Genetic Diversity of Campomanesia adamantium and Its Correlation with Land Use and Land Cover
Abstract
1. Introduction
2. Materials and Methods
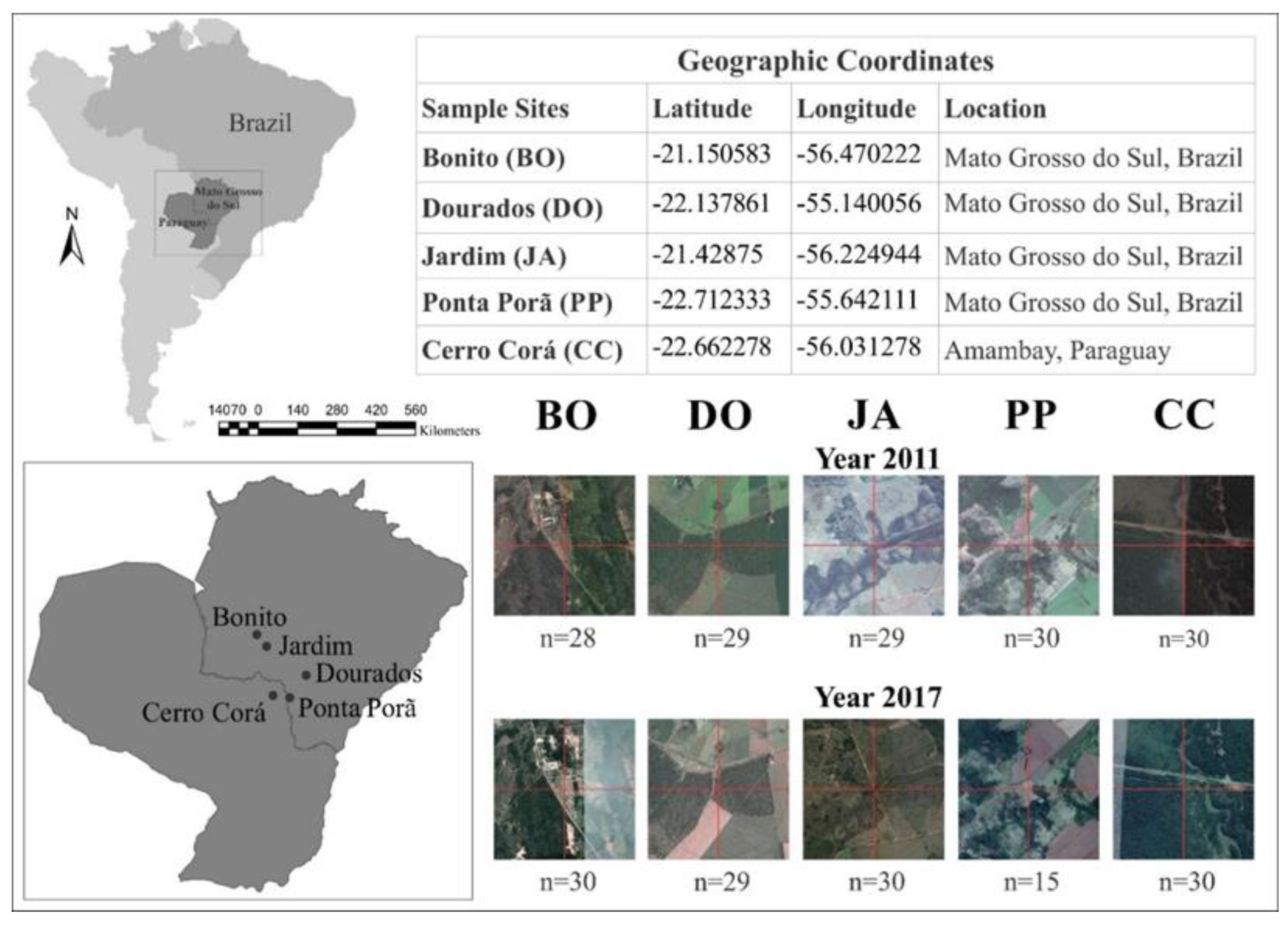
2.1. Study and Sampling Area
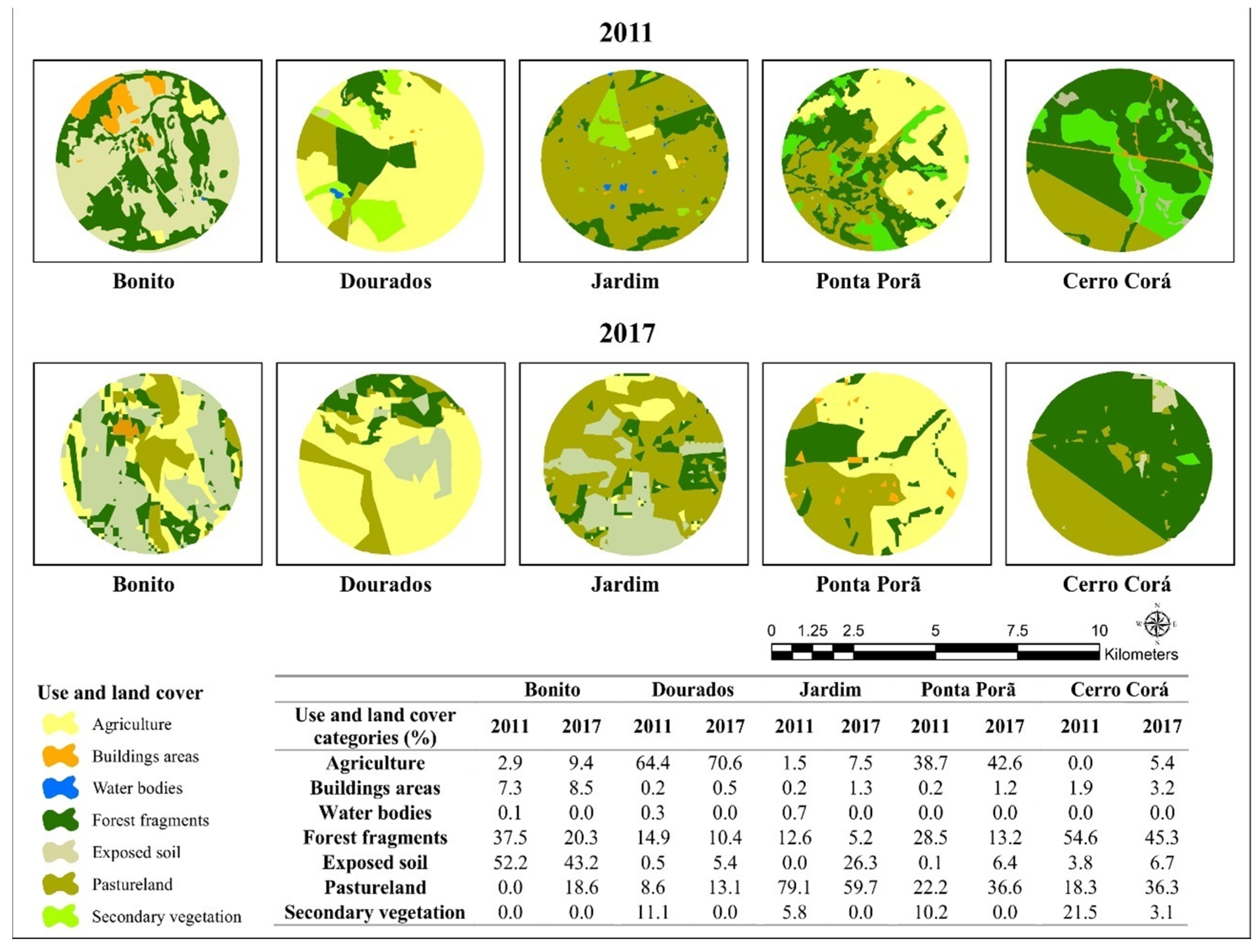
2.2. Characterization of the Collection Sites
2.3. DNA Extraction and Genotyping
2.4. Analysis of Genetic Diversity According to Year and Population
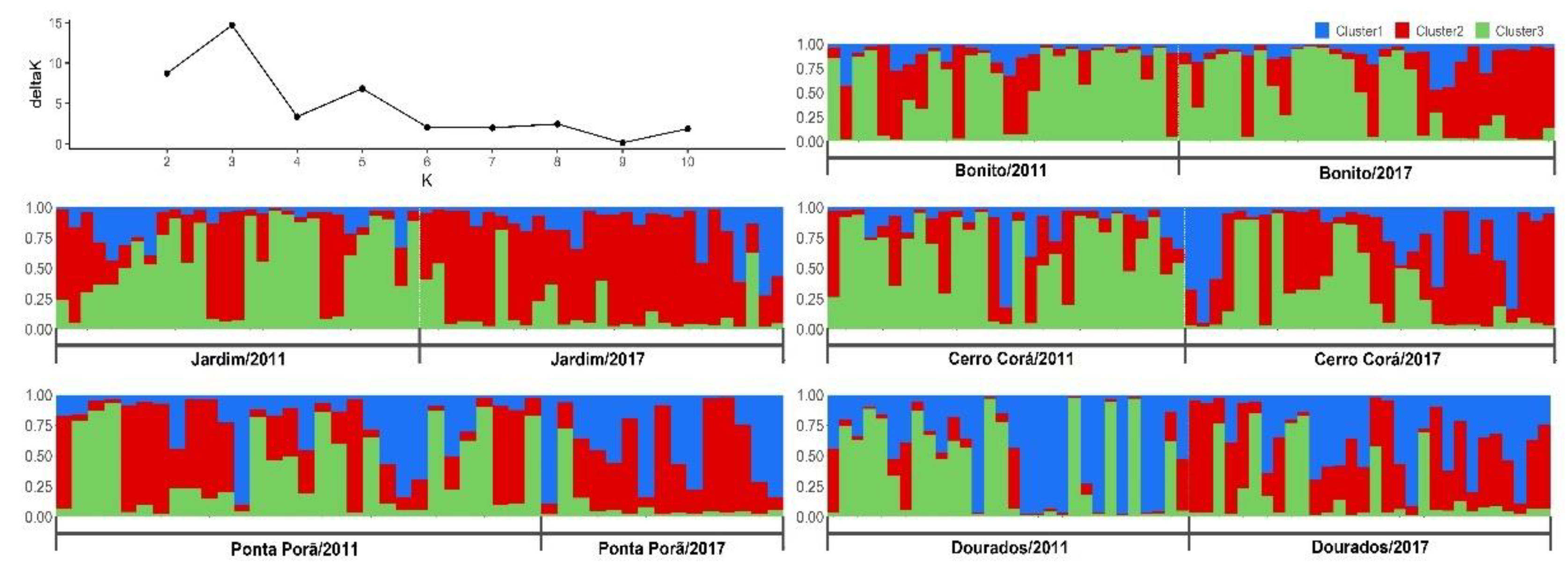
2.5. Population Structure
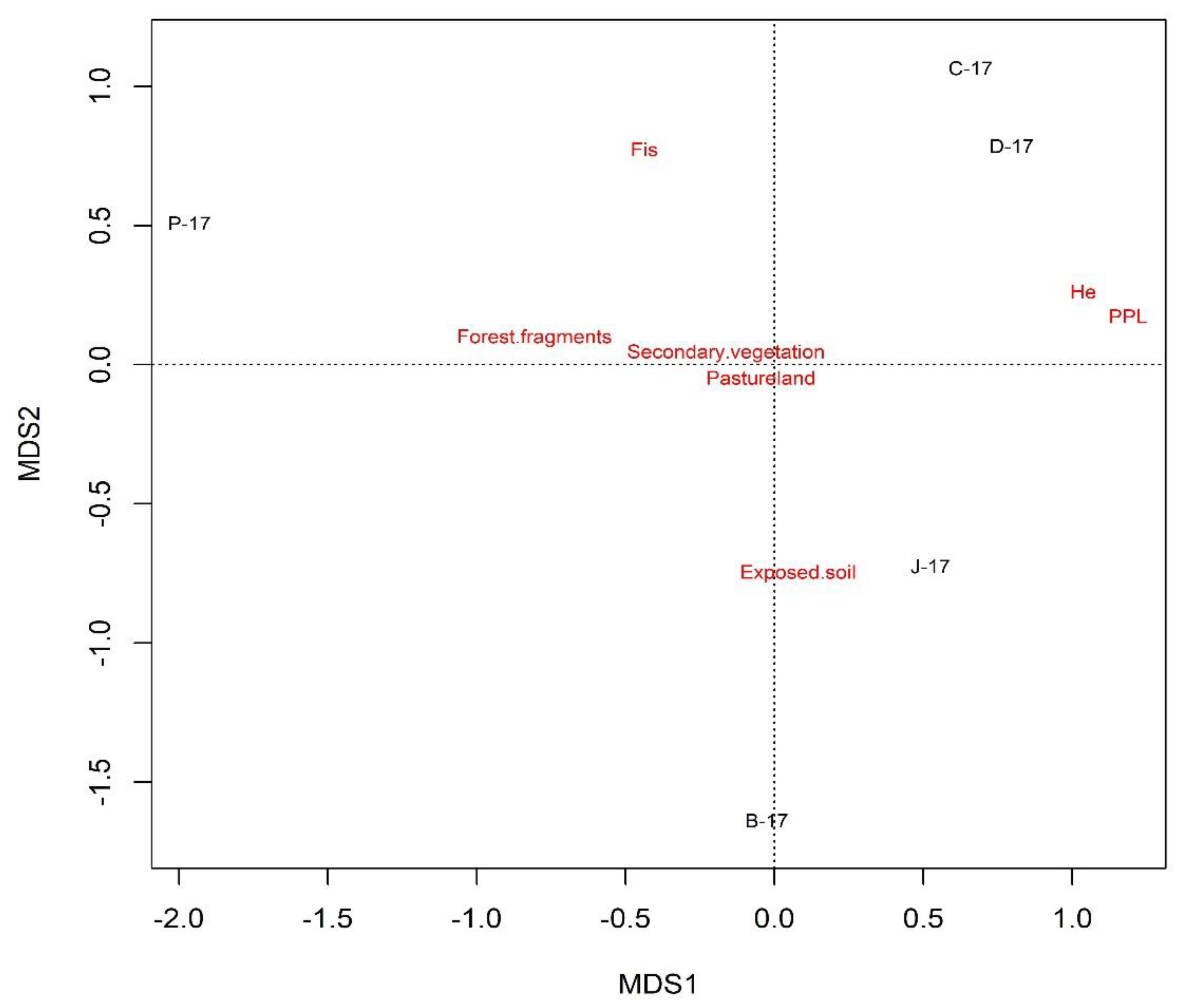
2.6. Correlation of Genetic Diversity with Land Use and Land Cover
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Forzza, R.C.; Baumgratz, J.F.A.; Bicudo, C.E.M.; Canhos, D.A.; Carvalho, A.A., Jr.; Coelho, M.A.N.; Costa, A.F.; Costa, D.P.; Hopkins, M.G.; Leitman, P.M.; et al. New Brazilian floristic list highlights conservation challenges. BioScience 2012, 62, 39–45. [Google Scholar] [CrossRef]
- Françoso, R.D.; Brandão, R.; Nogueira, C.C.; Salmona, Y.B.; Machado, R.B.; Colli, G.R. Habitat loss and the effectiveness of protected areas in the Cerrado Biodiversity Hotspot. Nat. Conserv. 2015, 13, 35–40. [Google Scholar] [CrossRef]
- Strassburg, B.B.; Brooks, T.; Feltran-Barbieri, R.; Iribarrem, A.; Crouzeilles, R.; Loyola, R.; Latawiec, A.E.; Oliveira Filho, F.J.B.; Scaramuzza, C.A.M.; Scarano, F.R.; et al. Moment of truth for the Cerrado hotspot. Nat. Ecol. Evol. 2017, 1, 0099. [Google Scholar] [CrossRef]
- Velazco, S.J.E.; Villalobos, F.; Galvão, F.; Júnior, P.M. A dark scenario for Cerrado plant species: Effects of future climate, land use and protected areas ineffectiveness. Divers. Distrib. 2019, 25, 660–673. [Google Scholar] [CrossRef]
- Nucci, M.; Alves-Junior, V.V. Biologia floral e sistema reprodutivo de Campomanesia adamantium (Cambess.) O. Berg-myrtaceae em área de Cerrado no sul do Mato Grosso do Sul, Brasil. Interciencia 2017, 42, 127–131. [Google Scholar]
- Barrett, S.C.H.; Harder, L.D. Ecology and evolution of plant mating. Trends. Ecol. Evol. 1996, 1173–1179. [Google Scholar] [CrossRef]
- Ghazoul, J. Pollen and seed dispersal among dispersed plants. Biol. Rev. 2005, 80, 413–443. [Google Scholar] [CrossRef]
- Dresch, D.M.; Masetto, T.E.; Scalon, S.P.Q. Campomanesia adamantium (Cambess.) O. Berg seed desiccation: Influence on vigor and nucleic acids. An. Acad. Bras. Ciênc. 2015, 87, 2217–2228. [Google Scholar] [CrossRef]
- Cardozo, C.M.L.; Inada, A.C.; Marcelino, G.; Figueiredo, P.S.; Arakaki, D.G.; Hiane, P.A.; Cardoso, C.A.L.; Guimarães, R.C.A.; Freitas, K.C. Therapeutic Potential of Brazilian Cerrado Campomanesia Species on Metabolic Dysfunctions. Molecules 2018, 23, 2336. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, I.D.; Cardoso, C.A.L.; Ré-Poppi, N.; Melo, A.M.; Vieira, M.C.; Honda, N.K.; Coelho, R.G. Gas Chromatography-Mass Spectrometry (GC-MS) and evaluation of antioxidant and antimicrobial activities of essential oil of Campomanesia adamantium (Cambess.) O. Berg (Guavira). Braz. J. Pharm. Sci. 2009, 45, 767–776. [Google Scholar] [CrossRef]
- Pavan, F.R.; Leite, C.Q.F.; Coelho, R.G.; Coutinho, I.D.; Honda, N.K.; Cardoso, C.A.L.; Vilegas, W.; Leite, S.R.A.; Sato, D.N. Evaluation of anti Mycobacterium tuberculosis activity of Campomanesia adamantium (Myrtaceae). Quím. Nova 2009, 32, 1222–1226. [Google Scholar] [CrossRef]
- Cardoso, C.A.L.; Salmazzo, G.R.; Honda, N.K.; Prates, C.B.; Vieira, M.C.; Coelho, R.G. Antimicrobial activity of the extracts and fractions of hexanic fruits of Campomanesia species (Myrtaceae). J. Med. Food 2010, 13, 1273–1276. [Google Scholar] [CrossRef] [PubMed]
- Souza, J.C.; Piccinelli, A.C.; Aquino, D.F.; de Souza, V.V.; Schmitz, W.O.; Traesel, G.K.; Cardoso, C.A.; Kassuya, C.A.; Arena, A.C. Toxicological analysis and antihyperalgesic, antidepressant, and anti-inflammatory effects of Campomanesia adamantium fruit barks. Nutr. Neurosci. 2017, 20, 23–31. [Google Scholar] [CrossRef]
- Coutinho, I.D.; Kataoka, V.M.F.; Honda, N.K.; Coelho, R.G.; Vieira, M.C.; Cardoso, C.A.L. The Influence of seasonal variation in levels of flavonoids and antioxidant activity of the leaves of Campomanesia adamantium. Rev. Bras. Farmacogn. 2010, 20, 322–327. [Google Scholar] [CrossRef]
- Espindola, P.P.T.; Rocha, P.S.; Carollo, C.A.; Schmit, W.O.; Pereira, Z.V.; Vieira, M.C.; Santos, E.L.; Souza, K.P. Antioxidant and antihyperlipidemic effects of Campomanesia adamantium O. Berg root. Oxid. Med. Cell Longev. 2016, 2016, 1–9. [Google Scholar] [CrossRef]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004, 5, 435–445. [Google Scholar] [CrossRef]
- Oliveira, E.J.; Pádua, J.G.; Zucchi, M.I.; Vencovsky, R.; Vieira, M.L.C. Origin, evolution and genome distribution of microsatellites. Genet. Mol. Res. 2006, 29, 294–307. [Google Scholar] [CrossRef]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; Munhoz, C.F. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef]
- Miranda, E.A.; Boaventura-Novaes, C.R.; Braga, R.S.; Reis, E.F.; Pinto, J.F.; Telles, M.P. Validation of EST-derived microsatellite markers for two Cerrado-endemic Campomanesia (Myrtaceae) species. Genet. Mol. Res. 2016, 15, 1–6. [Google Scholar] [CrossRef]
- Crispim, B.A.; Bajay, M.M.; Vasconcelos, A.A.; Deo, T.G.; Braga, R.S.; Telles, M.P.C.; Vieira, M.C.; Carnevali, T.O.; Solórzano, J.C.J.; Grisolia, A.B. Relationship between genetic variability and land use and land cover in populations of Campomanesia adamantium (Myrtaceae). Diversity 2018, 10, e106. [Google Scholar] [CrossRef]
- Crispim, B.A.; Déo, T.G.; Fernandes, J.S.; Vasconcelos, A.A.; Vieira, M.C.; Carnevali, T.O.; Bajay, M.M.; Zucchi, M.I.; Barufatti, A. Development and characterization of microsatellite markers in Campomanesia adamantium, a native plant of the Cerrado ecoregions of South America. Appl. Plant. Sci. 2019, 7, e11287. [Google Scholar] [CrossRef] [PubMed]
- Hansen, A.J.; DeFries, R.S.; Turner, W. Land Use Change and Biodiversity. In Land Change Science. Remote Sensing and Digital Image Processing; Gutman, G., Janetos, A.C., Justice, C.O., Moran, E.F., Mustard, J.F., Rindfuss, R.R., Skole, D., Turner, B.L., II, Cochrane, M.A., Eds.; Springer: Dordrecht, The Netherlands, 2012; Volume 6. [Google Scholar] [CrossRef]
- Blambert, L.; Mallet, B.; Humeau, L.; Pailler, T. Reproductive patterns, genetic diversity and in-mating depression in two closely related Jumellea species with contrasting patterns of commonness and distribution. Ann. Bot. 2016, 118, 93–103. [Google Scholar] [CrossRef]
- ESRI—Environmental Systems Research Institute. ArcGIS Desktop Trial Version®: Release 10.4. Redlands; Environmental Systems Research Institute: Redlands, CA, USA, 2015. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 39–40. [Google Scholar]
- R Development Core Team. R: A Language and Evironment for Statistical Computing, Version 3.4.4. R Core Team, Vienna, Austria. 2019. Available online: http://www.R-project.org/ (accessed on 23 September 2020).
- Jombart, T. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef] [PubMed]
- Keenan, K.; McGinnity, P.; Cross, T.F.; Crozier, W.W.; Prodöhl, P.A. diveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol. Evol. 2013, 4, 782–788. [Google Scholar] [CrossRef]
- Rioux Paquette, S. PopGenKit: Useful Functions for (Batch) File Conversion and Data Resampling in Microsatellite Datasets. R Package, Version 1.0. Available online: http://cran.r-project.org/web/packages/PopGenKit/index.html (accessed on 23 September 2020).
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 59–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Cavanzón-Medrano, L.; Machkour-M’Rabet, S.; Chablé-Iuit, L.; Pozo, C.; Hénaut, Y.; Legal, L. Effect of climatic conditions and land cover on genetic structure and diversity of Eunica tatila (Lepidoptera) in the Yucatan Peninsula, Mexico. Diversity 2018, 10, 79. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package, Version 2.5-4. 2019. Available online: https://cran.r-project.org/web/packages/vegan/vegan.pdf (accessed on 23 September 2019).
- Harrell, F.E., Jr. Hmisc: Harrell miscellaneous. R Package Version 4.2-0. 2019. Available online: https://cran.r-project.org/web/packages/Hmisc/Hmisc.pdf (accessed on 25 September 2020).
- Leberg, P.L. Estimating allelic richness: Effects of sample size and bottlenecks. Mol. Ecol. 2002, 11, 2445–2449. [Google Scholar] [CrossRef]
- Hughes, A.R.; Inouye, B.D.; Johnson, M.T.; Underwood, N.; Vellend, M. Ecological consequences of genetic diversity. Ecol. Lett. 2008, 11, 609–623. [Google Scholar] [CrossRef]
- Greenbaum, G.; Templeton, A.R.; Zarmi, Y.; Bar-David, S. Allelic richness following population founding events–a stochastic modeling framework incorporating gene flow and genetic drift. PLoS ONE 2014, 9, e115203. [Google Scholar] [CrossRef]
- Queirós, J.; Godinho, R.; Lopes, S.; Gortazar, C.; de la Fuente, J.; Alves, P.C. Effect of microsatellite selection on individual and population genetic inferences: An empirical study using cross-specific and species-specific amplifications. Mol. Ecol. Resour. 2015, 15, 747–760. [Google Scholar] [CrossRef]
- Carnevali, T.O.; Vieira, M.C.; Souza, N.H.; Ramos, D.D.; Heredia Zárate, N.A.; Cardoso, C.A.L. Espaçamentos entre plantas e adição de cama-de-frango na produção de biomassa das plantas e na composição química dos frutos da Campomanesia adamantium (Cambess.) O. Berg. Rev. Bras. Plantas Med. 2012, 14, 680–685. [Google Scholar] [CrossRef]
- Hedrick, P.W. Genetics of Populations, 4th ed.; Jones and Bartlett Publishers: Sudbury, MA, USA, 2011; 75p. [Google Scholar]
- Trang, N.T.P.; Triest, L. Genetic structure of the threatened Hopea chinensis in the Quang Ninh Province, Vietnam. Genet. Mol. Res. 2016, 15, e15028103. [Google Scholar] [CrossRef]
- Nucci, M.; Alves-Junior, V.V. Behavior and diversity of floral visitors to Campomanesia adamantium (Myrtaceae). Rev. Colom. Entomol. 2017, 43, 106–112. [Google Scholar] [CrossRef]
- Goulson, D.; Nicholls, E.; Botías, C.; Rotheray, E.L. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 2015, 347, 255957. [Google Scholar] [CrossRef] [PubMed]
- Giustina, L.D.; Luz, L.N.; Vieira, F.S.; Rossi, F.S.; Soares-Lopes, C.R.; Pereira, T.N.; Rossi, A.A. Population structure and genetic diversity in natural populations of Theobroma speciosum Willd. Ex Spreng (Malvaceae). Genet. Mol. Res. 2014, 13, 3510–3519. [Google Scholar] [CrossRef] [PubMed]
- Balloux, F.; Lugon-Moulin, N. The estimation of population differentiation with microsatellite markers. Mol. Ecol. 2002, 11, 155–165. [Google Scholar] [CrossRef] [PubMed]
- McGarigal, K.; Cushman, S.A. Comparative evaluation of experimental approaches to the study of habitat fragmentation effects. Ecol. Appl. 2002, 12, 335–345. [Google Scholar] [CrossRef]
- Fahrig, L. Effects of habitat fragmentation on biodiversity. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 487–515. [Google Scholar] [CrossRef]
- Wilson, M.C.; Chen, X.; Corlett, R.T.; Didham, R.K.; Ding, P.; Holt, R.D.; Holyoak, M.; Hu, G.; Hughes, A.C.; Jiang, L.; et al. Habitat fragmentation and biodiversity conservation: Key findings and future challenges. Landsc. Ecol. 2016, 31, 219–227. [Google Scholar] [CrossRef]
- Heinken, T.; Weber, E. Consequences of habitat fragmentation for plant species: Do we know enough? Perspect Plant. Ecol. Syst. 2013, 15, 205–216. [Google Scholar] [CrossRef]
- Pilon, N.A.L.; Durigan, G. Critérios para indicação de espécies prioritárias para a restauração da vegetação de cerrado. Sci. For. 2013, 41, 389–399. [Google Scholar]
- Lescano, C.H.; de Oliveira, I.P.; Zaminelli, T.; Baldivia, D.D.; da Silva, L.R.; Napolitano, M.; Silvério, C.B.; Lincopan, N.; Sanjinez-Argandoña, E.J. Campomanesia adamantium peel extract in antidiarrheal activity: The ability of inhibition of heat-stable enterotoxin by polyphenols. PLoS ONE 2016, 11, e0165208. [Google Scholar] [CrossRef]
- Prefeitura Municipal de Bonito. 15° Festival da Guavira Começa Hoje. Bonito: Mato Grosso do Sul. 2018. Available online: http://www.bonito.ms.gov.br/noticias/15-festival-da-guavira-comeca-hoje-confira-a-programacao (accessed on 20 September 2020).
- Weir, B.S. Genetic Data Analysis II: Methods for Discrete Poopulation Genetics, 2nd ed.; Sinauer Associates: Cary, NC, USA, 1996; 445p. [Google Scholar]
- Chapuis, M.P.; Estoup, A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 2007, 24, 621–631. [Google Scholar] [CrossRef]
| N | A | AR (95% CI) | HE (95% CI) | HO (95% CI) | FIS (95% CI) | HWE | ||
|---|---|---|---|---|---|---|---|---|
| Year | ||||||||
| 2011 | 146 | 15.5 | 14.34 (13.7–14.9) | 0.86 (0.85–0.87) | 0.49 (0.46–0.52) | 0.43 (0.39–0.47) | 0 | |
| 2017 | 134 | 15.3 | 14.33 (13.8–14.8) | 0.87 (0.85–0.87) | 0.54 (0.49–0.56) | 0.39 (0.36–0.43) | 0 | |
| Pop | ||||||||
| 2011 | BO | 28 | 9.9 | 7.54 (6.60–8.50) | 0.83 (0.80–0.86) | 0.55 (0.47–0.61) | 0.35 (0.26–0.43) | 5 |
| DO | 29 | 9.5 | 6.56 (5.60–7.50) | 0.81 (0.78–0.83) | 0.37 (0.30–0.43) | 0.54 (0.47–0.62) | 1 | |
| JA | 29 | 10.7 | 7.91 (6.80–8.90) | 0.85 (0.82–0.87) | 0.59 (0.52–0.65) | 0.31 (0.24–0.38) | 3 | |
| PP | 30 | 10.4 | 7.25 (6.20–8.20) | 0.83 (0.79–0.86) | 0.51 (0.43–0.57) | 0.40 (0.32–0.48) | 4 | |
| CC | 30 | 10.6 | 7.55 (6.50–8.50) | 0.84 (0.81–0.86) | 0.49 (0.41–0.56) | 0.42 (0.32–0.51) | 2 | |
| 2017 | BO | 30 | 10 | 7.31 (6.30–8.10) | 0.82 (0.78–0.85) | 0.60 (0.54–0.66) | 0.26 (0.18–0.32) | 2 |
| DO | 29 | 11.4 | 7.62 (6.50–8.70) | 0.84 (0.81–0.87) | 0.49 (0.40–0.54) | 0.38 (0.36–0.52) | 1 | |
| JA | 30 | 10.2 | 7.31 (6.30–8.20) | 0.84 (0.80–0.86) | 0.62 (0.53–0.64) | 0.30 (0.22–0.36) | 3 | |
| PP | 15 | 7.6 | 6.15 (5.1–7.10) | 0.79 (0.75–0.83) | 0.44 (0.33–0.52) | 0.47 (0.34–0.59) | 4 | |
| CC | 30 | 10.8 | 7.71 (6.70–8.60) | 0.85 (0.81–0.87) | 0.49 (0.43–0.55) | 0.42 (0.35–0.49) | 1 | |
| Population | ta | |
|---|---|---|
| 2011 | 2017 | |
| Bonito | 0.48 | 0.58 |
| Dourados | 0.29 | 0.45 |
| Jardim | 0.52 | 0.54 |
| Ponta Porã | 0.42 | 0.36 |
| Cerro Corá | 0.41 | 0.41 |
| Média | 0.42 | 0.47 |
| Source of Variation | Degree of Freedom | Sum of Squares | Mean Square | Estimated Variance | Variance Percentage |
|---|---|---|---|---|---|
| Among years | 1 | 22.211 | 22.211 | 0.056 | 1% |
| Among individuals within populations within years | 278 | 1786.790 | 6.427 | 2.067 | 47% |
| Among individuals within of each year | 280 | 642.000 | 2.293 | 2.293 | 52% |
| Total | 559 | 2451.002 | 4.417 | 100% | |
| FST = 0.013. p = 0.001 | |||||
| Correlation | Pearson Statistics | |
|---|---|---|
| r | p | |
| HE and Agriculture | −0.167 | 0.788 |
| HE and Buildings areas | −0.017 | 0.978 |
| HE and Forest fragments | 0.453 | 0.444 |
| HE and Exposed soil | −0.096 | 0.878 |
| HE and Pastureland | −0.081 | 0.898 |
| HE and Secondary vegetation | 0.874 | 0.05 * |
| FIS and Agriculture | 0.644 | 0.24 |
| FIS and Buildings areas | −0.676 | 0.211 |
| FIS and Forest fragments | 0.173 | 0.781 |
| FIS and Exposed soil | −0.957 | 0.011 * |
| FIS and Pastureland | −0.183 | 0.769 |
| FIS and Secondary vegetation | 0.558 | 0.328 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Crispim, B.d.A.; Fernandes, J.d.S.; Bajay, M.M.; Zucchi, M.I.; Batista, C.E.d.A.; Vieira, M.d.C.; Barufatti, A. Genetic Diversity of Campomanesia adamantium and Its Correlation with Land Use and Land Cover. Diversity 2021, 13, 160. https://doi.org/10.3390/d13040160
Crispim BdA, Fernandes JdS, Bajay MM, Zucchi MI, Batista CEdA, Vieira MdC, Barufatti A. Genetic Diversity of Campomanesia adamantium and Its Correlation with Land Use and Land Cover. Diversity. 2021; 13(4):160. https://doi.org/10.3390/d13040160
Chicago/Turabian StyleCrispim, Bruno do Amaral, Juliana dos Santos Fernandes, Miklos Maximiliano Bajay, Maria Imaculada Zucchi, Carlos Eduardo de Araújo Batista, Maria do Carmo Vieira, and Alexeia Barufatti. 2021. "Genetic Diversity of Campomanesia adamantium and Its Correlation with Land Use and Land Cover" Diversity 13, no. 4: 160. https://doi.org/10.3390/d13040160
APA StyleCrispim, B. d. A., Fernandes, J. d. S., Bajay, M. M., Zucchi, M. I., Batista, C. E. d. A., Vieira, M. d. C., & Barufatti, A. (2021). Genetic Diversity of Campomanesia adamantium and Its Correlation with Land Use and Land Cover. Diversity, 13(4), 160. https://doi.org/10.3390/d13040160






