Old and Cosmopolite: Molecular Phylogeny of Tropical–Subtropical Kites (Aves: Elaninae) with Taxonomic Implications
Abstract
1. Introduction
2. Materials and Methods
2.1. Laboratory Procedures
2.2. Phylogenetic Analysis
3. Results
4. Discussion
4.1. Morphological Features of Elanin Kites
4.2. Cytogenetics of Elanus caeruleus
4.3. Phylogenetics, Distribution, and Ecology of Elanins
4.4. Taxonomic Implications for Elanins and Other Accipitrids
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blyth, E. Conspectus of the ornithology of India, Burma, and the Malayan peninsula, inclusive of Sindh, Asám, Ceylon, and the Nicobar islands. J. Asiat. Soc. Bengal 1850, 19, 317–342. [Google Scholar]
- Peters, J.L. Check-List of Birds of the World; Harward University Press: Cambridge, MA, USA, 1931; Volume 1. [Google Scholar]
- Lerner, H.R.L.; Mindell, D.P. Phylogeny of eagles, Old World vultures, and other Accipitridae based on nuclear and mitochondrial DNA. Mol. Phylogenet. Evol. 2005, 37, 327–346. [Google Scholar] [CrossRef]
- Brown, L.; Amadon, D. Eagles, Hawks, and Falcons of the World; McGraw-Hill Book Company: New York, NY, USA, 1968; Volume 1. [Google Scholar]
- Ferguson-Lees, J.; Christie, D.A. Raptors of the World; Houghton Mifflin: Boston, MA, USA, 2001; ISBN 978-0-618-12762-7. [Google Scholar]
- Dickinson, E.C.; Remsen, J.V., Jr. The Howard and Moore Complete Checklist of the Birds of the World, 4th ed.; Volume 1, Non-Passerines; Aves Press: Eastbourne, UK, 2013; ISBN 978-0-9568611-0-8. [Google Scholar]
- IOC World Bird List 10.1. Available online: http://www.iucnredlist.org/details/22695042/0 (accessed on 25 June 2020). [CrossRef]
- Clements, J.F.; Schulenberg, T.S.; Iliff, M.J.; Billerman, S.M.; Fredericks, T.A.; Sullivan, B.L.; Wood, C.L. The eBird/Clements checklist of birds of the World: v2019. Available online: https://www.birds.cornell.edu/clementschecklist/download/ (accessed on 3 May 2020).
- Kemp, A.C.; Kirwan, G.M.; Marks, J.S.; Motis, A.; Garcia, E.F.J. Black-winged Kite (Elanus caeruleus). In Handbook of the birds of the World Alive; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D.A., de Juana, E., Eds.; Available online: https://www.hbw.com/node/52966 (accessed on 3 May 2020).
- BirdLife International. Elanus caeruleus. The IUCN Red List of Threatened Species 2019. Available online: https://www.iucnredlist.org/ (accessed on 18 June 2020). [CrossRef]
- BirdLife International. Elanus leucurus. The IUCN Red List of Threatened Species 2016. Available online: https://www.iucnredlist.org/ (accessed on 18 June 2020). [CrossRef]
- BirdLife International. Elanus axillaris. The IUCN Red List of Threatened Species 2016. Available online: https://www.iucnredlist.org/ (accessed on 18 June 2020). [CrossRef]
- BirdLife International. Elanus scriptus. The IUCN Red List of Threatened Species 2016. Available online: https://www.iucnredlist.org/ (accessed on 18 June 2020). [CrossRef]
- BirdLife International. Gampsonyx swainsonii. The IUCN Red List of Threatened Species 2016. Available online: https://www.iucnredlist.org/ (accessed on 18 June 2020). [CrossRef]
- BirdLife International. Chelictinia riocourii. The IUCN Red List of Threatened Species 2016. Available online: https://www.iucnredlist.org/ (accessed on 18 June 2020). [CrossRef]
- Ridgway, R. Catalogue of the ornithological collection of the Boston Society of Natural History. Part II. Falconidae. Proc. Boston Soc. Nat. Hist. 1873, 16, 43–72. [Google Scholar]
- Ridgway, R. Studies of the American Falconidae. Bull. U. S. Geol. Geogr. Surv. Terr. 1876, 2, 91–182. [Google Scholar]
- Shufeldt, R.W. Some comparative osteological notes on the North-American kites. IBIS 1891, 33, 228–232. [Google Scholar] [CrossRef]
- Pycraft, W.P. Contribution to the osteology of birds. Part V. Falconiformes. Proc. Zool. Soc. Lond. 1902, 1, 277–320. [Google Scholar]
- Friedmann, H. The Birds of North and Middle America; Unites States National Museum; U.S. Government Printing Office: Washington, DC, USA, 1950; Volume XI. [Google Scholar]
- Jollie, M. A contribution to the morphology and phylogeny of the Falconiformes (part II). Evol. Theory 1977, 2, 115–208. [Google Scholar]
- Jollie, M. A contribution to the morphology and phylogeny of the Falconiformes (part III). Evol. Theory 1977, 2, 209–300. [Google Scholar]
- Jollie, M. A contribution to the morphology and phylogeny of the Falconiformes (part IV). Evol. Theory 1977, 3, 1–141. [Google Scholar]
- Stresemann, E.; Amadon, D. Order Falconiformes. In Check-list of Birds of the World; Museum of Comparative Zoology: Cambridge, MA, USA, 1979; Volume 1, pp. 271–425. [Google Scholar]
- Wolters, H.E. Die Vogelarten der Erde: Eine Systematische Liste mit Verbreitungsangaben sowie Deutschen und Englischen Namen; Paul Parey: Hamburg, Deutschland; Berlin, Deutschland, 1982; ISBN 3-490-09118-3. [Google Scholar]
- Amadon, D.; Bull, J. Hawks and owls of the world: An annotated list of species. Proc. West. Found. Vertebr. Zool. 1988, 3, 297–330. [Google Scholar]
- Suschkin, P. Zur Morphologie des Vogelskelets: Vergleichende Osteologie der Normalen Tagraubvögel (Accipitres) und die Fragen der Classification; J.N. Kouchnéreff et C-ie: Moscou, Russie, 1905; Nouveaux mémoires de la Société impériale des naturalistes de Moscou, Tome XVI. [Google Scholar]
- Wink, M. Advances in DNA studies of diurnal and nocturnal raptors. In Raptors at Risk, Proceedings of the 5th World Conference on Birds of Prey and Owls, Midrand, South Africa, 4–11 August 1998; Chancellor, R.D., Meyburg, B.-U., Eds.; WWGBP, Hancock House: London, UK, 2000; pp. 831–844. [Google Scholar]
- Wink, M.; Sauer-Gürth, H. Phylogenetic relationships in diurnal raptors based on nucleotide sequences of Mitochondrial and Nuclear Marker Genes. In Raptors Worldwide, Proceedings of the 6th World Conference on Birds of Prey and Owls, Budapest, Hungary, 18–23 May 2003; Chancellor, R.D., Meyburg, B.-U., Eds.; WWGBP: Berlin, Deutschland, 2004; pp. 483–498. [Google Scholar]
- Starikov, I.J.; Sauer-Gürth, H.; Wink, M. Kites represent a polyphyletic group: Molecular phylogeny inferred from mitochondrial and nuclear DNA sequence. Vogelwarte 2018, 56, 331. [Google Scholar]
- Kocum, A. Phylogenie der Accipitriformes (Greifvögel) Anhand verschiedener nuklearer und mitochondrialer DNA-Sequenzen. Inauguraldissertation zur Erlangung des Akademischen Grades Doctor Rerum Naturalium (Dr. rer. nat.), Ernst-Moritz-Arndt-Universität Greifswald, Greifswald, Deutschland, 2006. [Google Scholar]
- Debus, S. Relationships of the Elanus kites. Boobook 2004, 22, 8. [Google Scholar]
- Griffiths, C.S.; Barrowclough, G.F.; Groth, J.G.; Mertz, L.A. Phylogeny, diversity, and classification of the Accipitridae based on DNA sequences of the RAG-1 exon. J. Avian Biol. 2007, 38, 587–602. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001; Volume 1, ISBN 0-87969-576-5. [Google Scholar]
- Kerr, K.C.R.; Lijtmaer, D.A.; Barreira, A.S.; Hebert, P.D.N.; Tubaro, P.L. Probing evolutionary patterns in neotropical birds through DNA barcodes. PLoS ONE 2009, 4, e4379. [Google Scholar] [CrossRef]
- Kerr, K.C.R.; Stoeckle, M.Y.; Dove, C.J.; Weigt, L.A.; Francis, C.M.; Hebert, P.D.N. Comprehensive DNA barcode coverage of North American birds. Mol. Ecol. Notes 2007, 7, 535–543. [Google Scholar] [CrossRef]
- Tavares, E.S.; Gonçalves, P.; Miyaki, C.Y.; Baker, A.J. DNA barcode detects high genetic structure within Neotropical bird species. PLoS ONE 2011, 6, e28543. [Google Scholar] [CrossRef]
- Schindel, D.; Stoeckle, M.; Milensky, C.; Trizna, M.; Schmidt, B.; Gebhard, C.; Graves, G. Project description: DNA barcodes of bird species in the National Museum of Natural History, Smithsonian Institution, USA. ZooKeys 2011, 152, 87–91. [Google Scholar] [CrossRef]
- Sorenson, M.D.; Ast, J.C.; Dimcheff, D.E.; Yuri, T.; Mindell, D.P. Primers for a PCR-based approach to mitochondrial genome sequencing in birds and other vertebrates. Mol. Phylogenet. Evol. 1999, 12, 105–114. [Google Scholar] [CrossRef]
- Wink, M.; Sauer-Gürth, H.; Heidrich, P.; Witt, H.-H.; Gwinner, E. A molecular phylogeny of stonechats and related turdids. In Stonechats. A Guide to the Genus Saxicola; Urquhart, E., Ed.; Helm: London, UK, 2002; pp. 22–29. ISBN 978-0-300-09970-6. [Google Scholar]
- Barth, D.; Bernhard, D.; Fritzsch, G.; Fritz, U. The freshwater turtle genus Mauremys (Testudines, Geoemydidae)—A textbook example of an east-west disjunction or a taxonomic misconcept? Zool. Scr. 2004, 33, 213–221. [Google Scholar] [CrossRef]
- Lohman, D.J.; Prawiradilaga, D.M.; Meier, R. Improved COI barcoding primers for Southeast Asian perching birds (Aves: Passeriformes). Mol. Ecol. Resour. 2009, 9, 37–40. [Google Scholar] [CrossRef]
- Groth, J.G.; Barrowclough, G.F. Basal divergences in birds and the phylogenetic utility of the nuclear RAG-1 gene. Mol. Phylogenet. Evol. 1999, 12, 115–123. [Google Scholar] [CrossRef]
- Irestedt, M.; Johansson, U.S.; Parsons, T.J.; Ericson, P.G.P. Phylogeny of major lineages of suboscines (Passeriformes) analysed by nuclear DNA sequence data. J. Avian Biol. 2001, 32, 15–25. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Jollie, M. A contribution to the morphology and phylogeny of the Falconiformes. Evol. Theory 1976, 1, 285–298. [Google Scholar]
- Gibb, G.C.; Kardailsky, O.; Kimball, R.T.; Braun, E.L.; Penny, D. Mitochondrial genomes and avian phylogeny: Complex characters and resolvability without explosive radiations. Mol. Biol. Evol. 2007, 24, 269–280. [Google Scholar] [CrossRef]
- do Amaral, F.R.; Sheldon, F.H.; Gamauf, A.; Haring, E.; Riesing, M.; Silveira, L.F.; Wajntal, A. Patterns and processes of diversification in a widespread and ecologically diverse avian group, the buteonine hawks (Aves, Accipitridae). Mol. Phylogenet. Evol. 2009, 53, 703–715. [Google Scholar] [CrossRef]
- Ong, P.S.; Luczon, A.U.; Quilang, J.P.; Sumaya, A.M.T.; Ibañez, J.C.; Salvador, D.J.; Fontanilla, I.K.C. DNA barcodes of Philippine accipitrids. Mol. Ecol. Resour. 2011, 11, 245–254. [Google Scholar] [CrossRef]
- Jeon, H.S.; Myeong, H.; Kang, S.-G.; Kim, J.A.; Lee, S.-H.; Lee, M.-Y.; An, J. The mitochondrial genome of Milvus migrans (Aves, Accipitriformes, Accipitridae), an endangered species from South Korea. Mitochondrial DNA Part B 2018, 3, 498–499. [Google Scholar] [CrossRef]
- Haring, E.; Kruckenhauser, L.; Gamauf, A.; Riesing, M.J.; Pinsker, W. The complete sequence of the mitochondrial genome of Buteo buteo (Aves, Accipitridae) indicates an early split in the phylogeny of raptors. Mol. Biol. Evol. 2001, 18, 1892–1904. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.-M.; Guan, Q.-X.; Shi, J.-P.; Hou, L.-X.; Qin, P.-S. Complete mitochondrial genome of the Spilornis cheela (Falconiformes, Accipitridae): Comparison of S. cheela and Spizaetus alboniger. Mitochondrial DNA 2013, 24, 255–256. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Liu, G.; Zhou, L.; Gu, C. Complete mitochondrial genome of Cinereous vulture Aegypius monachus (Falconiformes: Accipitridae). Mitochondrial DNA 2015, 26, 910–911. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Dou, H.; Yang, X.; Zhao, C.; Liu, G.; Zhang, J. The complete mitochondrial genome sequence of the Sparrowhawk (Accipiter nisus). Mitochondrial DNA 2014, 27, 1648–1649. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Sun, G.; Xia, T.; Zhao, C.; Wei, Q.; Sha, W.; Zhang, H. Complete mitochondrial genome sequence of the hen harrier (Circus cyaneus). Mitochondrial DNA Part B 2018, 3, 668–669. [Google Scholar] [CrossRef]
- Kim, J.A.; Kang, S.-G.; Jeon, H.S.; Jeon, J.H.; Jang, J.-H.; Kim, S.; An, J. Complete mitogenomes of two Accipitridae, Haliaeetus albicilla, and Pernis ptilorhynchus. Mitochondrial DNA Part B 2019, 4, 391–392. [Google Scholar] [CrossRef]
- Mahmood, M.T.; McLenachan, P.A.; Gibb, G.C.; Penny, D. Phylogenetic position of avian nocturnal and diurnal raptors. Genome Biol. Evol. 2014, 6, 326–332. [Google Scholar] [CrossRef][Green Version]
- Ryu, S.H.; Lee, J.H.; Hwang, U.W. Complete mitochondrial genome of the peregrine falcon Falco peregrinus (Aves, Falconiformes, Falconidae): Genetic differences between the two individuals. Mitochondrial DNA 2012, 23, 139–141. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Tavaré, S. Some probabilistic and statistical problems in the analysis of DNA sequences. In Some Mathematical Questions in Biology: DNA Sequence Analysis; Miura, R.M., Ed.; Lectures on Mathematics in the Life Sciences; American Mathematical Society: Providence, RI, USA, 1986. [Google Scholar]
- Shoemaker, J.S.; Fitch, W.M. Evidence from nuclear sequences that invariable sites should be considered when sequence divergence is calculated. Mol. Biol. Evol. 1989, 6, 270–289. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J. Mol. Evol. 1994, 39, 306–314. [Google Scholar] [CrossRef]
- Waddell, P.J.; Steel, M.A. General time-reversible distances with unequal rates across sites: Mixing gamma and inverse Gaussian distributions with invariant sites. Mol. Phylogenet. Evol. 1997, 8, 398–414. [Google Scholar] [CrossRef] [PubMed]
- Weir, J.T.; Schluter, D. Calibrating the avian molecular clock. Mol. Ecol. 2008, 17, 2321–2328. [Google Scholar] [CrossRef] [PubMed]
- Lavinia, P.D.; Kerr, K.C.R.; Tubaro, P.L.; Hebert, P.D.N.; Lijtmaer, D.A. Calibrating the molecular clock beyond cytochrome b: Assessing the evolutionary rate of COI in birds. J. Avian Biol. 2016, 47, 84–91. [Google Scholar] [CrossRef]
- Yule, G.U. A mathematical theory of evolution, based on the conclusions of Dr. Willis, J.C., Philos, F.R.S. Trans. R. Soc. Lond. B Biol. Sci. 1925, 213, 21–87. [Google Scholar]
- Gernhard, T. The conditioned reconstructed process. J. Theor. Biol. 2008, 253, 769–778. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Suschkin, P. Beiträge zur Classification der Tagraubvögel mit Zugrundelegung der osteologischen Merkmale. Zool. Anz. 1899, 22, 500–518. [Google Scholar]
- Negro, J.J.; Pertoldi, C.; Randi, E.; Ferrero, J.J.; López-Caballero, J.M.; Rivera, D.; Korpimäki, E. Convergent evolution of Elanus kites and the owls. J. Raptor Res. 2006, 40, 222–225. [Google Scholar] [CrossRef]
- Bed’Hom, B.; Coullin, P.; Guillier-Gencik, Z.; Moulin, S.; Bernheim, A.; Volobouev, V. Characterization of the atypical karyotype of the black-winged kite Elanus caeruleus (Falconiformes: Accipitridae) by means of classical and molecular cytogenetic techniques. Chromosome Res. 2003, 11, 335–343. [Google Scholar] [CrossRef]
- De Boer, L.E.M.; Sinoo, R.P. A karyological study of Accipitridae (Aves: Falconiformes), with karyotypic descriptions of 16 species new to cytology. Genetica 1984, 65, 89–107. [Google Scholar] [CrossRef]
- Ansari, H.A.; Kaul, D. Cytotaxonomic study in the order Falconiformes (Aves). Zool. Scr. 1986, 15, 351–356. [Google Scholar] [CrossRef]
- Harris, T.; Walters, C. Chromosomal sexing of the Black Shouldered Kite (Elanus caeruleus) (Aves: Accipitridae). Genetica 1982, 60, 19–20. [Google Scholar] [CrossRef]
- Nagy, J.; Tökölyi, J. Phylogeny, historical biogeography and the evolution of migration in accipitrid birds of prey (Aves: Accipitriformes). Ornis Hung. 2014, 22, 15–35. [Google Scholar] [CrossRef]
- Mindell, D.P.; Fuchs, J.; Johnson, J.A. Phylogeny, taxonomy, and geographic diversity of diurnal raptors: Falconiformes, Accipitriformes, and Cathartiformes. In Birds of Prey. Biology and Conservation in the XXI Century; Sarasola, J.H., Grande, J.M., Negro, J.J., Eds.; Springer: Cham, Switzerland, 2018; pp. 3–32. ISBN 978-3-319-73744-7. [Google Scholar]
- Farris, J.S. The retention index and rescaled consistency index. Cladistics 1989, 5, 417–419. [Google Scholar] [CrossRef]
- Sansom, R.S.; Wills, M.A. Differences between hard and soft phylogenetic data. Proc. R. Soc. B Biol. Sci. 2017, 284, 1–7. [Google Scholar] [CrossRef]
- Parkes, K.C. Specific relationships in the genus Elanus. Condor 1958, 60, 139–140. [Google Scholar]
- Mendelsohn, J.M. Social-behaviour and dispersion of Blackshouldered Kite. Ostrich 1983, 54, 1–18. [Google Scholar] [CrossRef]
- Oatley, T.B.; Oschadleus, H.D.; Navarro, R.A.; Underhill, T.G. Review of Ring Recoveries of Birds of Prey in Southern Africa: 1948–1998; Endangered Wildlife Trust: Johannesburg, South Africa, 1998; ISBN 978-0-620-22971-5. [Google Scholar]
- Bangs, O.; Penard, T.E. Two new American hawks. Proc. N. Engl. Zoöl. Club 1920, 7, 45–47. [Google Scholar]
- Clark, W.S.; Banks, R.C. The taxonomic status of the White-tailed Kite. Wilson Bull. 1992, 104, 571–579. [Google Scholar]
- Vigors, N.A., Jr. Sketches in ornithology; or observations on the leading affinities of some of the more extensive groups of birds. Zool. J. 1825, 2, 37–70. [Google Scholar]
- Pinto, O.M.d.O. Catalogo das aves do Brasil e lista dos exemplares que as representam no Museu Paulista; Museu Paulista: São Paulo, Brasil, 1938. [Google Scholar]
- Hellmayr, C.E.; Conover, B. Catalogue of Birds of the Americas and the Adjacent Islands in Field Museum of Natural History and Including all Species and Subspecies Known to Occur in North America, Mexico, Central America, South America, the West Indies, and Islands of the Caribbean Sea, the Galapagos Archipelago, and other Islands which May Be Included on Account of Their Faunal Affinities; Field Museum Press: Chicago, IL, USA, 1949; Volume 13. [Google Scholar]
- Meyer de Schauensee, R. The Birds of Colombia and Adjacent Areas of South and Central America; Livingston Publishing Company: Narberth, UK, 1964; Available online: http://globalraptors.org/grin/SpeciesResults.asp?specID=8035 (accessed on 18 June 2020).
- Stresemann, V. The wing molt and systematic position of the genus Gampsonyx. Auk 1959, 76, 360–361. [Google Scholar] [CrossRef]
- Brodkorb, P. The skeleton and systematic position of Gampsonyx. Auk 1960, 77, 88–89. [Google Scholar] [CrossRef]
- Meyer de Schauensee, R. A Guide to the Birds of South America; Oliver & Boyd: Edinburgh, UK, 1971; ISBN 978-0-05-002398-3. [Google Scholar]
- Chubb, C. Descriptions of new forms from South and Central American birds—Gampsonyx swainsonii magnus, G. s. leonæ, Falco rufogularis petoensis, F. r. pax. Bull. Br. Ornithol. Club 1918, 39, 21–23. [Google Scholar]
- Bierregaard, R.O., Jr.; Kirwan, G.M. Pearl Kite (Gampsonyx swainsonii). In Handbook of the Birds of the World Alive; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D.A., de Juana, E., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020; Available online: https://www.hbw.com/node/52965 (accessed on 3 May 2020).
- Clark, W.S.; Davies, R. African Raptors; Helm Identification Guides; Helm: London, UK, 2018; ISBN 978-0-7136-6538-3. [Google Scholar]
- Bierregaard, R.O., Jr.; Marks, J.S.; Boesman, P.; Kirwan, G.M. White-Tailed Kite (Elanus leucurus). In Handbook of the Birds of the World Alive; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D.A., de Juana, E., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020; Available online: https://www.hbw.com/node/52968 (accessed on 3 May 2020).
- Debus, S.; Kirwan, G.M.; Marks, J.S. Black-Shouldered Kite (Elanus axillaris). In Handbook of the Birds of the World Alive; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D.A., de Juana, E., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020; Available online: https://www.hbw.com/node/52967 (accessed on 3 May 2020).
- Debus, S.; Kirwan, G.M.; Christie, D.A.; Marks, J.S. Letter-Winged Kite (Elanus scriptus). In Handbook of the Birds of the World Alive; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D.A., de Juana, E., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020; Available online: https://www.hbw.com/node/52969 (accessed on 3 May 2020).
- Kemp, A.C.; Kirwan, G.M.; Marks, J.S. Scissor-Tailed Kite (Chelictinia riocourii). In Handbook of the Birds of the World Alive; del Hoyo, J., Elliott, A., Sargatal, J., Christie, D.A., de Juana, E., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020; Available online: https://www.hbw.com/node/52970 (accessed on 3 May 2020).
- Clay, T. Revisions of the Mallophaga genera. Degeeriella from the Falconiformes. Bull. Br. Mus. Nat. Hist. Entomol. 1958, 7, 121–207. [Google Scholar]
- Page, R.D.M.; Lee, P.L.M.; Becher, S.A.; Griffiths, R.; Clayton, D.H. A different tempo of mitochondrial DNA evolution in birds and their parasitic lice. Mol. Phylogenet. Evol. 1998, 9, 276–293. [Google Scholar] [CrossRef]
- Banks, J.C.; Palma, R.L.; Paterson, A.M. Cophylogenetic relationships between penguins and their chewing lice. J. Evol. Biol. 2006, 19, 156–166. [Google Scholar] [CrossRef]
- Hughes, J.; Kennedy, M.; Johnson, K.P.; Palma, R.L.; Page, R.D.M. Multiple cophylogenetic analyses reveal frequent cospeciation between pelecaniform birds and Pectinopygus Lice. Syst. Biol. 2007, 56, 232–251. [Google Scholar] [CrossRef] [PubMed]
- Štefka, J.; Hoeck, P.E.; Keller, L.F.; Smith, V.S. A hitchhikers guide to the Galápagos: Co-phylogeography of Galápagos mockingbirds and their parasites. BMC Evol. Biol. 2011, 11, 1–18. [Google Scholar] [CrossRef]
- Latham, J. Index Ornithologicus, Sive, Systema Ornithologiæ; Complectens Avium Divisionem in Classes, Ordines, Genera, Species, Ipsarumque Varietates: Adjectis Synonymis, Locis, Descriptionibus, &c.; Leigh & Sotheby: London, UK, 1790; Volume II. [Google Scholar]
- Cai, Y.; Yue, B.; Jiang, W.; Xie, S.; Li, J.; Zhou, M. DNA barcoding on subsets of three families in Aves. Mitochondrial DNA 2010, 21, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Dolinay, M. Genetická struktura populací dvou druhů sympatricky se vyskytujících luňáků (Milvus spp.); Diplomová práce; Masarykova univerzita: Brno, Czech Republic, 2015. [Google Scholar]
- Etherington, G.J.; Mobley, J.A. Molecular phylogeny, morphology and life-history comparisons within Circus cyaneus reveal the presence of two distinct evolutionary lineages. Avian Res. 2016, 7, 1–12. [Google Scholar] [CrossRef]
- Retnaningtyas, R.W.; Hermadhiyanti, W.; Listyorini, D. The phylogenetic study of the White-Bellied Sea Eagle [Haliaeetus leucogaster (Gmelin, 1788)] based on DNA barcoding cytochrome-c oxidase subunit I (COI). In Proceedings of the ICBS Conference, Yogyakarta, Indonesia, 18–19 September 2015; KnE Life Sciences: Dubai, UAE, 2017; pp. 208–212. [Google Scholar]
- Zein, M.S.A. Barkoding DNA burung Elang (Famili Accipitridae) di Indonesia. Ber. Biol. 2018, 17, 165–173. [Google Scholar] [CrossRef]
- Husain, K.Z. Notes on the taxonomy and zoogeography of the genus Elanus. Condor 1959, 61, 153–154. [Google Scholar]
- Mees, G.F. Birds from the lowlands of southern New Guinea (Merauke and Koembe). Zool. Verh. 1982, 191, 1–188. [Google Scholar]
- Rensch, B. Die Vogelwelt von Lombok, Sumbawa und Flores. Mitt. Zool. Mus. Berl. 1931, 17, 451–637. [Google Scholar]
- Mees, G.F. On some birds from southern Mexico. Zool. Meded. 1970, 44, 237–245. [Google Scholar]
- Mayr, E.; Short, L.L. Species Taxa of North American Birds. A Contribution to Comparative Systematics; Nuttall Ornithological Club: Cambridge, MA, USA, 1970. [Google Scholar]
- Handbuch der Vögel Mitteleuropas; Glutz von Blotzheim, U.N., Bauer, K.M., Bezzel, E., Eds.; Academische Verlagsgesellschaft: Frankfurt am Main, Deutschland, 1971; Volume 4. [Google Scholar]
- Sibley, C.G.; Monroe, B.L. Distribution and Taxonomy of Birds of the World; Yale University Press: New Haven, London, UK, 1990; ISBN 978-0-300-04969-5. [Google Scholar]
- Coues, E.; Prentiss, D.W. Avifauna Columbiana: Being a List of Birds Ascertained to Inhabit the District of Columbia, with the Times of Arrival and Departure of Such as Are Non-Residents, and Brief Notices of Habits, etc., 2nd ed.; U.S. Government Printing Office: Washington, DC, USA, 1883. [Google Scholar]
- Seebohm, H. Classification of Birds; An Attempt to Diagnose the Subclasses, Orders, Suborders, and Some of the Families of Existing Birds; R.H. Porter: London, UK, 1890. [Google Scholar]
- Sharpe, R.B. A Review of Recent Attempts to Classify Birds; An Address Delivered before the Second International Ornithological Congress on the 18th of May, 1891; International Ornithological Congress: Budapest, Austria-Hungary, 1891. [Google Scholar]
- Gadow, H. Dr. H.G. Bronn’s Klassen und Ordnungen des Thier-Reichs, Wissenschaftlich Dargestellt in Wort und Bild; C. F. Winter’sche Verlagshandlung: Leipzig, Sachsen, 1893; Volume 6, Abteilung 4, II. [Google Scholar]
- A Dictionary of Birds; Campbell, B., Lack, E., Eds.; T & AD Poyser Ltd.: Calton, UK, 2010. [Google Scholar]
- Cracraft, J. Toward a phylogenetic classification of the recent birds of the World (class Aves). Auk 1981, 98, 681–714. [Google Scholar]
- Wetmore, A. A systematic classification for the birds of the World. Smithson. Misc. Collect. 1940, 99, 1–11. [Google Scholar] [CrossRef]
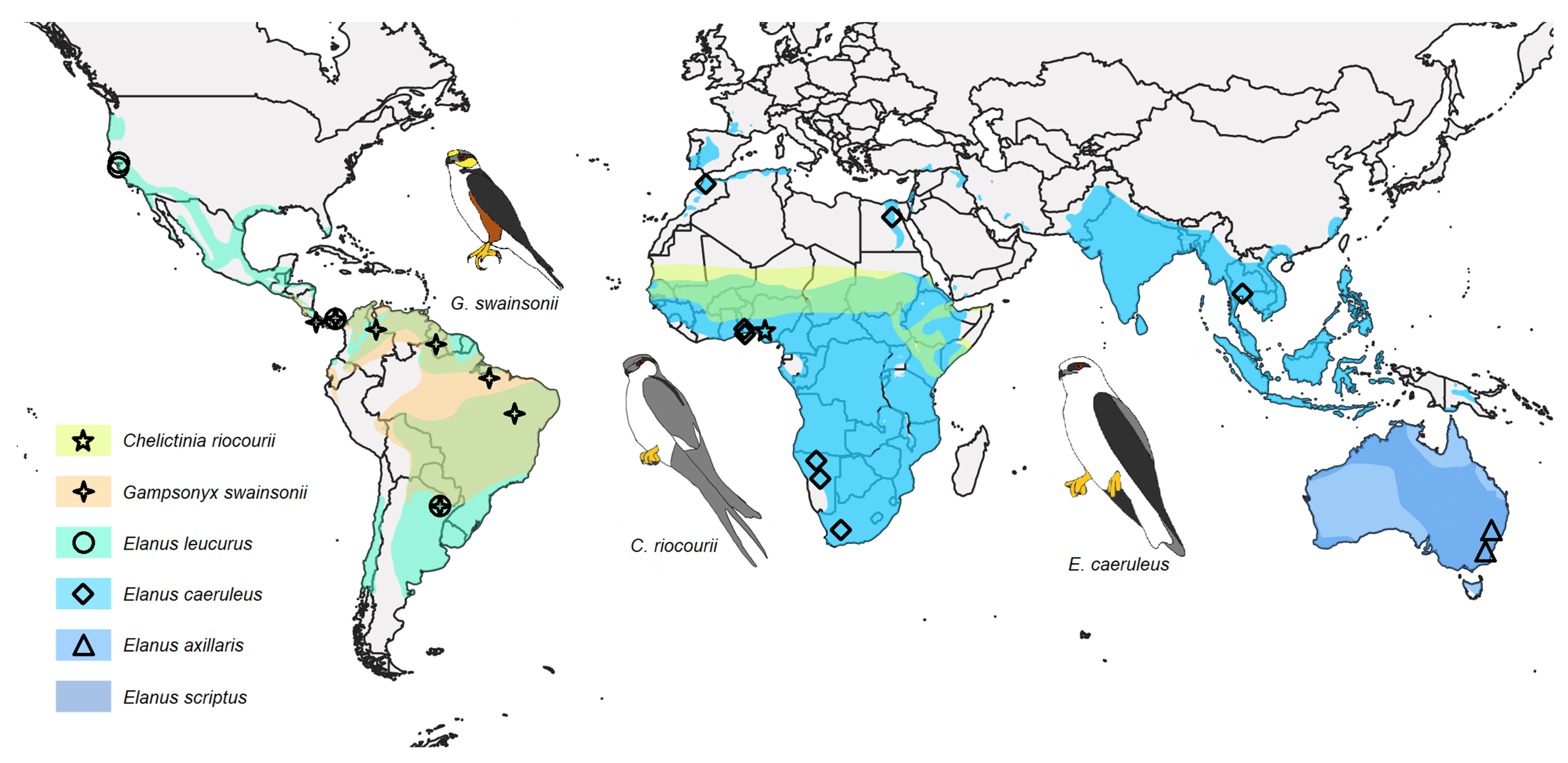
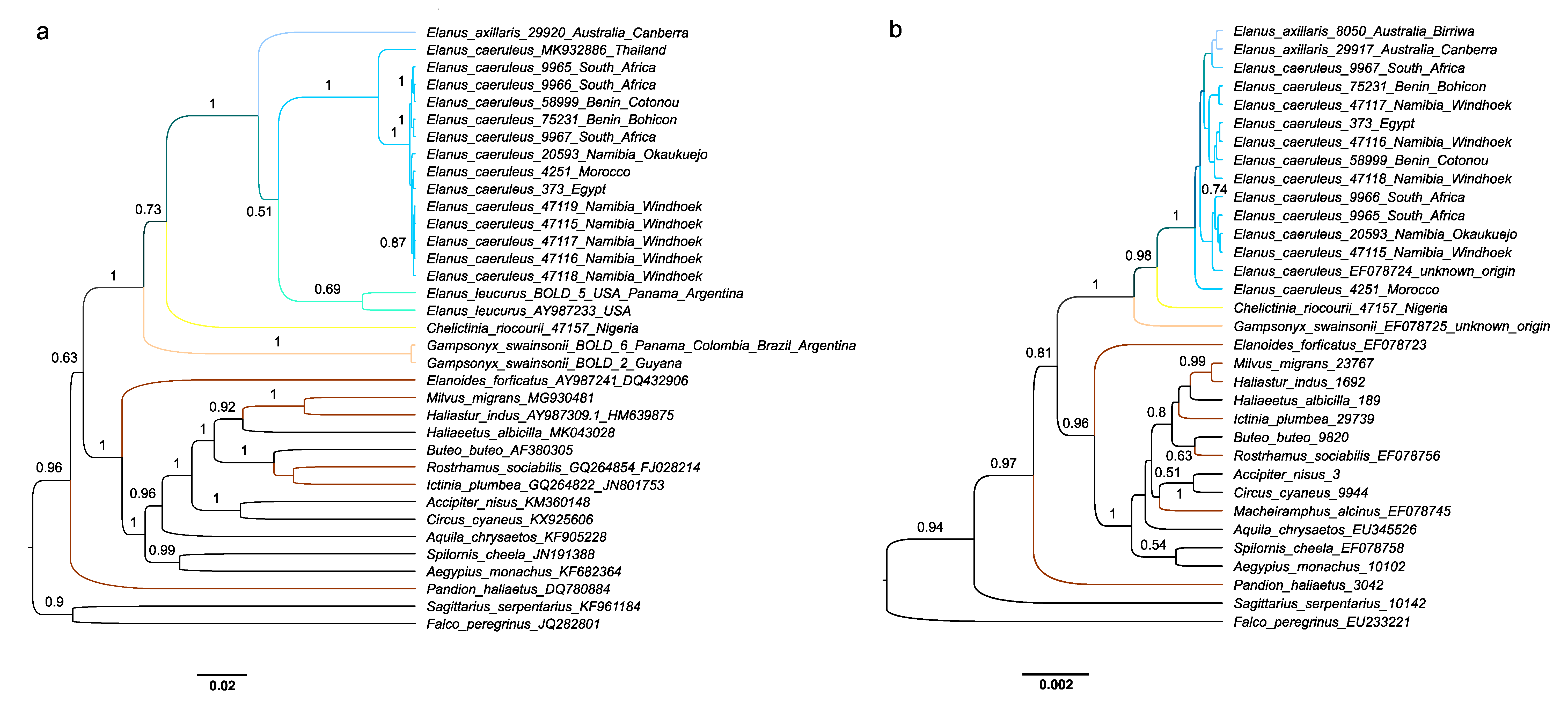
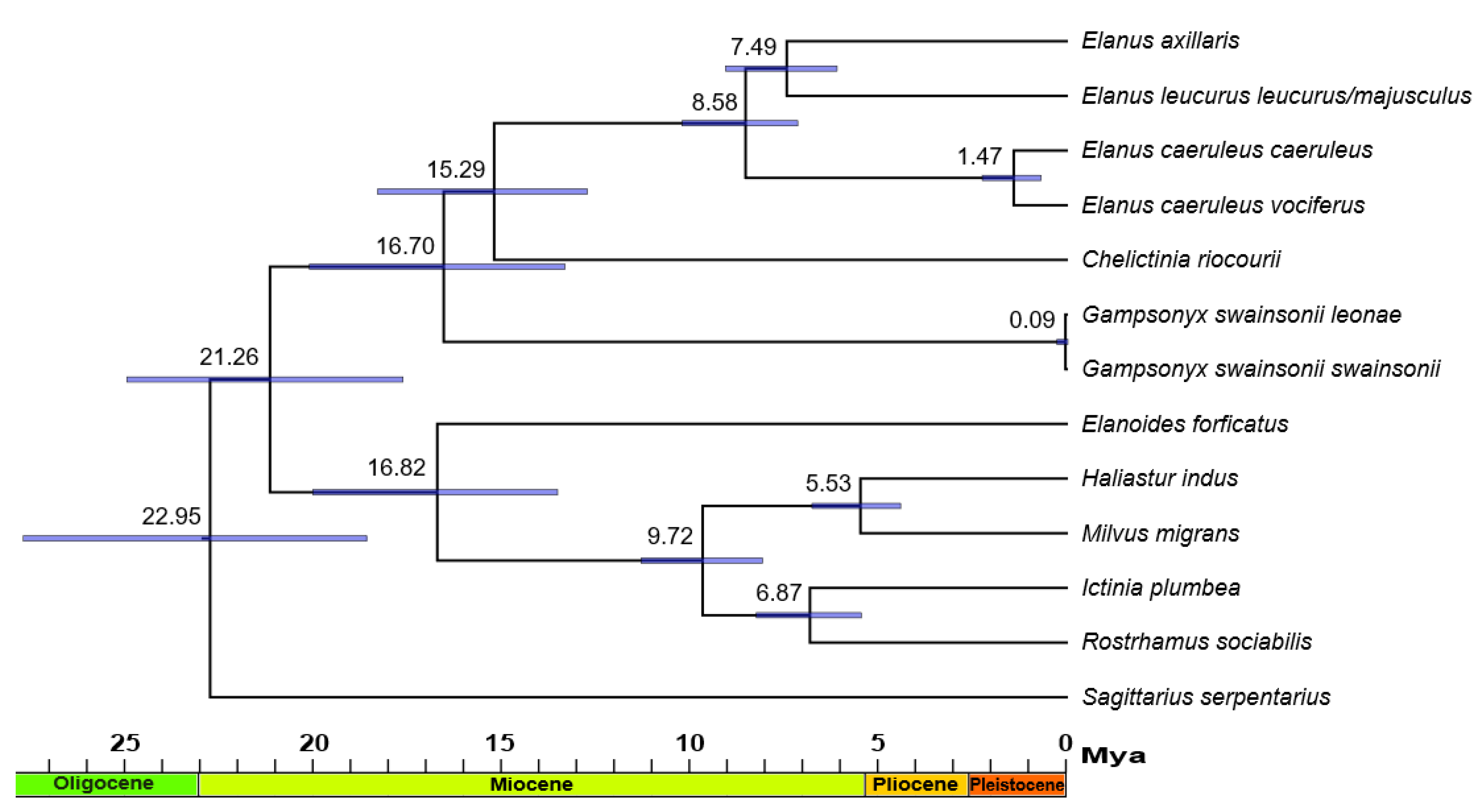
| Taxon | Origin | Source | IPMB Number/GenBank Accession Number/BOLD Process ID | ||
|---|---|---|---|---|---|
| Cyt b | COI | RAG-1 | |||
| Elanus caeruleus caeruleus | Egypt | This study | IPMB373/MT800519 | IPMB373/MT800534 | IPMB373/MT897851 |
| Elanus caeruleus caeruleus | Morocco | This study | IPMB4251/MT800520 | IPMB4251/MT800535 | IPMB4251/MT897852 |
| Elanus caeruleus caeruleus | South Africa, Cape Province | This study | IPMB9965/MT800521 | IPMB9965/MT800536 | IPMB9965/MT897853 |
| Elanus caeruleus caeruleus | South Africa, Cape Province | This study | IPMB9966/MT800522 | IPMB9966/MT800537 | IPMB9966/MT897854 |
| Elanus caeruleus caeruleus | South Africa, Cape Province | This study | IPMB9967/MT800523 | - | IPMB9967/MT897855 |
| Elanus caeruleus caeruleus | Namibia, Okaukuejo | This study | IPMB20593/MT800524 | - | IPMB20593/MT897856 |
| Elanus caeruleus caeruleus | Namibia, Windhoek | This study | IPMB47115/MT800525 | - | IPMB47115/MT897857 |
| Elanus caeruleus caeruleus | Namibia, Windhoek | This study | IPMB47116/MT800526 | IPMB47116/MT800538 | IPMB47116/MT897858 |
| Elanus caeruleus caeruleus | Namibia, Windhoek | This study | IPMB47117/MT800527 | - | IPMB47117/MT897859 |
| Elanus caeruleus caeruleus | Namibia, Windhoek | This study | IPMB47118/MT800528 | - | IPMB47118/MT897860 |
| Elanus caeruleus caeruleus | Namibia, Windhoek | This study | IPMB47119/MT800529 | - | - |
| Elanus caeruleus caeruleus | Benin, Cotonou | This study | IPMB58999/MT800530 | - | IPMB58999/MT897861 |
| Elanus caeruleus caeruleus | Benin, Bohicon | This study | IPMB75231/MT800531 | IPMB75231/MT800539 | IPMB75231/MT897862 |
| Elanus caeruleus vociferus | Thailand | Boonyaprakob and Kasorndorkbua, unpubl. | - | MK932886 | - |
| Elanus caeruleus ssp. | unknown | [33] | - | - | EF078724 |
| Elanus leucurus leucurus | Argentina, San Cayetano (Corrientes) | [35] | - | FJ027543/KBARG184-07 | - |
| Elanus leucurus majusculus | USA | [3] | AY987233 | - | - |
| Elanus leucurus majusculus | Panama, Pacora | Smithsonian Tropical Res. Inst., unpubl. | - | BSENC001-06 | - |
| Elanus leucurus majusculus | Panama, Pacora | Smithsonian Tropical Res. Inst., unpubl. | - | BSPAC008-14 | - |
| Elanus leucurus majusculus | USA, Rodeo (California) | [36] | - | DQ432907/CDMVZ014-05 | - |
| Elanus leucurus majusculus | USA, Berkeley | [36] | - | DQ432908/CDMVZ015-05 | - |
| Elanus axillaris | Australia, Birriwa vicinity | This study | - | - | IPMB8050/MT897863 |
| Elanus axillaris | Australia, Canberra | This study | - | - | IPMB29917/MT897864 |
| Elanus axillaris | Australia, Canberra | This study | IPMB29920/MT800532 | IPMB29920/MT800540 | - |
| Gampsonyx swainsonii swainsonii | Argentina, San Cayetano (Corrientes) | Mus. Argent. Cienc. Nat. Rivadavia, unpubl. | - | FJ027613/KBAR776-06 | - |
| Gampsonyx swainsonii swainsonii | Brazil, Serra das Confusoes | [37] | - | JN801680/LGEMA021-07 | - |
| Gampsonyx swainsonii swainsonii | Brazil, Tailândia | [37] | - | LGEMA404-08 | - |
| Gampsonyx swainsonii leonae | Guyana, Saddle Mountain vicinity | [38] | - | JQ174910/USNMI208-11 | - |
| Gampsonyx swainsonii leonae | Guyana, Saddle Mountain vicinity | [38] | - | JQ174911/USNMK306-11 | - |
| Gampsonyx swainsonii leonae | Panama, Pacora | Smithsonian Tropical Res. Inst., unpubl. | - | BSPBA002-07 | - |
| Gampsonyx swainsonii leonae | Panama, Puerto Armuelles | Smithsonian Tropical Res. Inst., unpubl. | - | BSPBA005-07 | - |
| Gampsonyx swainsonii leonae | Colombia, Caño Agua Verde (Arauca) vicinity | Inst. Invest. Recurs. Biol. von Humboldt, unpubl. | - | IAVHB082-13 | - |
| Gampsonyx swainsonii leonae | unknown | [33] | - | - | EF078725 |
| Chelictinia riocourii | Nigeria, Agbor | This study | IPMB47157/MT800533 | IPMB47157/MT800541 | IPMB47157/MT897865 |
| Gene | Primer | Sequence (5′-3′) | Reference |
|---|---|---|---|
| Cyt b | L14764 | F-TGRTACAAAAAAATAGGMCCMGAAGG | [39] |
| MT-c2 | F-TGAGGACAAATATCATTCTGAGG | [40,41] | |
| Mt-Fr | R-CTAAGAAGGGTGGAGTCTTCAGTTTTTGGTTTACAAGAC | [39], modified | |
| Mt-E | R-GCAAATAGGAAGTATCATTCTGG | [41] | |
| COI | Passer F1 | F-CCAACCACAAAGACATCGGAACC | [42] |
| Passer R1 | R-GTAAACTTCTGGGTGACCAAAGAATC | [42] | |
| RAG-1 | R17 | F-CCCTCCTGCTGGTATCCTTGCTT | [43] |
| R51 | R-GACCCTCTTTCTGCTATGAGGGGGC | [44] |
| № | Species | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Elanus caeruleus caeruleus | - | 0.109 | - | 0.103 | 0.130 | - | - | 0.167 | 0.153 | 0.159 | 0.169 | 0.151 | 0.155 | 0.167 | 0.157 | 0.164 | 0.153 | 0.163 | 0.170 | 0.162 | 0.162 | 0.177 | |
| 2 | Elanus caeruleus vociferus | 0.022 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| 3 | Elanus axillaris | 0.090 | 0.088 | - | 0.087 | 0.114 | - | - | 0.143 | 0.146 | 0.149 | 0.147 | 0.138 | 0.147 | 0.145 | 0.150 | 0.162 | 0.138 | 0.145 | 0.149 | 0.146 | 0.153 | 0.161 | |
| 4 | Elanus leucurus leucurus | 0.092 | 0.096 | 0.096 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| 5 | Elanus leucurus majusculus | 0.092 | 0.096 | 0.096 | 0.000 | 0.127 | - | - | 0.162 | 0.158 | 0.154 | 0.154 | 0.150 | 0.150 | 0.164 | 0.156 | 0.160 | 0.143 | 0.160 | 0.164 | 0.149 | 0.161 | 0.176 | |
| 6 | Chelictinia riocourii | 0.133 | 0.131 | 0.123 | 0.141 | 0.141 | - | - | 0.131 | 0.142 | 0.134 | 0.147 | 0.139 | 0.141 | 0.146 | 0.148 | 0.150 | 0.137 | 0.131 | 0.153 | 0.146 | 0.145 | 0.162 | |
| 7 | Gampsonyx swainsonii leonae | 0.137 | 0.136 | 0.151 | 0.119 | 0.119 | 0.128 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| 8 | Gampsonyx swainsonii swainsonii | 0.138 | 0.137 | 0.152 | 0.121 | 0.121 | 0.129 | 0.000 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| 9 | Elanoides forficatus | 0.163 | 0.164 | 0.159 | 0.164 | 0.164 | 0.149 | 0.153 | 0.154 | 0.145 | 0.137 | 0.134 | 0.127 | 0.136 | 0.144 | 0.137 | 0.138 | 0.141 | 0.138 | 0.140 | 0.157 | 0.158 | 0.159 | |
| 10 | Ictinia plumbea | 0.156 | 0.161 | 0.157 | 0.157 | 0.157 | 0.137 | 0.141 | 0.141 | 0.142 | 0.073 | 0.103 | 0.091 | 0.075 | 0.116 | 0.115 | 0.105 | 0.114 | 0.131 | 0.130 | 0.146 | 0.158 | 0.167 | |
| 11 | Rostrhamus sociabilis | 0.150 | 0.149 | 0.152 | 0.162 | 0.162 | 0.132 | 0.139 | 0.139 | 0.146 | 0.109 | 0.103 | 0.095 | 0.080 | 0.113 | 0.123 | 0.106 | 0.118 | 0.130 | 0.133 | 0.142 | 0.161 | 0.174 | |
| 12 | Haliastur indus | 0.153 | 0.161 | 0.156 | 0.161 | 0.161 | 0.154 | 0.159 | 0.159 | 0.137 | 0.124 | 0.114 | 0.075 | 0.100 | 0.124 | 0.121 | 0.113 | 0.117 | 0.132 | 0.144 | 0.157 | 0.168 | 0.174 | |
| 13 | Milvus migrans | 0.158 | 0.164 | 0.151 | 0.169 | 0.169 | 0.166 | 0.163 | 0.162 | 0.144 | 0.108 | 0.111 | 0.076 | 0.093 | 0.111 | 0.116 | 0.108 | 0.114 | 0.130 | 0.121 | 0.150 | 0.154 | 0.158 | |
| 14 | Buteo buteo | 0.160 | 0.159 | 0.151 | 0.147 | 0.147 | 0.127 | 0.139 | 0.139 | 0.139 | 0.089 | 0.098 | 0.113 | 0.106 | 0.113 | 0.119 | 0.109 | 0.121 | 0.130 | 0.133 | 0.146 | 0.160 | 0.176 | |
| 15 | Accipiter nisus | 0.138 | 0.151 | 0.144 | 0.167 | 0.167 | 0.152 | 0.154 | 0.156 | 0.154 | 0.123 | 0.121 | 0.127 | 0.123 | 0.136 | 0.123 | 0.133 | 0.138 | 0.134 | 0.137 | 0.161 | 0.174 | 0.177 | |
| 16 | Circus cyaneus | 0.155 | 0.156 | 0.144 | 0.164 | 0.164 | 0.146 | 0.159 | 0.159 | 0.156 | 0.119 | 0.126 | 0.127 | 0.113 | 0.119 | 0.108 | 0.131 | 0.134 | 0.138 | 0.135 | 0.162 | 0.167 | 0.172 | |
| 17 | Haliaeetus albicilla | 0.148 | 0.149 | 0.136 | 0.154 | 0.154 | 0.129 | 0.143 | 0.142 | 0.137 | 0.096 | 0.104 | 0.096 | 0.096 | 0.096 | 0.108 | 0.099 | 0.131 | 0.149 | 0.141 | 0.149 | 0.171 | 0.177 | |
| 18 | Aquila chrysaetos | 0.143 | 0.151 | 0.137 | 0.166 | 0.166 | 0.129 | 0.151 | 0.151 | 0.144 | 0.116 | 0.134 | 0.123 | 0.127 | 0.126 | 0.109 | 0.123 | 0.109 | 0.130 | 0.126 | 0.155 | 0.160 | 0.165 | |
| 19 | Spilornis cheela | 0.150 | 0.151 | 0.146 | 0.152 | 0.152 | 0.137 | 0.141 | 0.141 | 0.154 | 0.141 | 0.142 | 0.132 | 0.136 | 0.124 | 0.149 | 0.144 | 0.129 | 0.134 | 0.132 | 0.149 | 0.156 | 0.166 | |
| 20 | Aegypius monachus | 0.148 | 0.156 | 0.154 | 0.162 | 0.162 | 0.149 | 0.151 | 0.151 | 0.154 | 0.129 | 0.134 | 0.119 | 0.126 | 0.126 | 0.141 | 0.118 | 0.123 | 0.137 | 0.132 | 0.166 | 0.163 | 0.168 | |
| 21 | Pandion haliaetus | 0.145 | 0.144 | 0.154 | 0.157 | 0.157 | 0.121 | 0.148 | 0.149 | 0.139 | 0.139 | 0.156 | 0.147 | 0.149 | 0.141 | 0.142 | 0.149 | 0.132 | 0.136 | 0.142 | 0.161 | 0.150 | 0.179 | |
| 22 | Sagittarius serpentarius | 0.166 | 0.171 | 0.164 | 0.169 | 0.169 | 0.146 | 0.156 | 0.156 | 0.161 | 0.166 | 0.157 | 0.146 | 0.164 | 0.156 | 0.171 | 0.175 | 0.159 | 0.159 | 0.169 | 0.152 | 0.151 | 0.173 | |
| 23 | Falco peregrinus | 0.171 | 0.175 | 0.172 | 0.184 | 0.184 | 0.164 | 0.161 | 0.161 | 0.184 | 0.161 | 0.157 | 0.175 | 0.182 | 0.166 | 0.171 | 0.179 | 0.172 | 0.156 | 0.182 | 0.179 | 0.169 | 0.171 |
| Elanus | Chelictinia | Gampsonyx | |
|---|---|---|---|
| Length (cm) | 29–43 | 33–38 | 20–28 |
| Wing (cm) | 249–328 | 225–254 | 141–178 |
| Wingspan (cm) | 77–102 | 68–76 | 45–55 |
| Tail (cm) | 110–186 | 170–216 | 82–108 |
| Tarsus (mm) | 32–40 | 28–33 | 28–32 |
| Weight (g) | 160–427 | 100–140 | 94–140 |
| Plumage color | gray, black, and white | gray, black, and white | black, white, rufous, yellow |
| Eyes color | red; orange-rufous (E. leucurus) | red | Chestnut, or red |
| Range | tropical and subtropical biomes of the world | Subsaharan Africa | Central and South America |
| Habitat | open woodland, savanna, grassland, farmland, urban green areas, marsh, riverine vegetation; cultivated steppe, semi-desert, desert, rocks (E. caeruleus); coastal dunes (E. axillaris) | semi-desert, savanna, grassland | open woodland, savanna, scrub, urban green areas |
| Altitude (m) | 0–3000 (4200) | 0–500 | 0–1000+ |
| Movements | migratory or nomadic; also sedentary (E. axillaris) | migratory, nomadic | sedentary or locally nomadic |
| Feeding | small mammals, also small birds, reptiles and amphibians, large insects; occasionally dead fish (E. caeruleus) | insects, arachnids, lizards, also small snakes, sometimes rodents | lizards, large insects, spiders, also snakes, rodents |
| Social behavior | solitary or in pair; colonial (E. scriptus) | colonial or in pair | solitary or in pair |
| Nest place | trees; occasionally bush (E. caeruleus) or artificial structures | trees or bush | trees |
| Height of nest place (m) | 0.75–35 | 2–8 | 4–7 |
| Nest (across, cm) | 30–74 | 30–40 | 20 |
| Number of eggs | 3–5 (2–6) | 4 | 3–4 (2–4) |
| Egg measurements | 37–46 × 29–36 | 34–38 × 27–31 | 30 × 24 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Starikov, I.J.; Wink, M. Old and Cosmopolite: Molecular Phylogeny of Tropical–Subtropical Kites (Aves: Elaninae) with Taxonomic Implications. Diversity 2020, 12, 327. https://doi.org/10.3390/d12090327
Starikov IJ, Wink M. Old and Cosmopolite: Molecular Phylogeny of Tropical–Subtropical Kites (Aves: Elaninae) with Taxonomic Implications. Diversity. 2020; 12(9):327. https://doi.org/10.3390/d12090327
Chicago/Turabian StyleStarikov, Ivan J., and Michael Wink. 2020. "Old and Cosmopolite: Molecular Phylogeny of Tropical–Subtropical Kites (Aves: Elaninae) with Taxonomic Implications" Diversity 12, no. 9: 327. https://doi.org/10.3390/d12090327
APA StyleStarikov, I. J., & Wink, M. (2020). Old and Cosmopolite: Molecular Phylogeny of Tropical–Subtropical Kites (Aves: Elaninae) with Taxonomic Implications. Diversity, 12(9), 327. https://doi.org/10.3390/d12090327






