Mitogenomics and the Phylogeny of Mantis Shrimps (Crustacea: Stomatopoda)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. DNA Extraction and Sequencing
2.3. Mitochondrial Genome Assembly and Annotation
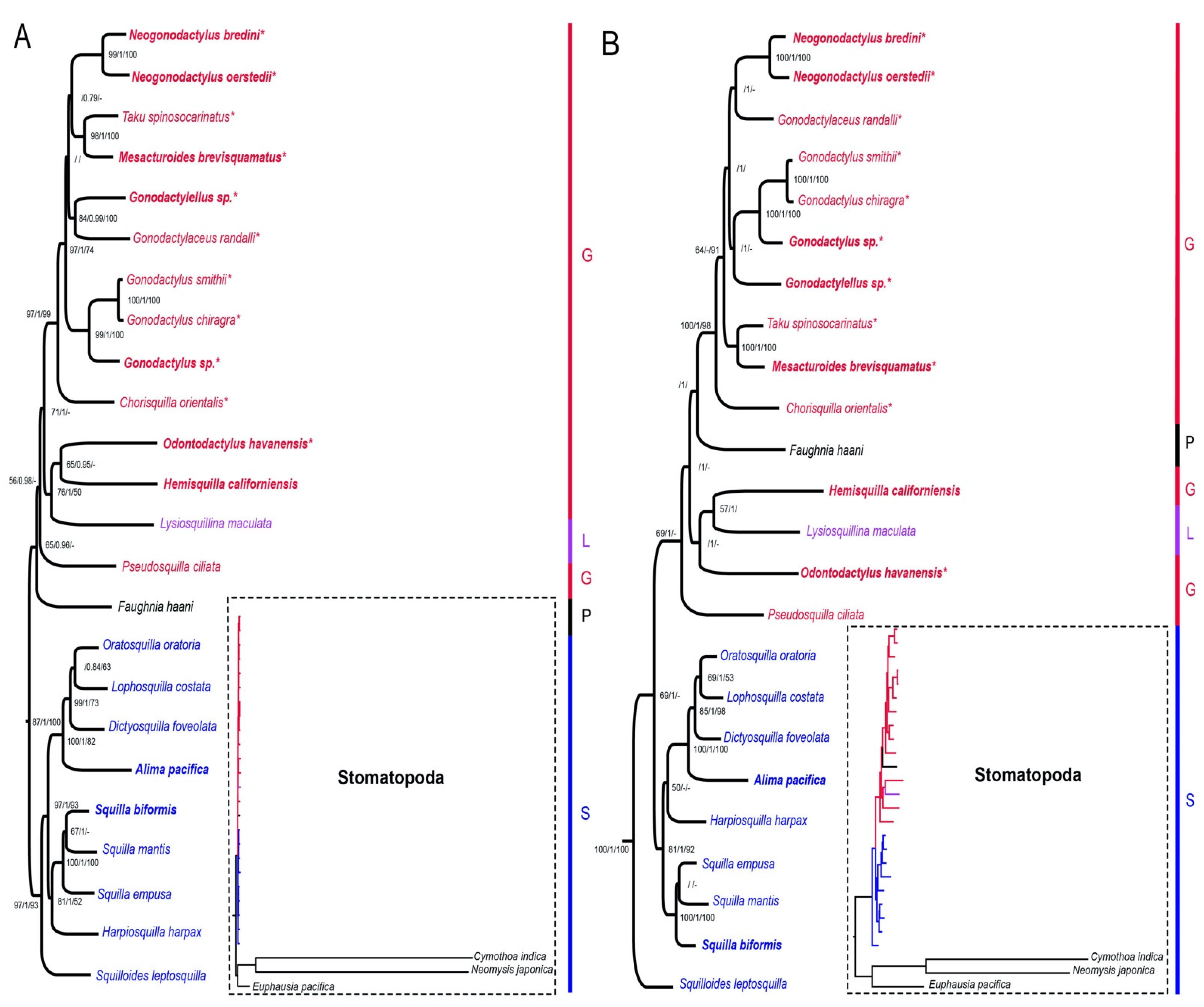
2.4. Phylogenetic Analyses
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Latreille, P.A. Les Crustacés, Les Arachnides et Les Insectes. In La Regne Animal Distribue d’Apres son Organisation, pour Servir de Base a l’Histoire Naturelle des Animaux et d’Introduction a l’Anatomie Compare; Chez Déterville: Paris, France, 1817; Volume 3, pp. 1–653. [Google Scholar]
- Caldwell, R.L.; Dingle, H. Ecology and evolution of agonistic behavior in stomatopods. Naturwissenschaften 1975, 62, 214–222. [Google Scholar] [CrossRef]
- Ahyong, S.T. Phylogenetic Analysis of the Stomatopoda (Malacostraca). J. Crustacean Biol. 1997, 17, 695–715. [Google Scholar] [CrossRef]
- Patek, S.N.; Korff, W.L.; Caldwell, R.L. Deadly strike mechanism of a mantis shrimp. Nature 2004, 428, 819–820. [Google Scholar] [CrossRef]
- DeVries, M.S.; Murphy, E.A.K.; Patek, S.N. Strike mechanics of an ambush predator: The spearing mantis shrimp. J. Exp. Biol. 2012, 215, 4374–4384. [Google Scholar] [CrossRef]
- Marshall, N.J. A unique colour and polarization vision system in mantis shrimps. Nature 1988, 333, 557–560. [Google Scholar] [CrossRef]
- Thoen, H.H.; How, M.J.; Chiou, T.H.; Marshall, J. A Different Form of Color Vision in Mantis Shrimp. Science 2014, 343, 411–413. [Google Scholar] [CrossRef]
- Sukumaran, K.K. Study on the fishery and biology of the mantis shrimp Oratosquilla nepa (Latreille) of south Kanara coast during 1979–1983. Indian J. Fish. 1987, 34, 292–305. [Google Scholar]
- Abelló, P.; Martín, P. Fishery dynamics of the mantis shrimp Squilla mantis (Crustacea: Stomatopoda) population off the Ebro delta (northwestern Mediterranean). Fish. Res. 1993, 16, 131–145. [Google Scholar] [CrossRef]
- Erdmann, M.V.; Caldwell, R.L. Stomatopod crustaceans as bioindicators of marine pollution stress on coral reefs. In Proceedings of the Eighth International Coral Reef Symposium, Panama City, Panama, 24–29 June 1996; Smithsonian Tropical Research Institute: Panama City, Panama, 1997; Volume 2, pp. 1521–1526. [Google Scholar]
- Kodama, K.; Shimizu, T.; Yamakawa, T.; Aoki, I. Changes in reproductive patterns in relation to decline in stock abundance of the Japanese mantis shrimp Oratosquilla oratoria in Tokyo Bay. Fish. Sci. 2006, 72, 568–577. [Google Scholar] [CrossRef]
- Ng, J.S.S.; Lui, K.K.Y.; Lai, C.-H.; Leung, K.M.Y. Harpiosquilla harpax (Crustacea, Stomatopoda) as a biomonitor of trace metal contamination in benthic sediments in Hong Kong waters. Mar. Pollut. Bull. 2007, 54, 1523–1529. [Google Scholar] [CrossRef]
- Antony, P.J.; Dhanya, S.; Lyla, P.S.; Kurup, B.M.; Ajmal Khan, S. Ecological role of stomatopods (mantis shrimps) and potential impacts of trawling in a marine ecosystem of the southeast coast of India. Ecol. Modell. 2010, 221, 2604–2614. [Google Scholar] [CrossRef]
- Latreille, P.A. Familles Naturelles Du Règne Animal: Exposées Succinctement et Dans Un Ordre Analytique, Avec Lindication de Leurs Genres; J.-B. Baillière: Paris, France, 1825; p. 283. [Google Scholar]
- Schram, F.R. Paleozoic proto-mantis shrimp revisited. J. Paleontol. 2007, 81, 895–916. [Google Scholar] [CrossRef]
- Haug, J.T.; Haug, C.; Maas, A.; Kutschera, V.; Waloszek, D. Evolution of mantis shrimps (Stomatopoda, Malacostraca) in the light of new Mesozoic fossils. BMC Evol. Biol. 2010, 10, 290. [Google Scholar] [CrossRef]
- Ahyong, S.T.; Harling, C. The phylogeny of the stomatopod Crustacea. Aust. J. Zool. 2000, 48, 607–642. [Google Scholar] [CrossRef]
- Van Der Wal, C.; Ahyong, S.T.; Ho, S.Y.W.; Lo, N. The evolutionary history of Stomatopoda (Crustacea: Malacostraca) inferred from molecular data. PeerJ 2017, 5, e3844. [Google Scholar] [CrossRef]
- Latreille, P.A. Histoire Naturelle, Générale et Particulière des Crustacés et des Insectes: Ouvrage Faisant Suite aux Owuvres de Leclerc de Buffon, et Partie du Cours Complet d’Histoire Naturelle Rédigé par C. S. Sonnini, Membre de Plusieurs Sociétés Savantes; Dufart: Paris, France, 1802; p. 56. [Google Scholar]
- Giesbrecht, W. Stomatopoden. Erster Theil. Fauna Flora Golf. Neapel 1910, 33, 1–239. [Google Scholar]
- Manning, R.B. Preliminary account of a new genus and a new family of Stomatopoda. Crustaceana 1967, 13, 238–239. [Google Scholar] [CrossRef]
- Manning, R.B. A Monograph of the West African Stomatopod Crustacea; Atlantide Rep. 12; Scandinavian Science Press: Copenhagen, Denmark, 1977; pp. 1–181. [Google Scholar]
- Manning, R.B.; Bruce, A.J. Erythrosquilla megalops, a remarkable new stomatopod from the western Indian Ocean. J. Crustacean Biol. 1984, 4, 329–332. [Google Scholar] [CrossRef]
- Manning, R.B. Stomatopod Crustacea of Vietnam: The legacy of Raoul Serène. Crustac. Res. 1995, 4, 1–339. [Google Scholar] [CrossRef]
- Ahyong, S.T.; Jarman, S.N. Stomatopod interrelationships: Preliminary results based on analysis of three molecular loci. Arthropod Syst. Phylogeny 2009, 67, 1864–8312. [Google Scholar]
- Manning, R.B. The superfamilies, families and genera of recent stomatopod Crustacea, with diagnoses of six new families. Proc. Biol. Soc. Wash. 1980, 93, 362–372. [Google Scholar]
- Porter, M.L.; Zhang, Y.; Desai, S.; Caldwell, R.L.; Cronin, T.W. Evolution of anatomical and physiological specialization in the compound eyes of stomatopod crustaceans. J. Exp. Biol. 2010, 213, 3473–3486. [Google Scholar] [CrossRef]
- Caldwell, R.L. Stomatopods: The better to see you with my dear. Aust. Nat. Hist. 1991, 23, 696–705. [Google Scholar]
- Trevisan, B.; Alcantara, D.M.C.; Machado, D.J.; Marques, F.P.L.; Lahr, D.J.G. Genome skimming is a low-cost and robust strategy to assemble complete mitochondrial genomes from ethanol preserved specimens in biodiversity studies. PeerJ 2019, 7, e7543. [Google Scholar] [CrossRef]
- Lin, F.J.; Liu, Y.; Sha, Z.; Tsang, L.M.; Chu, K.H.; Chan, T.Y.; Liu, R.; Cui, Z. Evolution and phylogeny of the mud shrimps (Crustacea: Decapoda) revealed from complete mitochondrial genomes. BMC Genom. 2012, 13, 631. [Google Scholar] [CrossRef]
- Cheng, J.; Chan, T.-Y.; Zhang, N.; Sun, S.; Sha, Z.-L. Mitochondrial phylogenomics reveals insights into taxonomy and evolution of Penaeoidea (Crustacea: Decapoda). Zool. Scr. 2018, 47, 582–594. [Google Scholar] [CrossRef]
- González-Castellano, I.; Pons, J.; González-Ortegón, E.; Martínez-Lage, A. Mitogenome phylogenetics in the genus Palaemon (Crustacea: Decapoda) sheds light on species crypticism in the rockpool shrimp P. elegans. LoS ONE 2020, 15, e0237037. [Google Scholar] [CrossRef]
- Kang, H.-E.; Kim, J.N.; Yoon, T.-H.; Park, K.D.; Park, W.G.; Park, H.; Kim, H.W. Total mitochondrial genome of mantis shrimp, Squilloides leptosquilla (Brooks, 1886) (Crustacea: Stomatopoda: Squillidae) in Korean waters. Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016, 27, 2842–2843. [Google Scholar] [CrossRef]
- Zhang, Y.; Bi, Y.; Feng, M. The complete mitochondrial genome of Lophosquillia costata (Malacostraca: Stomatopoda) from China and phylogeny of stomatopods. Mitochondrial DNA Part B 2020, 5, 2495–2497. [Google Scholar] [CrossRef]
- Hwang, H.-S.; Shin, J.; Jung, J. Complete mitochondrial genome of the mantis shrimp Taku spinosocarinatus (Fukuda, 1909) (Stomatopoda: Gonodactyloidea: Takuidae) in South Korea. Mitochondrial DNA Part B 2020, 5, 3609–3610. [Google Scholar] [CrossRef]
- Yang, M.; Liu, H.; Wang, R.; Tan, W. The complete mitochondrial genome of purple spot mantis shrimp Gonodactylus smithii (Pocock, 1893). Mitochondrial DNA Part B 2021, 6, 2028–2030. [Google Scholar] [CrossRef]
- Rafinesque, C.S. Analyse de la Nature, ou Tableau de l’Univers et des Corps Organisés; Self-Published: Palermo, Italy, 1815. [Google Scholar]
- Manning, R.B. Stomatopod Crustacea Collected by the Yale Seychelles Expedition, 1957–1958; Yale Peabody Museum of Natural History: Edinburgh, UK, 1962; Volume 68. [Google Scholar]
- Paul’son, O. Studies on Crustacea of the Red Sea with Notes Regarding Other Seas. Part I. Podophthalmata and Edriophthalmata (Cumacea); S.V. Kul’zhenko: Kiev, Ukraine, 1875; pp. 1–144. (In Russian) [Google Scholar]
- Ahyong, S.T. Revision of the Australian stomatopod Crustacea. Rec. Aust. Mus. 2001, 26, 1–326. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Allio, R.; Schomaker-Bastos, A.; Romiguier, J.; Prosdocimi, F.; Nabholz, B.; Delsuc, F. MitoFinder: Efficient automated large-scale extraction of mitogenomic data in target enrichment phylogenomics. Mol. Ecol. Resour. 2020, 20, 892–905. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Luo, R.; Liu, C.-M.; Leung, C.-M.; Ting, H.-F.; Sadakane, K.; Yamashita, H.; Lam, T.-W. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 2016, 102, 3–11. [Google Scholar] [CrossRef]
- Laslett, D.; Canbäck, B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2008, 24, 172–175. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Stephenson, W. A comparison of Australasian and American Specimens of Hemisquilla ensigera (Owen, 1832) (Crustacea: Stomatopoda). Proc. U. S. Natl. Mus. 1967, 120, 1–18. [Google Scholar] [CrossRef]
- Bigelow, R.P. Preliminary notes on the Stomatopoda of the Albatross collections and on other specimens in the National Museum. Johns Hopkins Univ. Circ. 1893, 12, 100–102. [Google Scholar]
- Hansen, J.H. Isopoden, Cumaceen Und Stomatopoden Der Plankton-Expedition. Ergeb. Plankton-Exped. Der Humboldt-Stift. 1895, 2, 1–105. [Google Scholar]
- Manning, R.B. Stomatopod Crustacea of the western Atlantic. Stud. Trop. Oceanogr. 1969, 8, 1–380. [Google Scholar]
- Pocock, R.I. Report upon the stomatopod crustaceans obtained by P. W. Bassett-Smith, Esq., Surgeon, R. N., during the cruise, in the Australian and China seas, of H.M.S. “Penguin,” Commander W. U. Moore. Ann. Mag. Nat. Hist. 1893, 11, 473–479. [Google Scholar] [CrossRef][Green Version]
- Fabricius, J.C. Species Insectorum Exhibentes Eorum Differentias Specificas, Synonyma Auctorum, Loca Natalia, Metamorphosin Adiectis, Observationibus, Descriptionibus; C.E. Bohnii: Hamburg, Germany; Kiel, Germany, 1781; Volume 1, pp. 1–552. [Google Scholar]
- Manning, R.B. Notes on some species of the Falcatus group of Gonodactylus (Crustacea: Stomatopoda: Gonodactylidae). Smithson. Contrib. Zool. 1978, 258, 1–15. [Google Scholar] [CrossRef]
- Fukuda, T. The Stomatopoda of Japan. Dobutsugaku Zasshi 1909, 21, 54–62. [Google Scholar]
- Fabricius, J.C. Mantissa Insectorum, Sistens Eorum Species Nuper Detectas, Adjectis Characteribus Genericis, Differentiis Specificis, Emendationibus Observationibus; C. G. Proft: Copenhagen, Germany, 1787; Volume 1, p. 348. [Google Scholar]
- Hwang, H.-S.; Ahyong, S.T.; Kim, W. A new species of Chorisquilla Manning, 1969 (Stomatopoda: Protosquillidae) from Korea and Japan with redescription of C. mehtae Erdmann Manning, 1998. Zootaxa 2018, 4483, 365–374. [Google Scholar] [CrossRef]
- De Haan, W. Crustacea. In Von Siebold, P.F., Fauna Japonica sive Descriptio Animalium, quae in Itinere per Japoniam, Jussu et Auspiciis Superiorum, qui Summum in India Batava Imperium Tenent, Suspecto, Annis 1823–1830 Collegit, Notis; Observationibus et Adumbrationibus Illustravit: Leiden, The Netherlands, 1844; pp. 1–243. [Google Scholar]
- Linnaeus, C. Systema Naturae per Regna Tria Naturae, Secundum Classes, Ordines, Genera, Species, cum Characteribus, Differentiis, Synonymis, Locis; Laurentius Salvius Holmiae: Stockholm, Sweden, 1758; Volume 1, p. 824. [Google Scholar]
- Say, T. An account of the Crustacea of the United States. J. Acad. Nat. Sci. Phila. 1818, 1, 445–458. [Google Scholar]
- Bigelow, R.P. Preliminary notes on some new species of Squilla. Johns Hopkins Univ. Circle 1891, 10, 93–94. [Google Scholar]
- Brooks, W.K. Report on the Stomatopoda collected by HMS Challenger during the years 1873–76. Rep. Sci. Results Voyag. HMS Chall. Zool. 1886, 16, 1–115. [Google Scholar]
- Wood-Mason, J. Figures and Descriptions of Nine Species of Squillidae from the Collection in the Indian Museum; Indian Museum: Calcutta, India, 1895; pp. 1–11. [Google Scholar]
- Fabricius, J.C. Entomologia Systematica Emendata et Aucta. Secundum Classes, Ordines, Genera, Species Adjectis Synonimis, Locis, Observationibus, Descriptionibus; Christ. Gottl. Proft: Copenhagen, Denmark, 1793; Volume 2, pp. 1–519. [Google Scholar]
- Holthuis, L.B. Stomatopod Crustacea of Suriname. Stud. Fauna Suriname Other Guyanas 1959, 3, 173–191. [Google Scholar]
- Hansen, H.J. The genera and species of the order Euphausiacea, with account of remarkable variation. Bull. L’institut Océanographique Monaco 1911, 210, 1–54. [Google Scholar]
- Nakazawa, K. Notes on Japanese Schizopoda. Annot. Zool. Jpn. 1910, 7, 247–261. [Google Scholar]
- Schioedte, J.C.; Meinert, F.W. Symbolae ad Monographiam Cymothoarum Isopodum Familiae 4. Cymothoidae. Trib. II. Cymothoinae. Trib. III. Livonecinae. Nat. Tidsskr. 1884, 3, 221–454. [Google Scholar]
- Schwentner, M.; Richter, S.; Rogers, D.C.; Giribet, G. Tetraconatan phylogeny with special focus on Malacostraca and Branchiopoda: Highlighting the strength of taxon-specific matrices in phylogenomics. Proc. Biol. Sci. 2018, 285, 20181524. [Google Scholar] [CrossRef]
- Lozano-Fernandez, J.; Giacomelli, M.; Fleming, J.F.; Chen, A.; Vinther, J.; Thomsen, P.F.; Glenner, H.; Palero, F.; Legg, D.A.; Iliffe, T.M.; et al. Pancrustacean Evolution Illuminated by Taxon-Rich Genomic-Scale Data Sets with an Expanded Remipede Sampling. Genome Biol. Evol. 2019, 11, 2055–2070. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Edler, D.; Klein, J.; Antonelli, A.; Silvestro, D. raxmlGUI 2.0: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol. Evol. 2021, 12, 373–377. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef]
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Version 3.61. 2019. Available online: http://www.mesquiteproject.org (accessed on 30 November 2021).
- Swofford, D.L. PAUP *. Phylogenetic Analysis Using Parsimony (* and Other Methods); Version 4; Sinauer Associates: Sunderland, MA, USA, 2003. [Google Scholar]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Shimodaira, H. An Approximately Unbiased test of phylogenetic tree selection. Syst. Biol. 2002, 51, 492–508. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Chernomor, O.; von Haeseler, A.; Minh, B.Q. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef]
- Schram, F.R.; Ahyong, S.T.; Patek, S.N.; Green, P.A.; Rosario, M.V.; Bok, M.J.; Cronin, T.W.; Vetter, K.S.M.; Caldwell, R.L.; Scholtz, G.; et al. Subclass Hoplocarida Calman, 1904: Order Stomatopoda Latreille, 18171. In Treatise on Zoology-Anatomy, Taxonomy, Biology. The Crustacea, Volume 4 Part A; Brill: Leiden, The Netherlands, 2013; pp. 179–355. [Google Scholar]
- Barber, P.H.; Erdmann, M.V. Molecular systematics of the Gonodactylidae (Stomatopoda) using mitochondrial cytochrome oxidase C (Subunit 1) DNA sequence data. J. Crustacean Biol. 2000, 20, 20–36. [Google Scholar] [CrossRef]
- Dana, J. United States Exploring Expedition. During the Year 1838, 1839, 1840, 1841, 1842; C. Sherman: Philadelphia, PA, USA, 1852; Volume 13, pp. 615–623. [Google Scholar]
- Caldwell, R.L.; Dingle, H. Stomatopods. Sci. Am. 1976, 234, 80–89. [Google Scholar] [CrossRef]
- DeVries, M.S.; Stock, B.C.; Christy, J.H.; Goldsmith, G.R.; Dawson, T.E. Specialized morphology corresponds to a generalist diet: Linking form and function in smashing mantis shrimp crustaceans. Oecologia 2016, 182, 429–442. [Google Scholar] [CrossRef] [PubMed]
- DeVries, M.S. The role of feeding morphology and competition in governing the diet breadth of sympatric stomatopod crustaceans. Biol. Lett. 2017, 13, 20170055. [Google Scholar] [CrossRef]
- Boore, J.L.; Lavrov, D.V.; Brown, W.M. Gene translocation links insects and crustaceans. Nature 1998, 392, 667–668. [Google Scholar] [CrossRef]
- Kilpert, F.; Podsiadlowski, L. The complete mitochondrial genome of the common sea slater, Ligia oceanica (Crustacea, Isopoda) bears a novel gene order and unusual control region features. BMC Genom. 2006, 7, 241. [Google Scholar] [CrossRef] [PubMed]
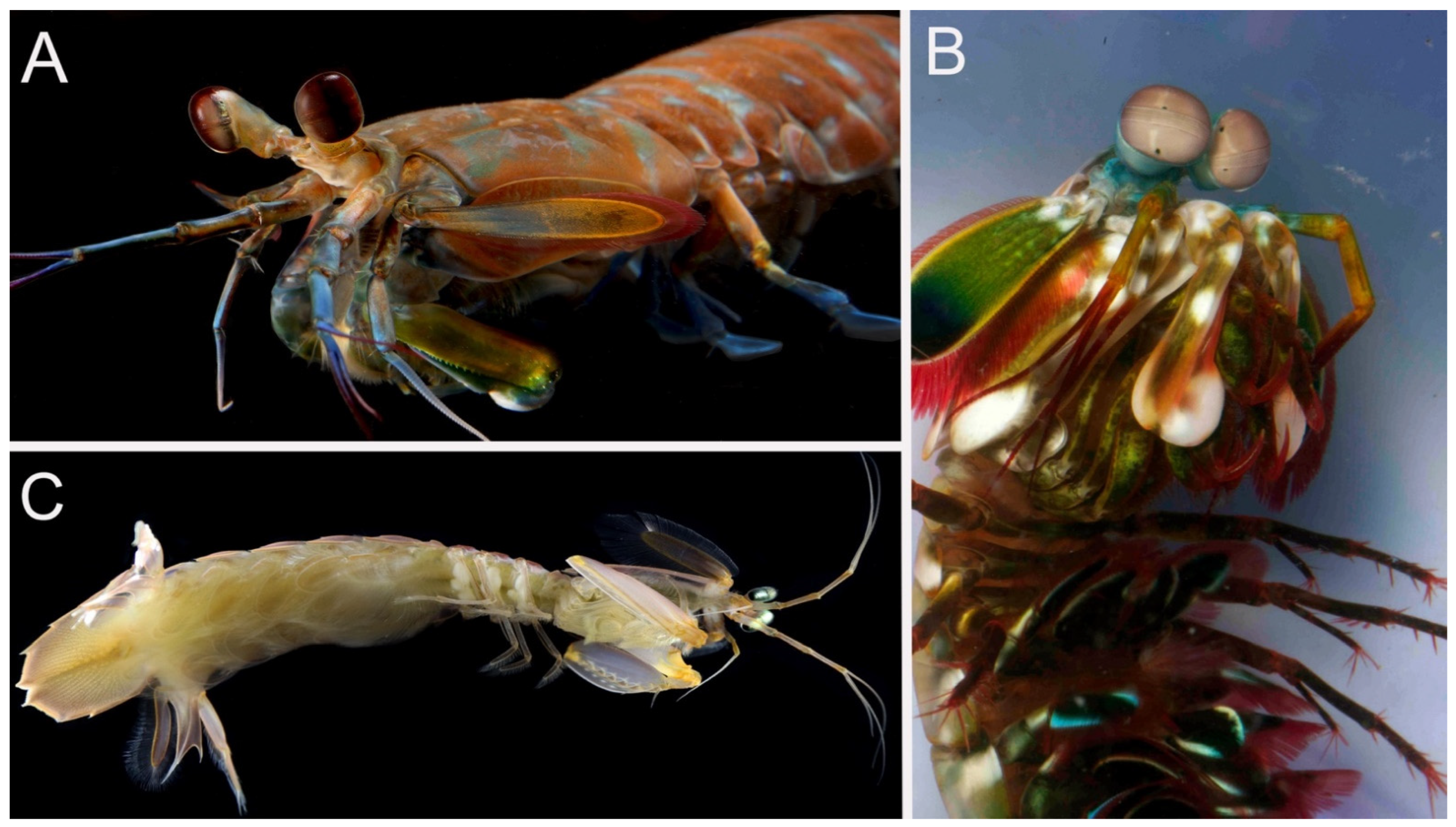




| Taxon | SIO-BIC Catalog Number | Locality | COI Accession Number | Mitogenome Accession Number | 18S rRNA Accession Number | Mitogenome Length |
|---|---|---|---|---|---|---|
| Gonodactyloidea | ||||||
| Hemisquillidae | ||||||
| Hemisquilla californiensis Stephenson, 1967 [49] | C14449 | California | MZ742104 | MW867302 | HM138876 | 16,030 |
| Odontodactylidae | ||||||
| Odontodactylus havanensis Bigelow, 1893 [50] | C14408 | Florida | MW867300 | HM138884 | 16,035 | |
| Gonodactylidae | ||||||
| Neogonodactylus oerstedii Hansen, 1895 [51] | C14405 | Florida | MW867303 | HM138882 | 16,327 | |
| Neogonodactylus bredini Manning, 1969 [52] | C14428 | Florida | MZ742108 | MW867301 | HM138881 | 16,342 |
| Gonodactylus smithii Pocock, 1893 [53] | - | MW574903 | HM138873 | 16,260 | ||
| Gonodactylus chiragra Fabricius, 1781 [54] | - | DQ191682 | HM138870 | 16,279 | ||
| Gonodactylaceus randalli Manning, 1978 [55] | - | MW019425 | - | 15,907 | ||
| Gonodactylus sp. | C12730 | Red Sea | MZ742105 | MW867306 | - | 16,032 |
| Gonodactylellus sp. | C12514 | Red Sea | MZ742107 | MW867308 | - | 16,011 |
| Takuidae | ||||||
| Mesacturoides brevisquamatus | C14383 | Red Sea | MZ742109 | MW867304 | - | 16,151 |
| Taku spinosocarinatus Fukuda, 1909 [56] | - | MT672285 | HM138899 | 15,960 | ||
| Pseudosquillidae | ||||||
| Pseudosquilla ciliata Fabricius, 1787 [57] | - | AY947836 | HM138888 | 14,621 (incomp.) | ||
| Protosquillidae | ||||||
| Chorisquilla orientalis Hwang et al., 2018 [58] | - | MT672286 | - | 15,880 | ||
| Squilloidea | ||||||
| Squillidae | ||||||
| Oratosquilla oratoria De Haan, 1844 [59] | - | GQ292769 | - | 15,783 | ||
| Squilla mantis Linnaeus, 1758 [60] | - | AY639936 | GQ328958 | 15,994 | ||
| Squilla empusa Say, 1818 [61] | - | DQ191684 | HM138897 | 15,828 | ||
| Squilla biformis Bigelow, 1891 [62] | C13808 | Costa Rica | MW867305 | - | 15,688 | |
| Squilloides leptosquilla Brooks, 1886 [63] | - | KR095170 | - | 16,376 | ||
| Harpiosquilla harpax De Haan, 1844 [59] | - | AY699271 | - | 15,714 | ||
| Lophosquilla costata De Haan, 1844 [59] | - | MT276143 | - | 15,771 | ||
| Alima pacifica Ahyong, 2001 [40] | C12719 | Red Sea | MZ742106 | MW867307 | HM138858 | 15,678 |
| Dictyosquilla foveolata Wood-Mason, 1895 [64] | - | MW864094 | - | 15,733 | ||
| Lysiosquilloidea | ||||||
| Lysiosquillidae | ||||||
| Lysiosquillina maculata Fabricius, 1793 [65] | - | DQ191683 | HM138878 | 16,325 | ||
| Parasquilloidea | ||||||
| Parasquillidae | ||||||
| Faughnia haani Holthuis, 1959 [66] | - | MW632159 | - | 16,089 | ||
| Outgroups | ||||||
| Euphausiacea | ||||||
| Euphausia pacifica Hansen, 1911 [67] | - | EU587005 | AY141010 | 16,898 | ||
| Mysida | ||||||
| Neomysis japonica Nakazawa, 1910 [68] | - | KR006340 | - | 17,652 | ||
| Isopoda | ||||||
| Cymothoa indica Schioedte & Meinert, 1884 [69] | - | MH396438 | - | 14,475 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koga, C.; Rouse, G.W. Mitogenomics and the Phylogeny of Mantis Shrimps (Crustacea: Stomatopoda). Diversity 2021, 13, 647. https://doi.org/10.3390/d13120647
Koga C, Rouse GW. Mitogenomics and the Phylogeny of Mantis Shrimps (Crustacea: Stomatopoda). Diversity. 2021; 13(12):647. https://doi.org/10.3390/d13120647
Chicago/Turabian StyleKoga, Cassandra, and Greg W. Rouse. 2021. "Mitogenomics and the Phylogeny of Mantis Shrimps (Crustacea: Stomatopoda)" Diversity 13, no. 12: 647. https://doi.org/10.3390/d13120647
APA StyleKoga, C., & Rouse, G. W. (2021). Mitogenomics and the Phylogeny of Mantis Shrimps (Crustacea: Stomatopoda). Diversity, 13(12), 647. https://doi.org/10.3390/d13120647







