Micractinium tetrahymenae (Trebouxiophyceae, Chlorophyta), a New Endosymbiont Isolated from Ciliates
Abstract
1. Introduction
2. Material and Methods
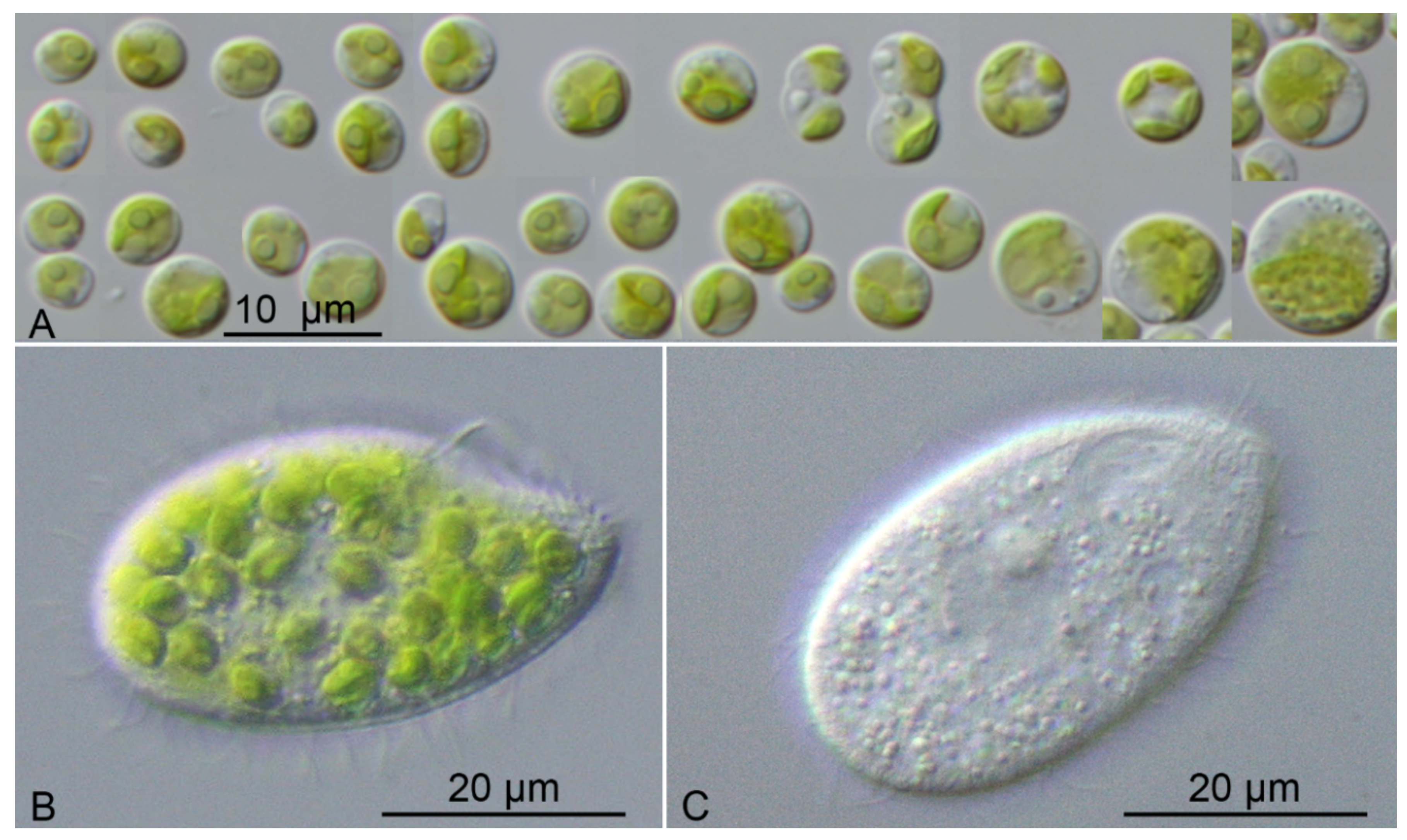
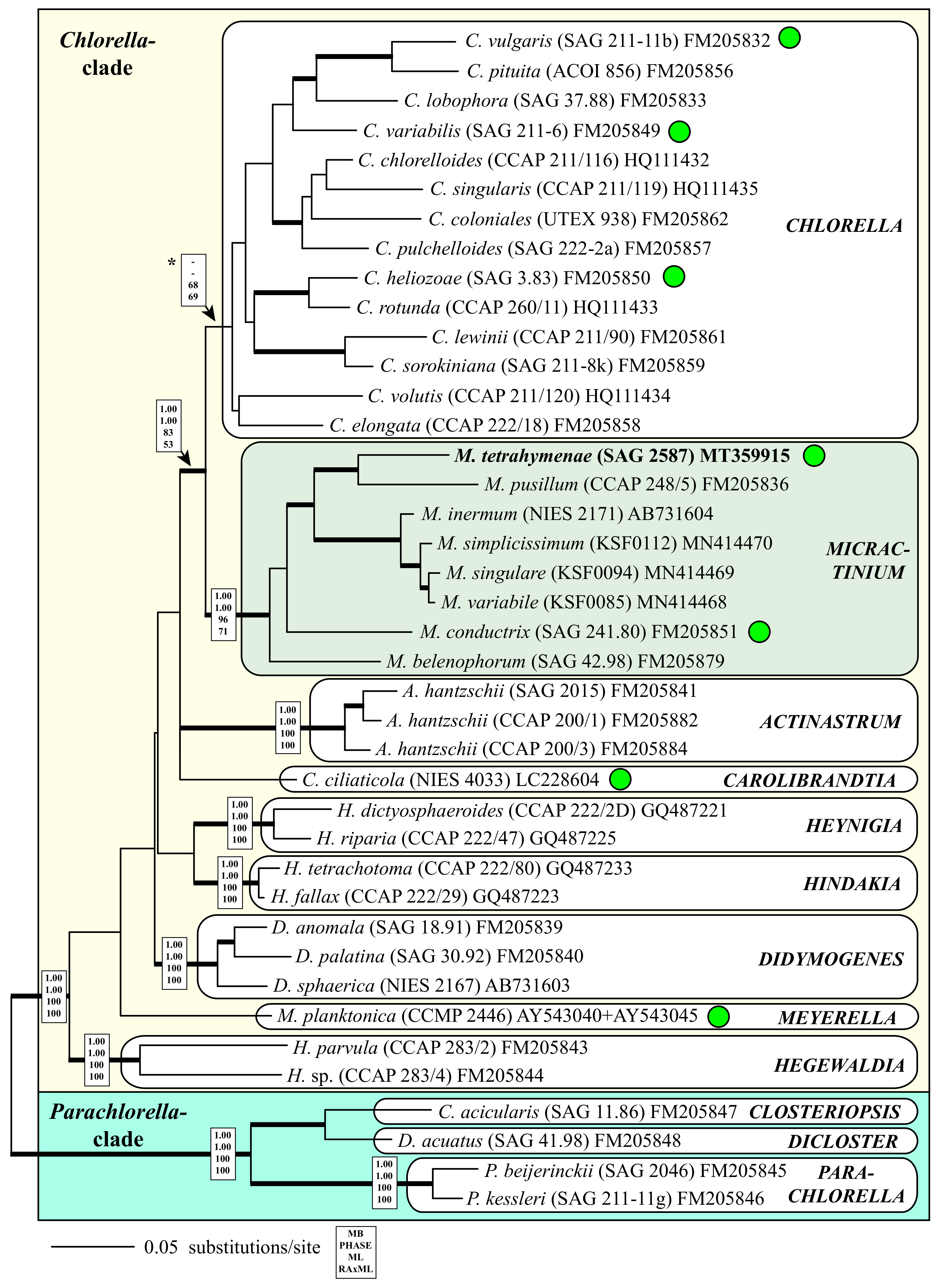
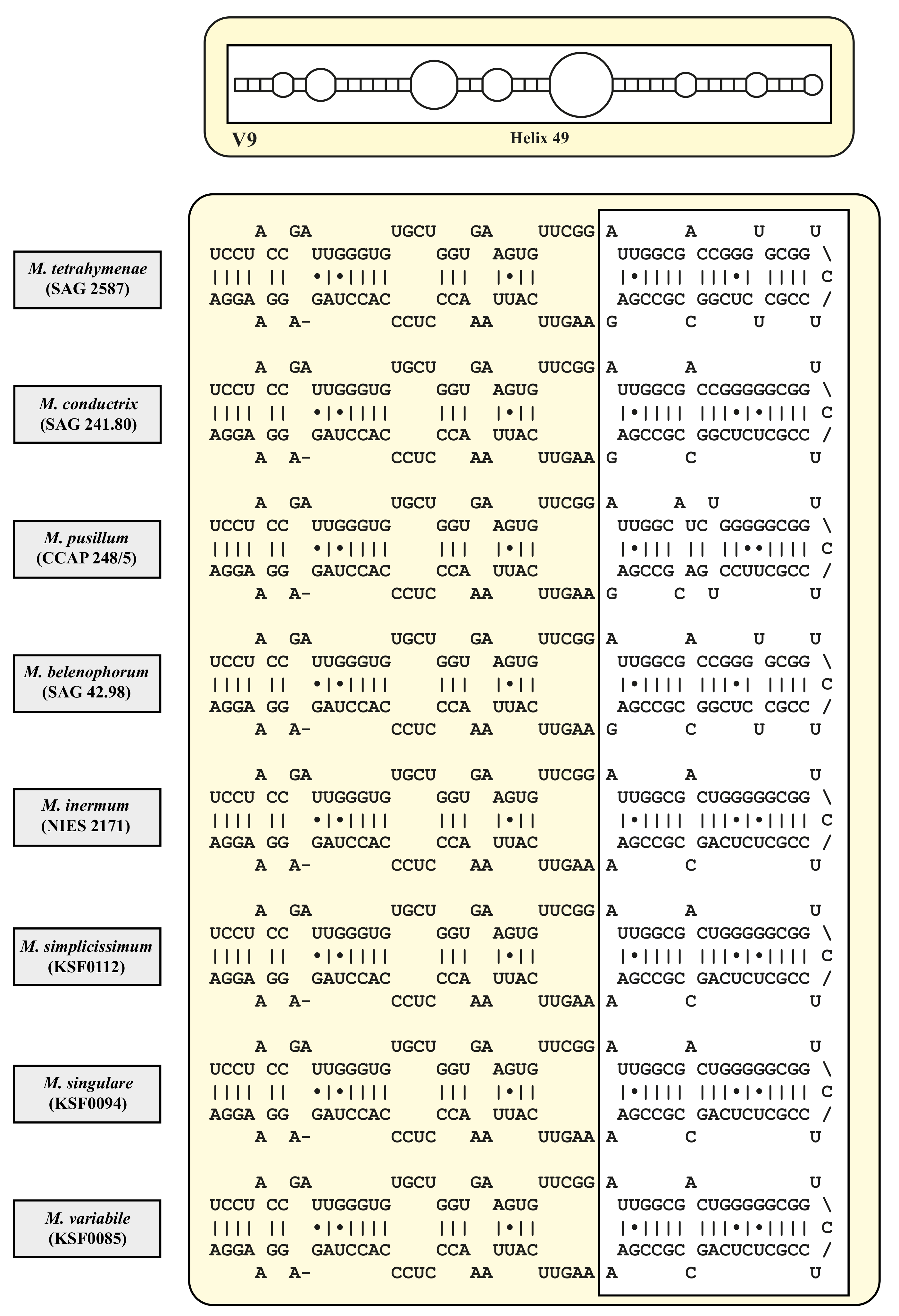
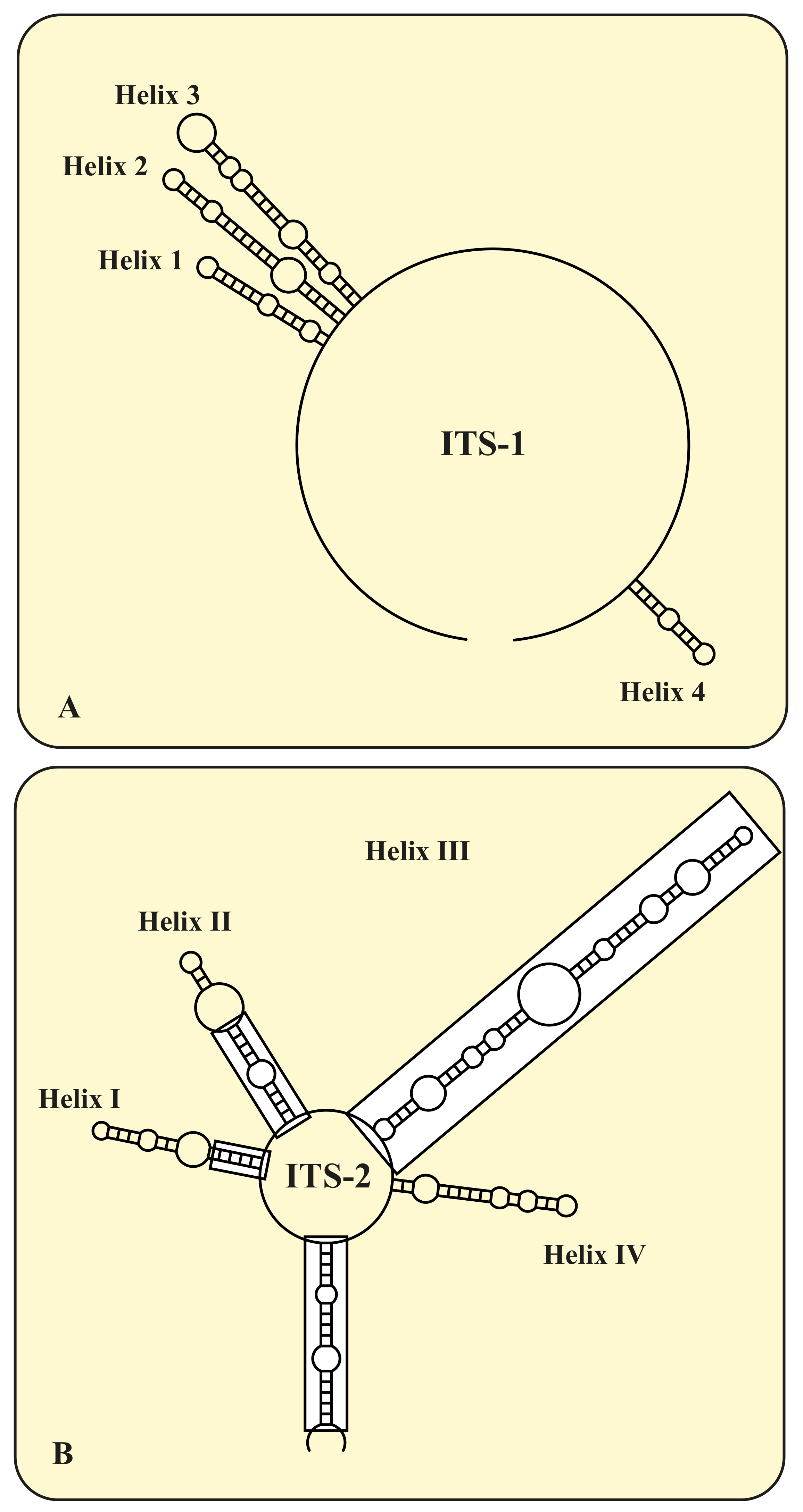
3. Results
4. Discussion
4.1. Green Algae in Endosymbiosis Belonging to the Chlorellaceae
4.2. Taxonomy and Systematics of the Genus Micractinium
4.3. Ecology and Distribution of Micractinium
4.4. Interactions between Tetrahymena Utriculariae and Micractinium Tetrahymenae
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Fresenius, G. Beiträge zur Kenntniss mikrokopischer Organismen. Abh. Senckenberg. Naturforsch. Ges. 1858, 2, 211–242. [Google Scholar]
- Komarek, J.; Fott, B. Chlorophyceae (Grünalgen) Ordnung: Chlorococcales. In Das Phytoplankton des Süßwassers 7. Teil, 1. Hälfte; Huber-Pestalozzi, G., Ed.; Schweizerbart: Stuttgart, Germany, 1983; pp. 1–1044. [Google Scholar]
- Luo, W.; Krienitz, L.; Pflugmacher, S.; Walz, N. Genus and species concept in Chlorella and Micractinium (Chlorophyta, Chlorellaceae): Genotype versus phenotypical variability under ecosystem conditions. VInternationale Vereinigung für theoretische und angewandte Limnologie: Verhandlungen 2005, 29, 170–173. [Google Scholar] [CrossRef]
- Luo, W.; Pflugmacher, S.; Pröschold, T.; Walz, N.; Krienitz, L. Genotype versus phenotype variability in Chlorella and Micractinium (Chlorophyta, Trebouxiophyceae). Protist 2006, 157, 315–333. [Google Scholar] [CrossRef]
- Luo, W.; Pröschold, T.; Bock, C.; Krienitz, L. Generic concept in Chlorella-related coccoid green algae (Chlorophyta, Trebouxiophyceae). Plant Biol. 2010, 12, 545–553. [Google Scholar] [CrossRef]
- Pröschold, T.; Bock, C.; Luo, W.; Krienitz, L. Polyphyletic distribution of bristle formation in Chlorellaceae: Micractinium, Diacanthos, Didymogenes and Hegewaldia gen. nov. (Trebouxiophyceae, Chlorophyta). Phycol. Res. 2010, 58, 1–8. [Google Scholar] [CrossRef]
- Pröschold, T.; Darienko, T.; Silva, P.C.; Reisser, W.; Krienitz, L. The systematics of "Zoochlorella" revisited employing an integrative approach. Environ. Microbiol. 2011, 13, 350–364. [Google Scholar] [CrossRef] [PubMed]
- Brandt, K. Über das Zusammenleben von Algen und Thieren. Biol. Centralbl. 1882, 1, 524–527. [Google Scholar]
- Kreutz, M.; Foissner, W. The Sphagnum ponds of Simmelried in Germany: A biodiversity hot-spot for microscopic organisms. Protozool. Monogr. 2006, 3, 1–267. [Google Scholar]
- Kreutz, M.; Stoeck, T.; Foissner, W. Morphological and molecular characterization of Paramecium (Viridoparamecium nov. subgen.) chlorelligerum Kahl 1935 (Ciliophora). J. Eukaryot. Microbiol. 2012, 59, 548–563. [Google Scholar] [CrossRef]
- Hoshina, R.; Kobayashi, M.; Suzaki, T.; Kusuoka, Y. Brandtia ciliaticola gen. et sp. nov. (Chlorellaceae, Trebouxiophyceae) a common symbiotic green coccoid of various ciliate species. Phycol. Res. 2018, 66, 76–81. [Google Scholar] [CrossRef]
- Hoshina, R.; Nakada, T. Carolibrandtia nom. nov. as a replacement name for Brandtia Hoshina (Chlorellaceae, Trebouxiophyceae). Phycol. Res. 2018, 66, 82–83. [Google Scholar] [CrossRef]
- Fujishima, M. Endosymbionts in Paramecium. Microbiol. Monogr. 2009, 12, 1–252. [Google Scholar]
- Pitsch, G.; Adamec, L.; Dirren, S.; Nitsche, F.; Simek, K.; Sirova, D.; Posch, T. The green Tetrahymena utriculariae n. sp. (Ciliophora, Oligohymenophorea) with its endosymbiotic algae (Micractinium sp.), living in traps of a carnivorous aquatic plant. J. Eukaryot. Microbiol. 2017, 64, 322–335. [Google Scholar] [CrossRef] [PubMed]
- Simek, K.; Pitsch, G.; Salcher, M.M.; Sirova, D.; Shabarova, T.; Adamec, L.; Posch, T. Ecological traits of the algae-bearing Tetrahymena utriculariae (Ciliophora) from traps of the aquatic carnivorous plant Utricularia reflexa. J. Eukaryot. Microbiol. 2017, 64, 336–348. [Google Scholar] [CrossRef] [PubMed]
- Schlösser, U.G. SAG-Sammlung von Algenkulturen at the University of Göttingen. Botanica Acta 1994, 107, 424–429. [Google Scholar]
- Marin, B.; Palm, A.; Klingberg, M.; Melkonian, M. Phylogeny and taxonomic revision of plastid-containing euglenophytes based on SSU rDNA sequence comparisons and synapomorphic signatures in the SSU rRNA secondary structure. Protist 2003, 154, 99–145. [Google Scholar] [CrossRef] [PubMed]
- Darienko, T.; Rad-Menendez, L.; Campbell, C.; Pröschold, T. Are there any true marine Chlorella species? Molecular phylogenetic assessment and ecology of marine Chlorella-like organisms, including a description of Droopiella gen. nov. Syst. Biodivers. 2019, 17, 811–829. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acid Res. 2003, 31, 3406–3615. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0b10; Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Jow, H.; Hudelot, C.; Rattray, M.; Higgs, P. Bayesian phylogenetics using an RNA substitution model applied to early mammalian evolution. Mol. Biol. Evol. 2002, 19, 1591–1601. [Google Scholar] [CrossRef] [PubMed]
- Higgs, P.; Jameson, D.; Jow, H.; Rattray, M. The evolution of tRNA-Leu genes in animal mitochondrial genomes. J. Mol. Evol. 2003, 57, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Hudelot, C.; Gowri-Shankar, V.; Jow, H.; Rattray, M.; Higgs, P. RNA-based phylogenetic methods: Application to mammalian mitochondrial RNA sequences. Mol. Phylogen. Evol. 2003, 28, 241–252. [Google Scholar] [CrossRef]
- Gibson, A.; Gowri-Shankar, V.; Higgs, P.; Rattray, M. A comprehensive analysis of mammalian mitochondrial genome base composition and improved phylogenetic methods. Mol. Biol. Evol. 2005, 22, 251–264. [Google Scholar] [CrossRef]
- Telford, M.J.; Wise, M.J.; Gowri-Shankar, V. Consideration of RNA secondary structure significantly improves likelihood-based estimates of phylogeny: Examples from the bilateria. Mol. Biol. Evol. 2005, 22, 1129–1136. [Google Scholar] [CrossRef]
- Coleman, A.W.; Mai, J.C. Ribosomal DNA ITS-1 and ITS-2 sequence comparisons as a tool for predicting genetic relatedness. J. Mol. Evol. 1997, 45, 168–177. [Google Scholar] [CrossRef]
- Hoshina, R.; Iwataki, M.; Imamura, N. Chlorella variabilis and Micractinium reisseri sp. nov. (Chlorellaceae, Trebouxiophyceae): Redescription of the endosymbiotic green algae of Paramecium bursaria (Peniculia, Oligohymenophorea) in the 120th year. Phycol. Res. 2010, 58, 188–201. [Google Scholar] [CrossRef]
- Pröschold, T.; Rieser, D.; Darienko, T.; Kammerlander, B.; Pitsch, G.; Bruni, E.P.; Qu, Z.; Forster, D.; Rad-Menendez, C.; Posch, T.; et al. An integrative approach sheds new light onto the systematics and ecology of the widespread ciliate genus Coleps (Ciliophora, Prostomatea). Environm. Microbiol. 2020, submitted. [Google Scholar]
- Hoshina, R.; Fujiwara, Y. Molecular characterization of Chlorella cultures of the National Institute for Environmental Studies culture collection with description of Micractinium inermum sp. nov., Didymogenes sphaerica sp. nov., and Didymogenes soliella sp. nov. (Chlorellaceae, Trebouxiophyceae). Phycol. Res. 2013, 31, 124–132. [Google Scholar]
- Chae, H.; Lim, S.; Kim, H.S.; Choi, H.-G.; Kim, J.H. Morphology and phylogenetic relationships of Micractinium (Chlorellaceae, Trebouxiophyceae) taxa, including three new species from Antarctica. Algae 2019, 34, 267–275. [Google Scholar] [CrossRef]
- Atkinson, A.W.; Gunning, B.E.S.; John, P.C.L. Sporopollenin in the cell wall of Chlorella and other algae: Ultrastructure, chemistry and incorporation of 14C-acetate, studied in synchronous cultures. Planta 1972, 107, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Hegewald, E.; Schnepf, E. Zur Struktur und Taxonomie bestachelter Chlorellales. Nova Hedwigia 1984, 39, 297–383. [Google Scholar]
- Schnepf, E.; Deichgräber, G.; Glaab, M.; Hegewald, E. Bristles and spikes in Chlorococcales: Ultrastructural studies in Acanthosphaera, Micractinium, Pediastrum, Polyedriopsis, Scenedesmus, and Siderocystopsis. J. Ultrastr. Res. 1980, 72, 367–379. [Google Scholar] [CrossRef]
- Krienitz, L.; Bock, C. Present state of the systematics of planktonic coccoid green algae of inland waters. Hydrobiologia 2012, 698, 295–326. [Google Scholar] [CrossRef]
- Hegewald, E.; Schnepf, E. Ergänzungen und Korrekturen zur Struktur und Taxonomie bestachelter Chlorellales. Nova Hedwigia 1987, 44, 537–541. [Google Scholar]
- Hegewald, E.; Schnepf, E. The ultrastructure and taxonomic placement of Diacanthos belenophorus Kors. (Chlorophyta, Trebouxiophyceae, Micractiniaceae). Constancea 2002, 83, 11. [Google Scholar]
- Nygaard, G. Hydrobiological studies on some Danish ponds and lakes. Kongl. Dansk Vid. Selskab. Biol. Skr. København 1949, 7, 1–293. [Google Scholar]
- Lund, J.W.G. Three new British algal records and spore formation in Micractinium pusillum Fres. Naturalist 1954, 1954, 81–85. [Google Scholar]
- Korshikov, A.A. Viznachnik prisnovodnihk vodorostey Ukrainsykoi RSR [Vyp] V. Pidklas Protokokovi (Protococcineae). In Vakuol’ni (Vacuolales) ta Protokokovi (Protococcales); Naukova Dumka: Kyiv, Ukraine, 1953. [Google Scholar]
- Hegewald, E. Interessante Algen aus dem Ischelandteich in Hagen. Dortmunder Beitr. Landesk. Naturw. Mitt. 1979, 11, 13–16. [Google Scholar]
- Iyengar, M.O.P.; Balakrishnan, M.S. On sexual reproduction in a new species of Golenkinia. J. Indian Bot. Soc. 1956, 35, 371–373. [Google Scholar]
- Starr, R.C. Homothallism in Golenkinia minutissima. In Studies on Microalgae and Photosynthetic Bacteria; Japanese Society of Plant Physiologists, Ed.; Univ. Tokyo Press: Tokyo, Japan, 1963; pp. 3–6. [Google Scholar]
- Ellis, R.J.; Machlis, L. Control of sexuality in Golenkinia. Am. J. Bot. 1968, 55, 600–610. [Google Scholar] [CrossRef]
- Moestrup, Ø. Observations on the fine structure of spermatozoids and vegetative cells of the green alga Golenkinia. Br. Phycol. J. 1972, 7, 169–183. [Google Scholar] [CrossRef][Green Version]
- Fucikova, K.; Pazoutova, M.; Rindi, F. Meiotic genes and sexual reproduction in the green algal class Trebouxiophyceae (Chlorophyta). J. Phycol. 2015, 51, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Andreyeva, V.M. New species of Chlorella Beijer. Bot. Zhurn. 1973, 58, 1735–1741. (in Russian). [Google Scholar]
- Hodac, L.; Hallman, C.; Spitzer, K.; Elster, J.; Faßhauer, F.; Brinkmann, N.; Lepka, D.; Diwan, V.; Friedl, T. Widespread green algae Chlorella and Stichococcus exhibit polar-temperate and tropical-temperate biogeography. FEMS Microbiol. Ecol. 2016, 93, 1–15. [Google Scholar]
- Cheng, C.-Y.; Chang, S.-L.; Lin, I.-T.; Yao, M.-C. Abundant and diverse Tetrahymena species living in the bladder traps of aquatic carnivorous Utricularia plants. Sci. Rep. 2019, 9, 13669. [Google Scholar] [CrossRef]
- Sirová, D.; Borovec, J.; Cerná, B.; Rejmánková, E.; Adamec, L.; Vrba, J. Microbial community development in the traps of aquatic Utricularia species. Aquat. Bot. 2009, 90, 129–136. [Google Scholar] [CrossRef]
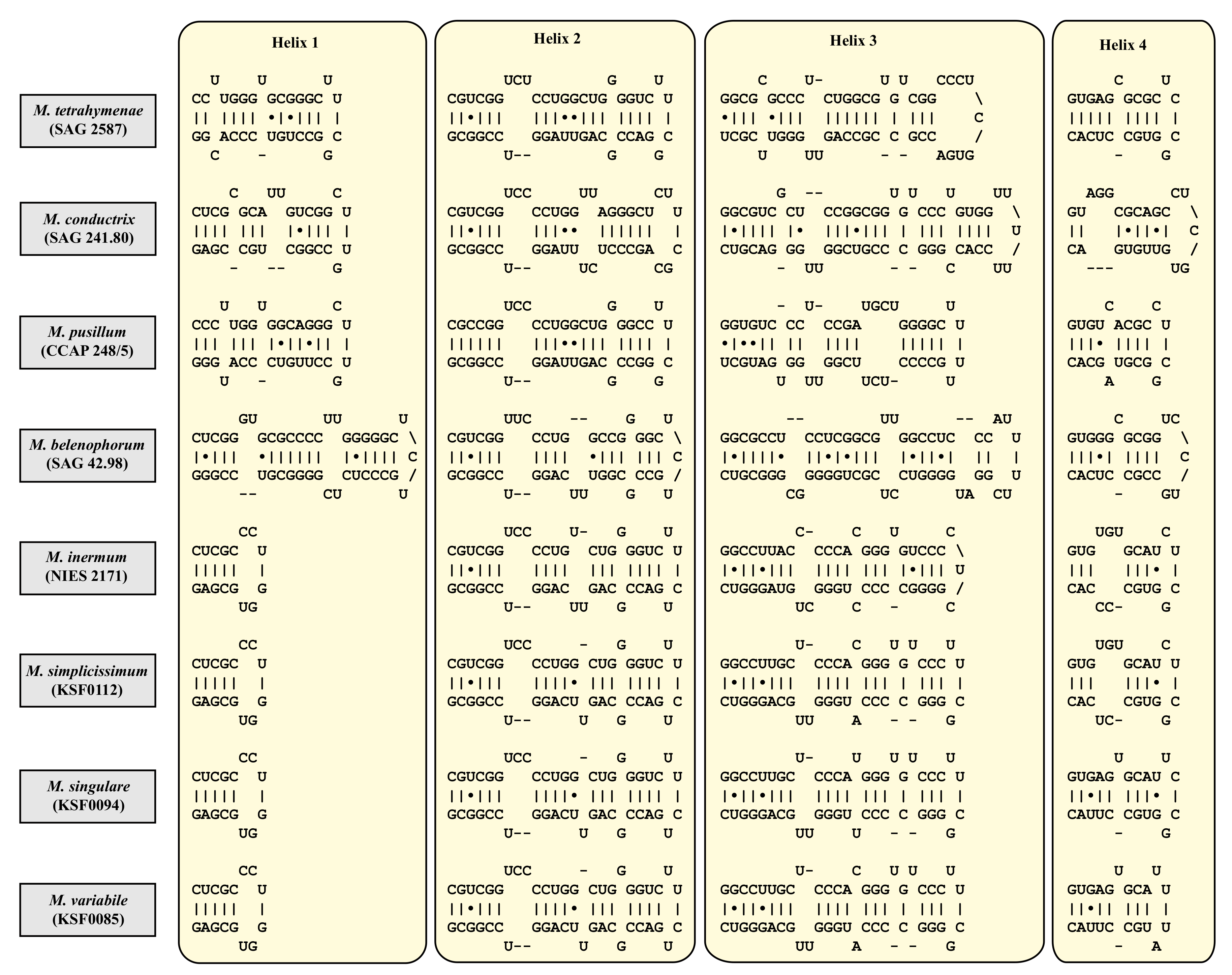
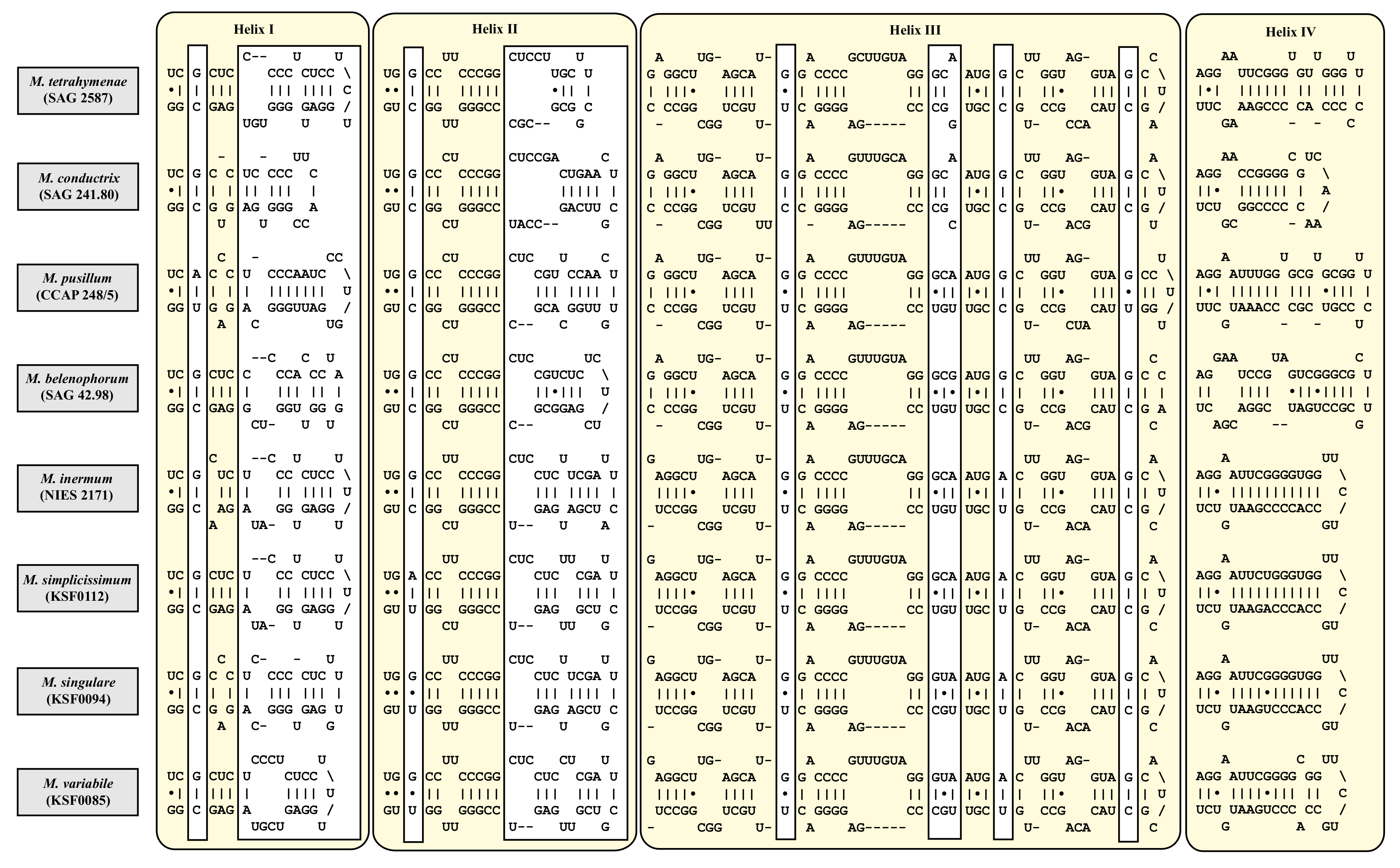
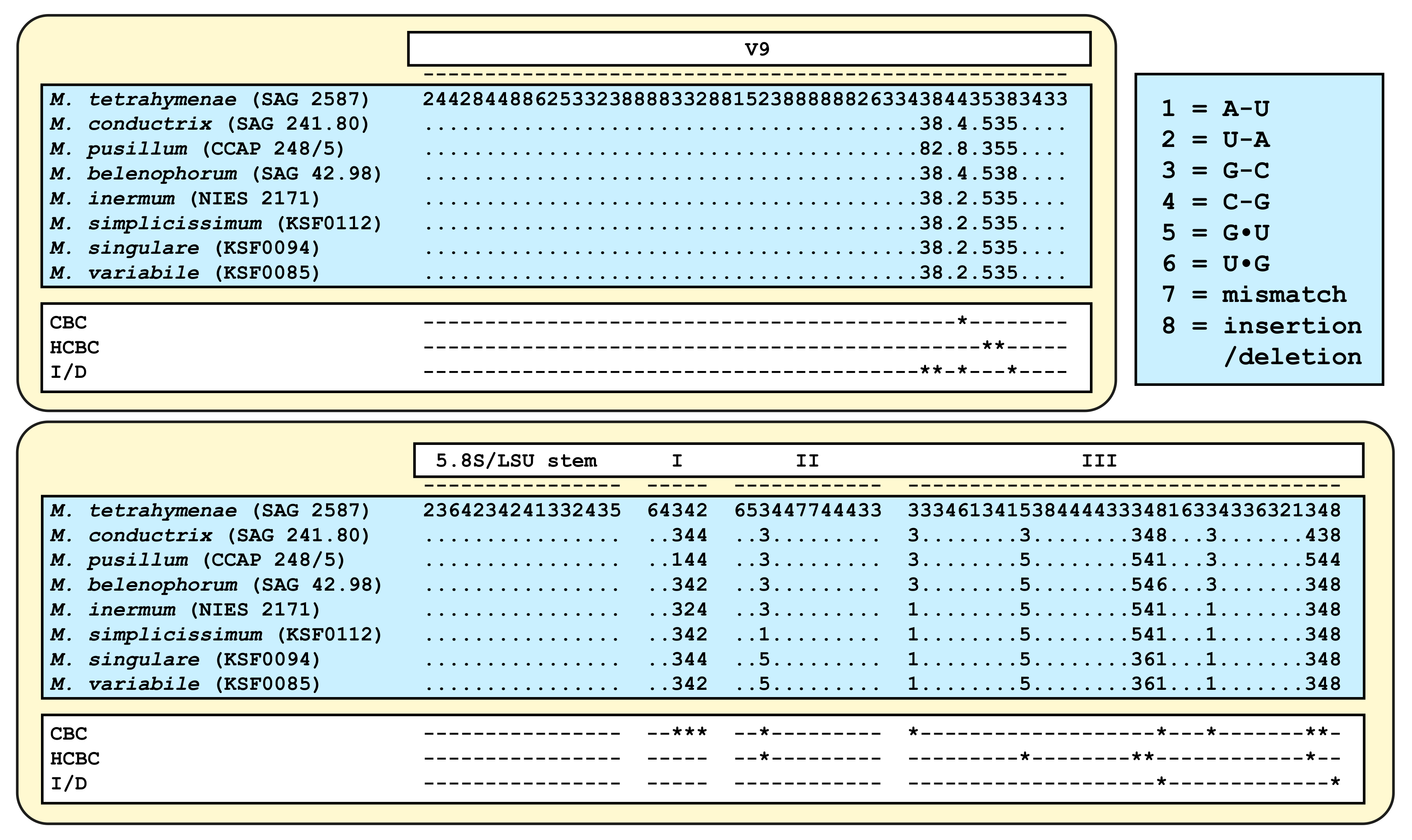
| Species | Cell Shape | Cell Size [µm] | Chloroplast Shape | Pyrenoid | Bristles | Length of Bristles [µm] | Colony Formation | Life Style | Habitat | Reference Strain |
|---|---|---|---|---|---|---|---|---|---|---|
| M.pusillum | spherical | 3.0–7.0–12.0 | cup-shaped | + | up to 8 per cell | 40–65–(100) | single, or up to 8–32 | free-living | plankton of ponds | CCAP 248/5 |
| M.elongatum | spherical | 6.0–7.0 | cup-shaped | + | 1–2 per cell | up to 50.0 | 4 - celled | free-living | plankton | - |
| M.appendiculatum | ellipsoidal, ovoid | 8.0 | parietal | + | 2–4 per cell | 28.0–70.0 | up to 64 | free-living | plankton of ponds | - |
| M.belenophorum | ellipsoidal, broadly ellipsoidal | 8.0–10.0 × 4.5–5.5 | parietal | + | 2 per cell at the poles | up to 55.0 | up to 4 | free-living | plankton of ponds and rivers | SAG 42.98 |
| M.conductrix | spherical | 4.0 -10.0 | cup-shaped | + | - | - | - | endosymbiotic | Paramecium bursaria | SAG 241.80 |
| M.extremum | spherical | 5.2–6.4 | cup-shaped | + | 1–2 per cell | up to 30.0 | 8 celled | free-living | plankton | - |
| M.inermum | ellipsoidal up to spherical | 5.0–5.4–3.2–3.7 | cup-shaped | + | - | - | - | free-living | NIES 2171 | |
| M.quadrisetum | ovoid, ellipsoidal | 6.0- 10.0 × 4.0–7.0 | cup-shaped | + | 1–4 | 23.0–50.0 | 16 celled and more | free-living | Plankton of ponds and rivers | - |
| M.simplicissmum | ellipsoidal up to spherical | 5.5–5.7 × 3.3–3.9 | cup-shaped | + | - | - | - | free-living | KSF0112 | |
| M.singulare | ellipsoidal up to spherical | 7.2–7.4 × 4.5–4.7 | cup-shaped | + | - | - | - | free-living | KSF0094 | |
| M.tetrahymenae | spherical | cup-shaped | + | - | - | - | endosymbiotic | Tetrahymena utriculariae | SAG 2587 | |
| M.variabile | ellipsoidal up to sphaerical | 8.2–8.6 × 5.0–5.4 | cup-shaped | + | 4–8 (?) | 10–30 | solitary,+ | free-living | plankton | KSF0085 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pröschold, T.; Pitsch, G.; Darienko, T. Micractinium tetrahymenae (Trebouxiophyceae, Chlorophyta), a New Endosymbiont Isolated from Ciliates. Diversity 2020, 12, 200. https://doi.org/10.3390/d12050200
Pröschold T, Pitsch G, Darienko T. Micractinium tetrahymenae (Trebouxiophyceae, Chlorophyta), a New Endosymbiont Isolated from Ciliates. Diversity. 2020; 12(5):200. https://doi.org/10.3390/d12050200
Chicago/Turabian StylePröschold, Thomas, Gianna Pitsch, and Tatyana Darienko. 2020. "Micractinium tetrahymenae (Trebouxiophyceae, Chlorophyta), a New Endosymbiont Isolated from Ciliates" Diversity 12, no. 5: 200. https://doi.org/10.3390/d12050200
APA StylePröschold, T., Pitsch, G., & Darienko, T. (2020). Micractinium tetrahymenae (Trebouxiophyceae, Chlorophyta), a New Endosymbiont Isolated from Ciliates. Diversity, 12(5), 200. https://doi.org/10.3390/d12050200





