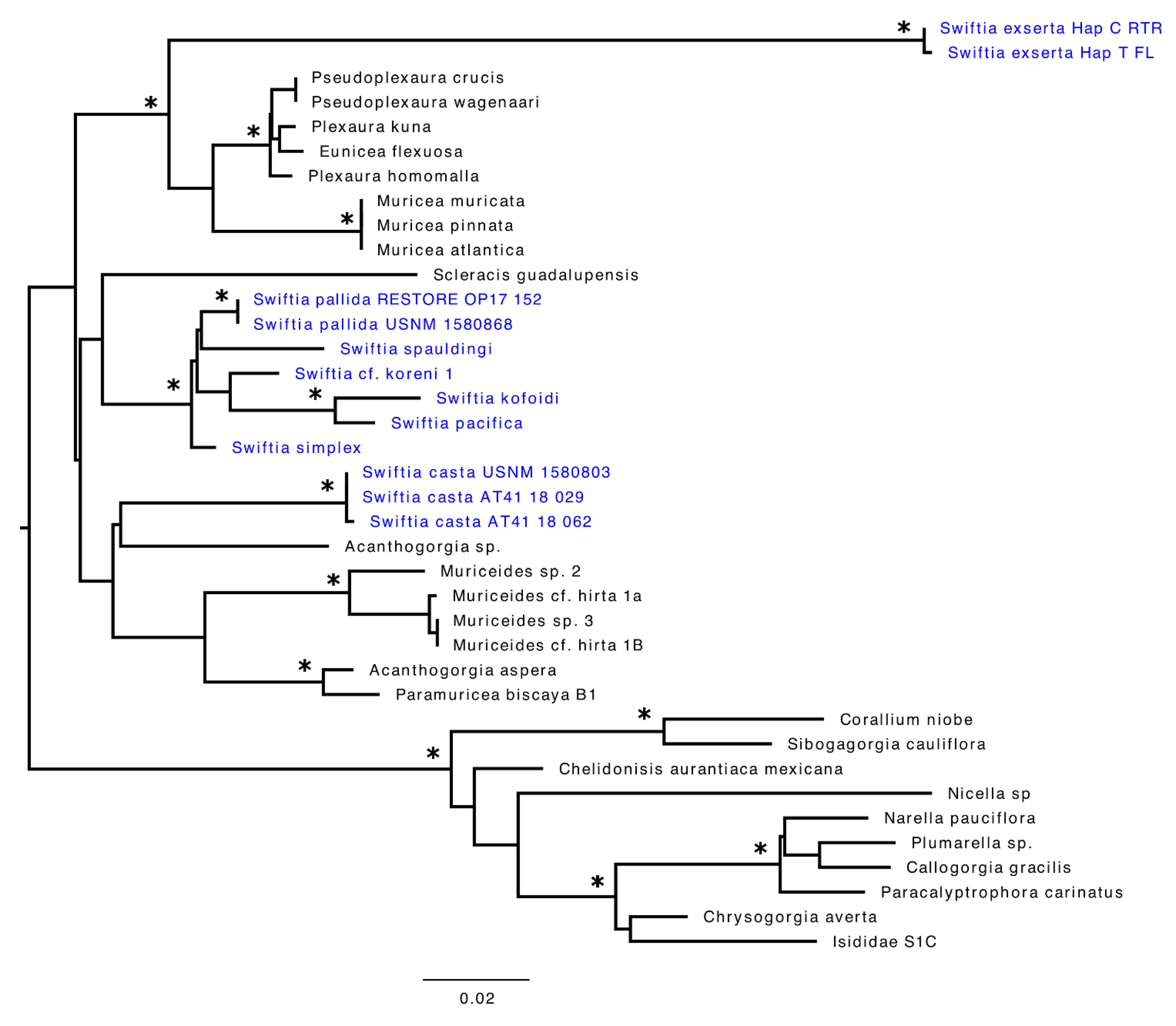
3.3. Phylogenetic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
NOAA Disclaimer
References
- Lesser, M.P.; Slattery, M.; Leichter, J.J. Ecology of mesophotic coral reefs. J. Exp. Mar. Biol. Ecol. 2009, 375, 1–8. [Google Scholar] [CrossRef]
- Etnoyer, P.J.; Wickes, L.N.; Silva, M.; Dubick, J.D.; Balthis, L.; Salgado, E.; MacDonald, I.R. Decline in condition of gorgonian octocorals on mesophotic reefs in the northern Gulf of Mexico: Before and after the Deepwater Horizon oil spill. Coral Reefs 2016, 35, 77–90. [Google Scholar] [CrossRef]
- Gardner, J.V.; Dartnell, P.; Sulak, K.J.; Calder, B.; Hellequin, L. Physiography and Late Quaternary-Holocene Processes of Northeastern Gulf of Mexico Outer Continental Shelf off Mississippi and Alabama. Gulf Mex. Sci. 2001, 19, 6. [Google Scholar] [CrossRef]
- Gittings, S.R.; Bright, T.J.; Schroeder, W.W.; Sager, W.W.; Laswell, J.S.; Rezak, R. Invertebrate assemblages and ecological controls on topographic features in the northeast Gulf of Mexico. Bull. Mar. Sci. 1992, 50, 435–455. [Google Scholar]
- Prouty, N.G.; Fisher, C.R.; Demopoulos, A.W.J.; Druffel, E.R.M. Growth rates and ages of deep-sea corals impacted by the Deepwater Horizon oil spill. Deep Sea Res. Part. Ii Top. Stud. Oceanogr. 2016, 129, 196–212. [Google Scholar] [CrossRef]
- Choy, E.; Watanabe, K.; Williams, B.; Stone, R.; Etnoyer, P.; Druffel, E.; Lorenson, T.; Knaak, M. Understanding growth and age of red tree corals (Primnoa pacifica) in the North Pacific Ocean. PLoS ONE 2020, 15, e0241692. [Google Scholar] [CrossRef]
- Mistri, M.; Ceccherelli, V.U. Growth and secondary production of the Mediterranean gorgonian Paramuricea clavata. Mar. Ecol. Prog. Ser. 1994, 103, 291. [Google Scholar] [CrossRef]
- Sulak, K.; Berg, J.; Randall, M.; Dennis, G.D., III; Brooks, R.A. Dualcarbon sources fuel the OCS deep-reef community, a stable isotope investigation. In Proceedings of the 11th International Coral Reef Symposium Proceedings, Ft. Lauderdale, FL, USA, 7–11 July 2008; Volume 2, pp. 945–949. [Google Scholar]
- Sherwood, O.A.; Heikoop, J.M.; Scott, D.B.; Risk, M.J.; Guilderson, T.P.; McKinney, R.A. Stable isotopic composition of deep-sea gorgonian corals Primnoa spp.: A new archive of surface processes. Mar. Ecol. Prog. Ser. 2005, 301, 135–148. [Google Scholar] [CrossRef]
- Silva, M.; Etnoyer, P.J.; MacDonald, I.R. Coral injuries observed at Mesophotic Reefs after the Deepwater Horizon oil discharge. Deep Sea Res. Part. Ii Top. Stud. Oceanogr. 2016, 129, 96–107. [Google Scholar] [CrossRef]
- Duchassaing, P. Mémoire sur les Coralliaires des Antilles. Mem. Della R. Accad. Delle Sci. Torinoserie Second 1860, 19, 56–87. [Google Scholar]
- Ellis, J.; Solander, D. The Natural History of Many Curious and Uncommon Zoophytes: Collected from Various Parts of The Globe; Benjamin White and Son: London, UK, 1786; p. 208. [Google Scholar]
- Goldberg, W.M. The sclerites and geographic distribution of the gorgonian Swiftia exserta (Coelenterata: Octocorallia: Holaxonia). Bull. Biol. Soc. Wash. 2001, 10, 100–109. [Google Scholar]
- Cairns, S.D.; Bayer, F.M. Octocorallia (Cnidaria) of the Gulf of Mexico. Gulf Mex. Orig. Waters Biota 2009, 1, 321–331. [Google Scholar]
- Cordeiro, R.; McFadden, C.; van Ofwegen, L.; Williams, G. World List of Octocorallia. Swiftia Duchassaing & Michelotti. 1864. Available online: http://www.marinespecies.org/aphia.php?p=taxdetails&id=125314 (accessed on 23 October 2020).
- Williams, G.C. New taxa and revisionary systematics of alcyonacean octocorals from the Pacific coast of North America (Cnidaria, Anthozoa). Zookeys 2013, 15–42. [Google Scholar] [CrossRef]
- Williams, G.C.; Breedy, O. A New Species of Gorgonian Octocoral from the Mesophotic Zone off the Central Coast of California, Eastern Pacific with a Key to Related Regional Taxa (Anthozoa, Octocorallia, Alcyonacea). Proc. Calif. Acad. Sci. 2019, 65, 143–158. [Google Scholar]
- Shuler, A.; Etnoyer, P.J. Alcyonacean octocorals of the Pinnacle Trend: A photo-identification guide. Noaa Tech. Memo. Nos Nccos 282 2020, 38–39. [Google Scholar] [CrossRef]
- Frometa, J.; DeLorenzo, M.E.; Pisarski, E.C.; Etnoyer, P.J. Toxicity of oil and dispersant on the deep water gorgonian octocoral Swiftia exserta, with implications for the effects of the Deepwater Horizon oil spill. Mar. Pollut Bull. 2017, 122, 91–99. [Google Scholar] [CrossRef]
- Frometa, J. Effects of Oil and Dispersants on Swiftia Exserta, A Structure-Forming Deepwater Gorgonian Octocoral from Mesophotic Reefs in The Gulf Of Mexico. Master’s Thesis, College of Charleston, Charleston, SC, USA, 2016. [Google Scholar]
- McFadden, C.S.; France, S.C.; Sanchez, J.A.; Alderslade, P. A molecular phylogenetic analysis of the Octocorallia (Cnidaria: Anthozoa) based on mitochondrial protein-coding sequences. Mol. Phylogenet Evol 2006, 41, 513–527. [Google Scholar] [CrossRef] [PubMed]
- McFadden, C.S.; Benayahu, Y.; Pante, E.; Thoma, J.N.; Nevarez, P.A.; France, S.C. Limitations of mitochondrial gene barcoding in Octocorallia. Mol. Ecol. Resour. 2011, 11, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Shearer, T.L.; Van Oppen, M.J.; Romano, S.L.; Worheide, G. Slow mitochondrial DNA sequence evolution in the Anthozoa (Cnidaria). Mol. Ecol. 2002, 11, 2475–2487. [Google Scholar] [CrossRef]
- Hellberg, M.E. No variation and low synonymous substitution rates in coral mtDNA despite high nuclear variation. BMC Evol. Biol. 2006, 6, 24. [Google Scholar] [CrossRef]
- Bilewitch, J.P.; Degnan, S.M. A unique horizontal gene transfer event has provided the octocoral mitochondrial genome with an active mismatch repair gene that has potential for an unusual self-contained function. BMC Evol. Biol. 2011, 11, 1–15. [Google Scholar] [CrossRef]
- Fukami, H.; Omori, M.; Hatta, M. Phylogenetic relationships in the coral family acroporidae, reassessed by inference from mitochondrial genes. Zool. Sci 2000, 17, 689–696. [Google Scholar] [CrossRef] [PubMed]
- van Oppen, M.J.; McDonald, B.J.; Willis, B.; Miller, D.J. The evolutionary history of the coral genus Acropora (Scleractinia, Cnidaria) based on a mitochondrial and a nuclear marker: Reticulation, incomplete lineage sorting, or morphological convergence? Mol. Biol. Evol. 2001, 18, 1315–1329. [Google Scholar] [CrossRef]
- Quattrini, A.M.; Wu, T.; Soong, K.; Jeng, M.S.; Benayahu, Y.; McFadden, C.S. A next generation approach to species delimitation reveals the role of hybridization in a cryptic species complex of corals. BMC Evol. Biol. 2019, 19, 116. [Google Scholar] [CrossRef]
- McFadden, C.S.; Sanchez, J.A.; France, S.C. Molecular phylogenetic insights into the evolution of Octocorallia: A review. Integr Comp. Biol. 2010, 50, 389–410. [Google Scholar] [CrossRef]
- McFadden, C.S.; van Ofwegen, L.P. Molecular phylogenetic evidence supports a new family of octocorals and a new genus of Alcyoniidae (Octocorallia, Alcyonacea). Zookeys 2013, 59–83. [Google Scholar] [CrossRef]
- McFadden, C.S.; Brown, A.S.; Brayton, C.; Hunt, C.B.; van Ofwegen, L.P. Application of DNA barcoding in biodiversity studies of shallow-water octocorals: Molecular proxies agree with morphological estimates of species richness in Palau. Coral Reefs 2014, 33, 275–286. [Google Scholar] [CrossRef]
- Herrera, S.; Baco, A.; Sanchez, J.A. Molecular systematics of the bubblegum coral genera (Paragorgiidae, Octocorallia) and description of a new deep-sea species. Mol. Phylogenet Evol. 2010, 55, 123–135. [Google Scholar] [CrossRef]
- McFadden, C.S.; van Ofwegen, L.P. Stoloniferous octocorals (Anthozoa, Octocorallia) from South Africa, with descriptions of a new family of Alcyonacea, a new genus of Clavulariidae, and a new species of Cornularia (Cornulariidae). Invertebr. Syst. 2012, 26, 331–356. [Google Scholar] [CrossRef]
- Quattrini, A.M.; Georgian, S.E.; Byrnes, L.; Stevens, A.; Falco, R.; Cordes, E.E. Niche divergence by deep-sea octocorals in the genus Callogorgia across the continental slope of the Gulf of Mexico. Mol. Ecol. 2013, 22, 4123–4140. [Google Scholar] [CrossRef] [PubMed]
- Ament-Velasquez, S.L.; Breedy, O.; Cortes, J.; Guzman, H.M.; Worheide, G.; Vargas, S. Homoplasious colony morphology and mito-nuclear phylogenetic discordance among Eastern Pacific octocorals. Mol. Phylogenet Evol. 2016, 98, 373–381. [Google Scholar] [CrossRef]
- Baums, I.B. A restoration genetics guide for coral reef conservation. Mol. Ecol. 2008, 17, 2796–2811. [Google Scholar] [CrossRef]
- Bostrom-Einarsson, L.; Babcock, R.C.; Bayraktarov, E.; Ceccarelli, D.; Cook, N.; Ferse, S.C.A.; Hancock, B.; Harrison, P.; Hein, M.; Shaver, E.; et al. Coral restoration–A systematic review of current methods, successes, failures and future directions. PLoS ONE 2020, 15, e0226631. [Google Scholar] [CrossRef]
- Population Connectivity of Deepwater Corals in the Northern Gulf of Mexico. Available online: https://restoreactscienceprogram.noaa.gov/projects/deepwater-corals (accessed on 5 December 2020).
- Program, N.D.S.C.R.T. NOAA National Database for Deep-Sea Corals and Sponges (version 20201021-0). NOAA Deep Sea Coral Research & Technology Program. Available online: https://deepseacoraldata.noaa.gov/ (accessed on 5 January 2020).
- France, S.C.; Hoover, L.L. DNA sequences of the mitochondrial COI gene have low levels of divergence among deep-sea octocorals (Cnidaria: Anthozoa). Hydrobiologia 2002, 471, 149–155. [Google Scholar] [CrossRef]
- Sánchez, J.A.; McFadden, C.S.; France, S.C.; Lasker, H.R. Molecular phylogenetic analyses of shallow-water Caribbean octocorals. Mar. Biol. 2003, 142, 975–987. [Google Scholar] [CrossRef]
- Gene Codes Corporation. Sequencher® Version 5.4.6 DNA Sequence Analysis Software; Gene Codes Corporation: Ann Arbor, MI, USA, 2017. [Google Scholar]
- Geneious Prime. Available online: https://www.geneious.com (accessed on 5 January 2020).
- Quattrini, A.M.; Etnoyer, P.J.; Doughty, C.; English, L.; Falco, R.; Remon, N.; Rittinghouse, M.; Cordes, E.E. A phylogenetic approach to octocoral community structure in the deep Gulf of Mexico. Deep Sea Res. Part. Ii Top. Stud. Oceanogr. 2014, 99, 92–102. [Google Scholar] [CrossRef]
- Quattrini, A.M.; Gomez, C.E.; Cordes, E.E. Environmental filtering and neutral processes shape octocoral community assembly in the deep sea. Oecologia 2016, 183, 221–236. [Google Scholar] [CrossRef]
- Wirshing, H.H.; Messing, C.G.; Douady, C.J.; Reed, J.; Stanhope, M.J.; Shivji, M.S. Molecular evidence for multiple lineages in the gorgonian family Plexauridae (Anthozoa: Octocorallia). Mar. Biol. 2005, 147, 497–508. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucl. Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.L. PAUP*. Phylogenetic Analysis Using Parsimony (Version 4. Sinauer Associates, Sunderland, MA, USA). 2003. Available online: https://paup.phylosolutions.com/ (accessed on 5 January 2020).
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; Drummond, A.J.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Herrera, S.; Shank, T.M. RAD sequencing enables unprecedented phylogenetic resolution and objective species delimitation in recalcitrant divergent taxa. Phylogenet. Evol. 2016, 100, 70–79. [Google Scholar] [CrossRef]
- Untiedt, C.B.; Quattrini, A.M.; McFadden, C.S.; Alderslade, P.A.; Pante, E.; Burridge, C.P. Phylogenetic Relationships Within Chrysogorgia (Alcyonacea: Octocorallia), a Morphologically Diverse Genus of Octocoral, Revealed Using a Target Enrichment Approach. Front. Mar. Sci. 2021, 7, hal-03154113. [Google Scholar] [CrossRef]
- Calixto Botia, I.; Sanchez, J.A. A case of modular phenotypic plasticity in the depth gradient for the gorgonian coral Antillogorgia bipinnata (Cnidaria: Octocorallia). BMC Evol. Biol. 2017, 17, 1–8. [Google Scholar] [CrossRef]
- Sanchez, J.A.; Aguilar, C.; Dorado, D.; Manrique, N. Phenotypic plasticity and morphological integration in a marine modular invertebrate. BMC Evol. Biol. 2007, 7, 122. [Google Scholar] [CrossRef]
- Goldberg, W.M. The ecology of the coral-octocoral communities of the Southeast Florida Coast: Geomorphology, species composition, and zonation. Bull. Mar. Sci. 1973, 23, 465–488. [Google Scholar]
- Breedy, O.; Cairns, S.D.; Haussermann, V. A new alcyonacean octocoral (Cnidaria, Anthozoa, Octocorallia) from Chilean fjords. Zootaxa 2015, 3919, 327–334. [Google Scholar] [CrossRef][Green Version]
- Breedy, O.; Rouse, G.W.; Stabbins, A.; CortÉS, J.; Cordes, E.E. New records of Swiftia (Cnidaria, Anthozoa, Octocorallia) from off the Pacific Costa Rican margin, including a new species from methane seeps. Zootaxa 2019, 4671, 407–419. [Google Scholar] [CrossRef]
- Bayer, F.M. The shallow-water Octocorallia of the West Indian region. Stud. Fauna Curacao Caribb. Isl. 1961, 12, 1–373. [Google Scholar]
- Deichmann, E. The Alcyonaria of the Western Part of the Atlantic Ocean. Mem. Mus. Comp. Zool. Harv. Coll. 1936, 53, 1–317. [Google Scholar]
- Bayer, F.M. Key to the genera of Octocorallia exclusive of Pennatulacea (Coelenterata: Anthozoa), with diagnosis of new taxa. Proc. Biol. Soc. Wash. 1981, 49, 363–373. [Google Scholar]
- Pante, E.; Abdelkrim, J.; Viricel, A.; Gey, D.; France, S.C.; Boisselier, M.C.; Samadi, S. Use of RAD sequencing for delimiting species. Heredity (Edinb) 2015, 114, 450–459. [Google Scholar] [CrossRef]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Maliet, O.; Gascuel, F.; Lambert, A. Ranked Tree Shapes, Nonrandom Extinctions, and the Loss of Phylogenetic Diversity. Syst. Biol. 2018, 67, 1025–1040. [Google Scholar] [CrossRef] [PubMed]
- Purvis, A.; Agapow, P.M.; Gittleman, J.L.; Mace, G.M. Nonrandom extinction and the loss of evolutionary history. Science 2000, 288, 328–330. [Google Scholar] [CrossRef]
- White, H.K.; Hsing, P.Y.; Cho, W.; Shank, T.M.; Cordes, E.E.; Quattrini, A.M.; Nelson, R.K.; Camilli, R.; Demopoulos, A.W.; German, C.R.; et al. Impact of the Deepwater Horizon oil spill on a deep-water coral community in the Gulf of Mexico. Proc. Natl. Acad. Sci. USA 2012, 109, 20303–20308. [Google Scholar] [CrossRef] [PubMed]

| Swiftia exserta | Swiftia casta | Swiftia kofoidi | Swiftia koreni | Swiftia pacifica | Swiftia pallida | Swiftia simplex | |
|---|---|---|---|---|---|---|---|
| (a) | |||||||
| S. casta | 14.9 | ||||||
| S. kofoidi | 15.1 | 9.2 | |||||
| S. koreni | 13.5 | 7.1 | 3.9 | ||||
| S. pacifica | 14.6 | 8.8 | 2.1 | 3.2 | |||
| S. pallida | 13.3 | 6.4 | 4.5 | 2.1 | 3.7 | ||
| S. simplex | 13.3 | 6.4 | 4.2 | 1.8 | 3.4 | 1.1 | |
| S. spauldingi | 14.4 | 7.1 | 5.9 | 3.5 | 4.9 | 2.7 | 2.7 |
| (b) | |||||||
| S. casta | 13.7 | ||||||
| S. koreni | 12.2 | 8.1 | |||||
| S. pallida | 13.2 | 8.2 | ---- | 1.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frometa, J.; Etnoyer, P.J.; Quattrini, A.M.; Herrera, S.; Greig, T.W. Genetic Divergence and Polyphyly in the Octocoral Genus Swiftia [Cnidaria: Octocorallia], Including a Species Impacted by the DWH Oil Spill. Diversity 2021, 13, 172. https://doi.org/10.3390/d13040172
Frometa J, Etnoyer PJ, Quattrini AM, Herrera S, Greig TW. Genetic Divergence and Polyphyly in the Octocoral Genus Swiftia [Cnidaria: Octocorallia], Including a Species Impacted by the DWH Oil Spill. Diversity. 2021; 13(4):172. https://doi.org/10.3390/d13040172
Chicago/Turabian StyleFrometa, Janessy, Peter J. Etnoyer, Andrea M. Quattrini, Santiago Herrera, and Thomas W. Greig. 2021. "Genetic Divergence and Polyphyly in the Octocoral Genus Swiftia [Cnidaria: Octocorallia], Including a Species Impacted by the DWH Oil Spill" Diversity 13, no. 4: 172. https://doi.org/10.3390/d13040172
APA StyleFrometa, J., Etnoyer, P. J., Quattrini, A. M., Herrera, S., & Greig, T. W. (2021). Genetic Divergence and Polyphyly in the Octocoral Genus Swiftia [Cnidaria: Octocorallia], Including a Species Impacted by the DWH Oil Spill. Diversity, 13(4), 172. https://doi.org/10.3390/d13040172






