Abstract
Tardigrades are microscopic invertebrates that can withstand complete desiccation, but their interspecies interactions with prokaryotes and eukaryotes within their microhabitat remain relatively unexplored. Here, I utilized combined metabarcoding of eukaryotes and prokaryotes to simultaneously identify entire community structures within xeric and mesic mosses that harbor tardigrades. The populations of organisms within the microecosystems were successfully determined in 45 xeric moss samples and 47 mesic moss samples. Organismal composition was largely consistent regardless of the moss/lichen substrate, but significantly varied in the two tested locations, possibly because of the differences in environmental humidity. Xeric mosses containing xerophilic tardigrades and other anhydrobiotic invertebrates tended to have significantly limited biological diversity and prokaryotic population dominated by cyanobacteria, suggesting a selection due to extreme desiccation. A combined metabarcoding approach to identify both eukaryotes and prokaryotes can successfully elucidate community structures within microscopic ecosystems, and this can be a potential approach to study the microecology of meiofauna, including tardigrades.
1. Introduction
Upon dehydration of the habitat, the time required for migration to another hydrated environment often exceeds the time it takes for the surroundings to dry up for limno-terrestrial meiofauna, and sometimes the primary strategy for tolerating these adverse conditions is to stay dormant [1,2]. Many tardigrades are known to withstand almost complete desiccation through a mechanism called anhydrobiosis [3,4]. Ecological studies of tardigrades thus far have been predominantly focused on biogeographic scales [5,6,7], and studies on interspecies interactions and community structures within the microenvironments of tardigrade habitats are still limited; however, factors within the tardigrade habitat, such as humidity, food source, and the presence of antagonists, are suggested to play important roles in determining tardigrade distributions, possibly more so than geographical or abiotic factors such as altitude or substrate and plant type [8,9,10]. Humidity is also reported to sometimes affect the reproductive traits of tardigrades [11].
Interspecies interactions between tardigrades and other organisms within the microhabitats, such as xeric moss or lichens, have often been studied in detail for one or several specific counterparts. Within tardigrades, negative associations between Milnesium and Minibiotus species have been observed, which can possibly be explained by predation, or may be due to inverse substrate specificities for mosses or lichens, where significant interspecific associations between tardigrade species and cryptogam substrates seems evident [12]. Similar interspecific associations have also been reported and presumably linked to direct predation between Milnesium and Hypsibius species, and negative associations were suggested to be as a result of competitive exclusion between Macrobiotus and Hypsibius species [13]. Carnivorous tardigrades, including Milnesium and several Macrobiotus species, prey on nematodes; therefore, the population density of predatory tardigrades and prey nematodes are directly associated [14,15]. Likewise, the artificial addition of predatory arthropods that prey on both tardigrades and nematodes have been shown to reduce the tardigrade population [16]. In laboratory culture, eutardigrades are commonly fed microalgae, rotifers, or nematodes, and the availability of such organisms would likely affect the size of the tardigrade population in a similar manner [17]. The rearing of heterotardigrades in the laboratory environment is still challenging, but those living in moss are reported to feed on plant materials at least in laboratory settings, suggesting substrate preferences for mosses [7].
Ecological interactions between tardigrades and microscopic organisms, including fungi, protists, and microbes, remain mostly unexplored. Xerophilic tardigrades do not favor prolonged hydration of their habitat [13], suggested to be partly due to an increase in fungi and their predation or parasitization on tardigrades in high relative humidity environments, resulting in the reduction of the tardigrade population [18]. Symbiotic microbes have been reported in the specialized cephalic vesicles of several marine tardigrades of the family Halechiniscidae, and the protist Pyxidium tardigradum is known as a specialist that selectively attaches to eutardigrades (see [19] for a comprehensive review on this subject). Observations based on the isolation of specific organisms are, however, limited in the analysis of the overall community structures within these microhabitats. Amplicon sequencing of DNA barcodes, such as 16S rRNA for microbes and 18S rRNA for eukaryotes, allows for the investigation of the comprehensive community structures of these organisms. This approach has been successfully applied to study geographic and ecological distributions of tardigrades using clade-specific primers to exclusively amplify DNA barcodes of eutardigrades and bdelloid rotifers from environmental samples [20], and to observe microbial communities associated with tardigrades both in natural habitats and in cultured environments, to find consistent and distinct microbiota associated with tardigrades [21].
Xeric mosses, such as those that grow on concrete surfaces, are particularly intriguing as microecosystems because they are partly self-contained, and each is separated by an almost uninhabitable drained area. This microenvironment harbors the substrate moss plant, a variety of meiofauna including tardigrades, nematodes, arthropods, and rotifers, and communities of fungi, protozoans, and bacteria. To study such a complex ecosystem and to elucidate the ecology surrounding tardigrades as recently being explored [21,22], I employed an amplicon sequencing approach using a set of universal primers for both microbes and eukaryotes.
2. Materials and Methods
2.1. Sample Collection
Xeric mosses, including Bryum argenteum [23], were sampled at the type locality of Macrobiotus shonaicus [24] in Otsuka-machi, Tsuruoka city, Japan (38°44′25″ N, 139°48′26″ E; 13 m asl) in June 2015, from the side of a concrete fence base approximately 10 cm from the ground (Figure 1). A total of 48 samples were collected at approximately 50 cm intervals, where the mosses were separated by a bare concrete surface. A small subset of the samples was inspected for tardigrades using a SZ61 stereo microscope (Olympus), and the presence of the following species was confirmed: Echiniscus testudo, Viridiscus perviridis [25], Milnesium inceptum [26], Macrobiotus shonaicus, and Hypsibius spp.
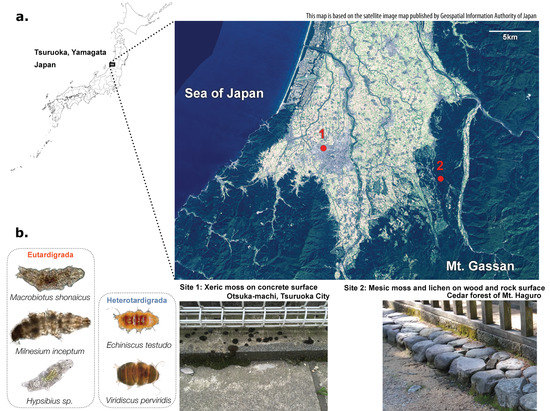
Figure 1.
Details of the sampling location. (a) Map of sampling location (above) and photos of the sampling site (below). Site 1 is within Tsuruoka city, and site 2 is in the cedar forest of Mt. Haguro. This map is based on the satellite image map published by the Geospatial Information Authority of Japan. (b) Microscope images of five tardigrades morphologically identified in site 1: xeric moss.
Mesic mosses and lichens (species were not identified) on rock and tree surfaces were sampled in the cedar forest at the foot of Mt. Haguro, Tsuruoka city, Japan (38°42′21″ N, 139°57′52″ E; 114 m asl) in June 2015; rock or pavement surfaces were approximately 10 cm from the ground. A total of 41 samples were collected at approximately 2 m intervals. Additionally, eight samples of semi-xeric mosses on the rock surfaces were collected from near the road just outside of the forest. Tardigrades were not found on the mesic moss samples upon observation under a microscope, as generally coniferous forests have less tardigrade diversity than hardwood forests. The samples collected at the above two locations were immediately brought back to the lab, where they were kept at −20 °C for approximately one week until they were processed for DNA extraction.
2.2. DNA Extraction and PCR Amplification
The basic protocol for DNA extraction and PCR amplification followed that of the Earth Microbiome Project (EMP) [27]. DNA was extracted from about 1 cm3 of moss samples with a PowerSoil DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA, USA), following the manufacturer’s protocol with a centrifuge-based method, and bead maceration was performed with a Multi-Beads Shocker (Yasui Kikai, Osaka, Japan). After quality checking with a Qubit fluorometer (Life Technologies, Grand Island, NY, USA) and TapeStation 2200 (Agilent Technologies, Santa Clara, CA, USA), the bacterial 16S rRNA region was amplified using EMP 16S V4 (515f-926r) amplification primers for Illumina (http://press.igsb.anl.gov/earthmicrobiome/protocols-and-standards/16s/), and the eukaryotic 18S rRNA region was amplified using EMP Euk1391f-EukBr amplification primers (V9 hypervariable region) for Illumina (http://www.earthmicrobiome.org/protocols-and-standards/18s/). As the majority of DNA was derived from the moss, amplification was performed using TKS Gflex polymerase (Takara Bio, Shiga, Japan) with the addition of peptide nucleic acid (PNA) oligomers designed to block the amplification of plant host plastids and mitochondrial 16S contamination, as described in the literature [28]. Amplicons were then run through E-Gel EX (Life Technologies, Grand Island, NY, USA) for electrophoresis, and bands corresponding to the target sizes (390 bp for 16S and 260 bp for 18S) were cut out and purified using NucleoSpin Gel and PCR Clean-up Kit (Takara Bio, Shiga, Japan).
2.3. Sequencing and Analysis
Amplicons were quantified using a Qubit fluorometer (Life Technologies, Grand Island, NY, USA), normalized and multiplexed, and sequenced on a MiSeq sequencer (Illumina, San Diego, CA, USA) using a MiSeq Reagent Kit v3 (600 cycles) (Illumina, San Diego, CA, USA) with EMP sequencing primers. Sequenced reads were demultiplexed using MiSeq Reporter (Illumina, San Diego, CA, USA) and then taxonomically assigned with the UPARSE pipeline (USEARCH v.8.1.1861) with default options [29] against the SILVA database (Release 123) [30], both for 16S and 18S sequences. Paired read merging (-fastq_mergepairs), read quality filtering (-fastq_filter), length trimming (-fastq_truncate –trunclength 250), dereplication (-derep_fulllength –minseqlength 250), singletons discarding (-sortbysize –minsize 2), out (Operational Taxonomic Unit) clustering (-cluster_otus –relabel OUT –minsize 2, -usearch_global –id 0.97, closed reference approach), and taxonomic identification of the OTUs was performed for each sample by a BLAST search against the SILVA database (BLASTN, -e 1e-25). The top match of the BLAST search may not always be accurate, but as the primary focus of this work is the classification at the phylum level, the uncertainty in the OUT classification should be minimal. Tardigrade OTUs were further confirmed by BLAST searches against the National Center for Biotechnology Information (NCBI) GenBank database to assign them to a genus and species level. One sample from mesic mosses (S38) and three from xeric mosses (H8, H10, and H24) were discarded because of insufficient sequenced reads (less than 100,000 raw reads; a strict criterion was set here because a sufficient number of samples >48 was sampled in each location), but the raw data were available for the omitted samples as well. Read counts were normalized by the total reads obtained for the compositional analyses (Figure 2 and Figure 3). The full list of identified OTUs are provided in Supplemental Data.
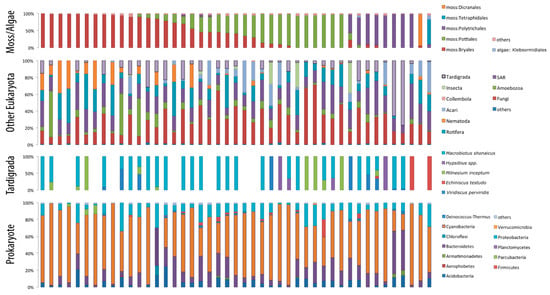
Figure 2.
Amplicon abundance ratios in xeric mosses. Relative abundances of amplicon reads within OTUs assigned to moss/lichen, other Eukaryota, tardigrades, and bacteria are shown separately. The moss samples were mostly from Bryum argenteum (Bryales), and as the sampled location was the type locality of Macrobiotus shonaicus, we see several moss contained 18S amplicons from this species. The majority of bacteria consisted of Cyanobacteria, Proteobacteria, and Bacteroidetes.
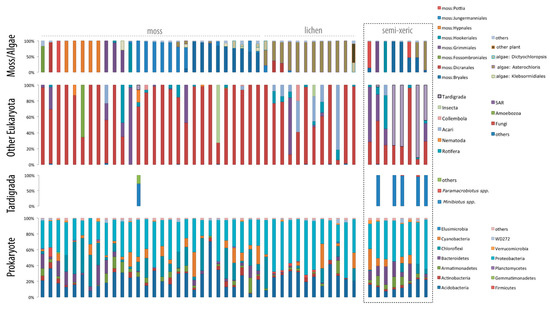
Figure 3.
Amplicon abundance ratios in mesic mosses. Relative abundances of amplicon reads within OTUs assigned to moss/lichen, other Eukaryota, tardigrades, and bacteria are shown separately. Forty samples were collected from mosses or lichens on rocks or tree surfaces within a cedar forest, and eight samples were collected from semi-xeric mosses near the road just outside of the forest. Moss species varied among samples, but lichen samples were dominated by the green algal genus Asterochloris. Fungi was dominant in almost all of the samples among eukaryotes, and almost no tardigrades were detected in the mesic samples. The largest bacterial clades were Proteobacteria and Acidobacteria. The full list of OTUs is available in the Supplemental Data.
2.4. Statistics
All of the statistical analyses were performed on JMP 14.1.0, except for Simpson’s Diversity Index calculated from bacterial abundances using a custom Perl script. Two-sided Student’s t-test was performed for a comparison of the abundance ratios between xeric and mesic samples. For correlation clustering, organism groups were clustered based on Pearson correlations between abundance ratios among 45 xeric and 48 mesic samples. Principal component analysis (PCA) was performed using all samples with OTUs appearing in more than ten samples (653 OTUs) using the Wide method. Partial least squares modeling was performed for 45 xeric moss samples using a nonlinear iterative partial least squares (NIPALS) algorithm, setting tardigrade abundance of each species as the response variable. The initial modeling using all of the variables did not converge within 15 factors, therefore the variables below variable importance for projection value of 0.8 were omitted from the model. After variable selection, the model with six factors was tested to be significant (p < 0.01) by leave-one-out cross validation with van der Voet T2.
2.5. Data Availability
The sequence data obtained in this work were deposited in the NCBI Sequence Read Archive (SRA) under accession number PRJNA524973.
3. Results
Metabarcode sequencing produced a total of 45 xeric moss samples and 47 mesic moss samples resulting in a sufficient number of reads (>100,000 reads), which successfully captured the diversity of microecosystems, including moss/lichen substrates; eukaryotes, including fungi, arthropods, nematodes, and tardigrades; and a variety of microbes (Figure 2 and Figure 3). On average, 279 OTUs were identified in each moss/lichen sample. The barcode sequence used in this study was relatively short (390 bp for 16S and 260 bp for 18S), and in order to observe the overview of the microecosystem, compositions were mainly analyzed at the phylum level. The reads classified to moss or lichen were assigned to the closest matching families, but the accuracy of such taxonomic assignment was not confirmed. However, in the xeric moss samples, tardigrade barcodes could be matched to each of the five species identified by visual observation (Figure 2).
The population distribution of prokaryotes and eukaryotes within xeric or mesic moss samples was mostly consistent regardless of the type of substrate (whether moss or lichen), but the samples from different humidity conditions showed striking variations. This is confirmed by PCA as shown in Figure 4. Figure 5 shows comparisons between the two humidity conditions in which the differences were highly significant (p < 0.0001 with t-test). Firstly, xeric mosses were much less diverse in terms of species diversity (Figure 5a), which is partly mirrored by the ratio of organellar DNA within the prokaryotic barcodes (Figure 5b). A significant amount (up to approximately 50% on average) of organellar DNA contamination remained in the xeric moss samples even though inhibitory peptide nucleic acid (PNA) successfully suppressed organellar DNA in mesic moss samples, suggesting the limited abundance of bacteria within the xeric samples. While different moss species could have different numbers of organelles and thus influence the PNA effectiveness, the overall difference among the xeric and mesic mosses was significant. Therefore, bacteria seem to be limited both in abundance and diversity in extremely desiccated moss environments. For eukaryotes, fungi were strongly overrepresented in the mesic samples (Figure 5c), and cyanobacteria dominated the prokaryotic community in xeric moss (Figure 5d).
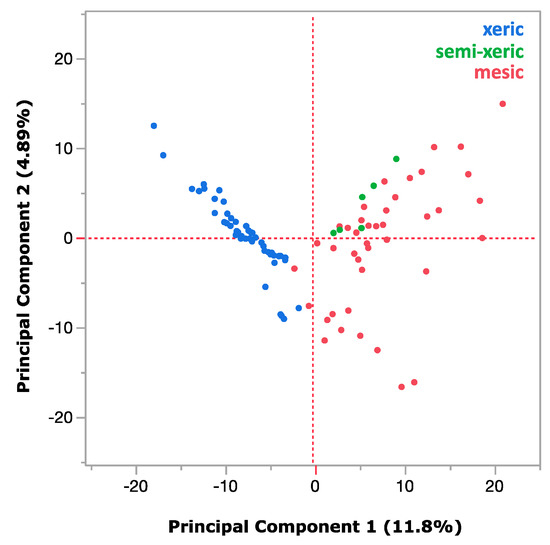
Figure 4.
Principal component analysis of all of the moss samples analyzed in this work (blue: xeric mosses; green: semi-xeric mosses; red: mesic moss and lichens). Xeric and mesic mosses show distinct clusters, and semi-xeric mosses tend to shift from the mesic mosses and lichens towards the xeric mosses.
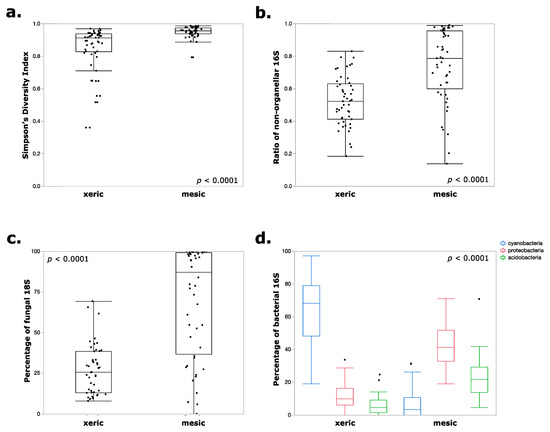
Figure 5.
Comparisons between xeric and mesic and semi-xeric samples. (a) Simpson’s diversity index distribution calculated for bacterial amplicon abundances. The xeric samples were much less diverse than the mesic samples. (b) The xeric samples contained a large amount of organellar 16S rDNA (both from chloroplasts and mitochondria), with as much as 50% on average, even with the presence of inhibitory PNAs. Together with the lower Simpson’s diversity, these results suggest a limited bacterial population within xeric mosses. (c) Fungi was highly abundant in mesic samples, whereas (d) Cyanobacteria was much more abundant in xeric mosses, and the mesic samples contained more Proteobacteria and Acidobacteria. All of the comparisons between xeric and mesic samples were significant (p < 0.0001) in two-tailed Student’s t-tests.
To observe the interspecies associations of tardigrades in more detail, barcode abundance profiles for all organisms, including eukaryotes and prokaryotes, were used to observe the correlations between the groups of organisms (Figure 6A) and to model the predicted abundance of each tardigrade species (Figure 6B). Because of the predominance of M. shonaicus, data for other species may not have been sufficient for such observations, but M. shonaicus had 36 profiles that were sufficient for these analyses. In terms of correlations, M. shonaicus abundance was most correlated with the abundances of Armatimonadetes, Bacteroidetes, Deinococcus–Thermus, and Acidobacteria.
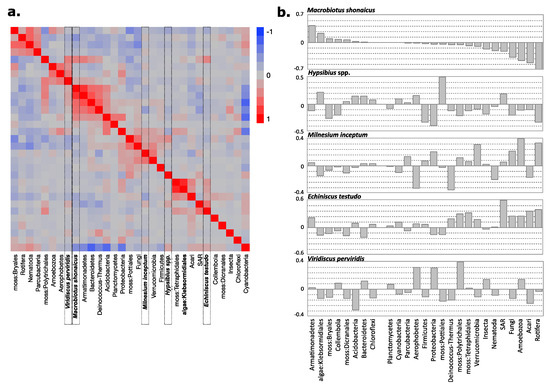
Figure 6.
Correlations between species abundances within mesic samples. (a) Pearson correlation clustering of species abundances. Macrobiotus shonaicus is clustered with Armatimonadetes, Bacteroidetes, Deinococcus–Thermus, and Acidobacteria but is negatively correlated with Cyanobacteria and Rotifera. (b) Normalized coefficients for the partial least squares (PLS) model. Similar to the Pearson correlation, Armatimonadetes has the highest positive coefficient for M. shonaicus, and Rotifera has the highest negative coefficient.
4. Discussion
Simultaneous metabarcoding of the eukaryote and prokaryote community structure of the tardigrade habitat was successfully performed by suppressing the amplification of moss-derived DNA by the use of inhibitory PNA. This approach was effective as demonstrated by the high percentage of non-organellar 16S barcode sequenced in the mesic samples. This sequence-based approach is useful to identify the diversity of organisms and their compositions in the study of microscopic ecology, where manual observations are difficult to reveal the entirety of the community [31]. The relatively high frequency of M. shonaicus within the xeric moss samples is in agreement with manual observation at this type locality of this species.
Taxonomic resolution and coverage is, however, likely limited in the current approach, especially for eukaryotes. While the methods in this work followed the protocols and primers of the Earth Microbiome Project (EMP), which was contributed by hundreds of international scientists [27,32], the focus of EMP was in prokaryotes, and eukaryotic metabarcoding is less optimized in this respect. Moreover, although the scientific community is slowly converging towards the use of the V9 hypervariable region of 18S rDNA used by EMP and in this work for eukaryotic metabarcoding, it still underestimates the true number of metazoan species, and the resolution of taxonomic identification is at its best at the genus level [31,33]. The optimization of the primers or the use of multiple markers would be a possible future direction to better understand the microecosystems within mosses, but at the least it was possible to identify all five genera of tardigrades morphologically identified in the xeric moss samples. It should also be noted that the number of reads in eukaryotic metabarcoding does not mirror the abundance of species, because of the diverse copy number of 18S rDNA genes (including polyploidy and multinucleation), as well as the number of cells within an organism. However, it was also shown in a previous study that if the reads were normalized for the cell count and copy number of genes, the V9 region of 18S rDNA actually shows a good correlation with species abundance in marine plankton [34].
The main aim of this work is to test the feasibility of the simultaneous metabarcoding approach in analyzing the tardigrade habitat, and the current data are too limited to discuss ecological implications of the community structures. Nevertheless, the general eukaryotic and prokaryotic community composition was highly consistent within the sampled location (xeric or mesic), representing a high reproducibility of this approach. It is worth noting that the community composition was consistent regardless of the moss/lichen substrate. Xeric mosses, mostly represented by the family Bryales, contained a larger fraction of meiofauna barcodes, and 80% of the samples contained tardigrades. On the other hand, tardigrades were rarely observed (2.5%) within the mesic moss/lichen samples, confirming microscopic observations. It is unusual to have less tardigrade diversity in mesic mosses and lichens than in xeric ones, but few tardigrade species have been reported from mosses or lichens on the bark of coniferous trees [7], so the substrates may not have been appropriate for tardigrades. The horizontal distribution of tardigrades that are reported to be important in tardigrade sampling is another factor to be considered in future analysis [35]. Mirroring previous findings, xerophilic tardigrades Milnesium, Macrobiotus, and Echiniscus species were found in the xeric environment, and hygrophilic tardigraes Minibiotus and Paramacrobiotus species were found in the mesic environment [13]. The contrasting population distribution in eukaryotes was predominantly represented by the significant dominance of fungi within the high-humidity samples. Again, taxonomic coverage and resolution is limited by the current approach with V9 primers, and while causation is not clear from the current data, there seems to be a clear negative correlation between invertebrates (including tardigrades, but also other meiofauna) and fungal abundance. Although the current analysis did not have the resolution to identify specific parasites, several fungi are known to be parasitic on tardigrades [19], which may partly explain the reason for the negative correlations. Fungal diversity is also known to be correlated sometimes with the severity of the environment [36], and further studies covering multiple locations rather than the two sites used in this work would provide further insights into the biological interactions between them. Even within the xeric environment, Bryales moss samples corresponding to Bryum argenteum tended to be extremely dry and contained an even lower fraction of fungi reads and inversely greater fractions of nematodes and rotifers, which are known to be anhydrobiotic species. These results also support the hypothesis that the severity of the environment results in less biological diversity [36]. This is actually contradictory to the tardigrade diversity observed in this work, and to previous findings that there is generally a higher tardigrade diversity in the mesic environment. This could be the result of the small number of samples collected in the forest. Each of the xeric samples usually contain only one or two species, displaying patchy distribution as previously suggested [37], so the species diversity was nonetheless very low in the xeric samples tested in this work.
Prokaryotic diversity also showed contrasting variations among the two environments. As with fungi, prokaryotes were significantly more diverse in the mesic samples (Figure 3). Moreover, the total abundance of bacteria also seemed to have been limited in the xeric samples. While mesic mosses were significantly enriched in Proteobacteria and Acidobacteria, reflecting the typical microbial composition seen in soil environments [38], xeric mosses were dominated by Cyanobacteria. Cyanobacteria colonizing mosses contribute up to 50% of the total nitrogen fixation within the microecosystem [39], but more importantly, many Cyanobacteria are known to be highly resistant to desiccation [40,41]. In a very dry environment, strong selection is exerted on the xeric moss environment, limiting the diversity of the organisms dwelling within such microecosystems to desiccation-tolerant invertebrates, such as xerophilic tardigrades, and desiccation-tolerant bacteria, such as cyanobacteria.
Inter-species association of tardigrades with other organisms needs further analyses to be more informative, but in the partial least squares (PLS) fitting, the normalized contribution was the most positive with Armatimonadetes and algae Klebsormidiales and the most negative with Fungi, Amoebozoa, Acari, and Rotifera. Armatimonadetes are a diverse group of Gram-negative bacteria found in numerous metagenomic samples, and their desiccation-tolerance or biological role in the moss environment is unknown. M. shonaicus can feed both on algae and rotifers in laboratory culture [24], and M. inceptum exclusively feeds on rotifers in laboratory conditions [42]; however, the contribution was positive for M. inceptum, whereas it was negative for M. shonaicus. Coupled with strong negative contributions of Fungi, Amoebozoa, and Acari, these data may suggest interspecies interaction preferences of M. shonaicus. These data provide an ecosystem-wide perspective on tardigrade inter-specific association, which could complement more detailed analyses on tardigrade symbiosis [21,22].
5. Conclusions
To the best of my knowledge, this is the first work that simultaneously studied the abundance of both eukaryotes and prokaryotes within xeric and mesic mosses through the use of two universal barcodes and PNA inhibitors for organellar DNA. Although the number of sites was limited, a large number of samples in each of the humidity conditions allowed for a statistical analysis of the interspecies interactions, and humidity was shown to limit the diversity of both eukaryotes and prokaryotes, selecting for desiccation-tolerant species.
Supplementary Materials
The following are available online at https://www.mdpi.com/1424-2818/12/3/110/s1: Data S1: full list of OTUs and corresponding FASTA sequences.
Funding
This work was supported by KAKENHI Grant-in-Aid for Scientific Research (B), by the Japan Society for the Promotion of Science (JSPS; grant no. 17H03620), and partly by research funds from the Yamagata Prefectural Government and Tsuruoka City, Japan.
Acknowledgments
The author thanks Nozomi Abe for technical assistance and Piotr Gąsiorek, Witek Morek, and Łukasz Michalczyk for helping with the identification of Echiniscus testudo, Viridiscus perviridis, and Milnesium inceptum.
Conflicts of Interest
The author declares no conflict of interest.
References
- Greven, H.; Schuttler, L. How to crawl and dehydrate on moss. Zool. Anz. 2001, 240, 341–344. [Google Scholar] [CrossRef]
- Nelson, D.R.; Adkins, R.G. Distribution of tardigrades within a moss cushion: Do tardigrades migrate in response to changing moisture conditions? Zool. Anz. 2001, 240, 493–500. [Google Scholar] [CrossRef]
- Guidetti, R.; Altiero, T.; Rebecchi, L. On dormancy strategies in tardigrades. J. Insect. Physiol. 2011, 57, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Keilin, D. The Leeuwenhoek Lecture, The problem of anabiosis or latent life: History and current concept. Proc. Royal. Soc. London Series B 1959, 150, 149–191. [Google Scholar]
- Bartels, P.J.; Nelson, D.R. An evaluation of species richness estimators for tardigrades of the Great Smoky Mountains National Park, TN and NC, USA. J. Limnol. 2007, 66, 104–110. [Google Scholar] [CrossRef]
- McInnes, S.J.; Pugh, P.J.A. Tardigrade biogeography. In Water Bears: The Biology of Tardigrades; Schill, R.O., Ed.; Springer: Basel, Switzerland, 2019; pp. 115–129. [Google Scholar]
- Nelson, D.R.; Bartels, P.J.; Fegley, S.R. Environmental correlates of tardigrade community structure in mosses and lichens in the Great Smoky Mountains National Park (Tennessee and North Carolina, USA). Zool. J. Linn. Soc. 2019. [Google Scholar] [CrossRef]
- Kathman, D.; Cross, S. Ecological distribution of moss-dwelling tardigrades on Vancouver Island, British Columbia, Canada. Can. J. Zool. 2011, 69, 122–129. [Google Scholar] [CrossRef]
- Guil, N.; Hortal, J. Effects of macro and micro-environmental factors on the species richness of terrestrial tardigrade assemblages in an Iberian mountain environment. Landsc. Ecol. 2009, 24, 375–390. [Google Scholar] [CrossRef]
- Kaczmarek, Ł.; Goldyn, B.; Welnic, W.; Michalczyk, Ł. Ecological factors determining Tardigrada distribution in Costa Rica. J. Zool. Syst. Evol. Res. 2011, 49, 78–83. [Google Scholar] [CrossRef]
- Schuster, R.; Greven, H. Reproductive traits of Macrobiotus hufelandi during a long-term field study with notes on Paramacrobiotus richtersi and Diphascon pingue (Eutardigrada). J. Limnol. 2013, 72, 166–174. [Google Scholar] [CrossRef]
- Meyer, H. Interspecific association and substrate specificity in tardigrades from Florida, Southeastern United States. Hydrobiologia 2006, 558, 129–132. [Google Scholar] [CrossRef]
- Wright, J.C. The significance of four xeric parameters in the ecology of terrestrial Tardigrada. J. Zool. 1991, 224, 59–77. [Google Scholar] [CrossRef]
- Jönsson, K. Long-term experimental manipulation of moisture conditions and its impact on moss-living tardigrades. J. Limnol. 2007, 66, 119–125. [Google Scholar] [CrossRef]
- Sanchez-Moreno, S.; Ferris, H.; Guil, N. Role of tardigrades in the suppressive service of a soil food web. Agric. Ecosyst. Environ. 2008, 124, 187–192. [Google Scholar] [CrossRef]
- Hyvönen, R.; Persson, T. Effects of fungivorous and predatory arthropods on nematodes and tardigrades in microcosms with coniferous forest soil. Biol. Fertil. Soils 1996, 21, 121–127. [Google Scholar] [CrossRef]
- Altiero, T.; Rebecchi, L. Rearing tardigrades: Results and problems. Zool. Anz. 2001, 240, 217–221. [Google Scholar] [CrossRef]
- Morgan, C.I. Population dynamics of two species of Tardigrada, Macrobiotus hufelandii (Schultze) and Echiniscus testudo (Doyère), in roof moss from Swansea. J. Anim. Ecol. 1977, 46, 263–279. [Google Scholar] [CrossRef]
- Vecchi, M.; Vicente, F.; Guidetti, R.; Bertolani, R.; Rebecchi, L.; Cesari, M. Interspecific relationships of tardigrades with bacteria, fungi and protozoans, with a focus on the phylogenetic position of Pyxidium tardigradum (Ciliophora). Zool. J. Linn. Soc. 2016, 178, 846–855. [Google Scholar] [CrossRef]
- Robeson, M.S., II; Costello, E.K.; Freeman, K.R.; Whiting, J.; Adams, B.; Martin, A.P.; Schmidt, S.K. Environmental DNA sequencing primers for eutardigrades and bdelloid rotifers. BMC Ecol. 2009, 9, 25. [Google Scholar] [CrossRef]
- Vecchi, M.; Newton, I.L.G.; Cesari, M.; Rebecchi, L.; Guidetti, R. The microbial community of tardigrades: Environmental influence and species specificity of microbiome structure and composition. Microb. Ecol. 2018, 76, 467–481. [Google Scholar] [CrossRef]
- Kaczmarek, L.; Roszkowska, M.; Poprawa, I.; Janelt, K.; Kmita, H.; Gawlak, M.; Fialkowska, E.; Mioduchowska, M. Integrative description of bisexual Paramacrobiotus experimentalis sp. nov. (Macrobiotidae) from republic of Madagascar (Africa) with microbiome analysis. Mol. Phylogenet. Evol. 2020, 145, 106730. [Google Scholar] [CrossRef] [PubMed]
- Greenwood, J.L.; Stark, L.R.; Chiquoine, L.P. Effects of rate of drying, life history phase, and ecotype on the ability of the moss Bryum argenteum to survive desiccation events and the influence on conservation and selection of material for restoration. Front. Ecol. Evol. 2019, 7. [Google Scholar] [CrossRef]
- Stec, D.; Arakawa, K.; Michalczyk, L. An integrative description of Macrobiotus shonaicus sp. nov. (Tardigrada: Macrobiotidae) from Japan with notes on its phylogenetic position within the hufelandi group. PLoS ONE 2018, 13, e0192210. [Google Scholar] [CrossRef]
- Gąsiorek, P.; Morek, W.; Stec, D.; Michalczyk, Ł. Untangling the Echiniscus Gordian knot: Paraphyly of the “arctomys group” (Heterotardigrada: Echiniscidae). Cladistics 2019, 35, 633–653. [Google Scholar] [CrossRef]
- Morek, W.; Suzuki, A.C.; Schill, R.O.; Georgiev, D.; Yankova, M.; Marley, N.J.; Michalczyk, Ł. Reinstituting Milnesium alpigenum Ehrenberg, 1853 (Tardigrada: Apochela) after decades of oblivion, with a description of the laboratory model species Milnesium inceptum sp. nov. Zootaxa 2018, 4586, 35–64. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Yourstone, S.; Mieczkowski, P.; Jones, C.D.; Dangl, J.L. Practical innovations for high-throughput amplicon sequencing. Nat. Methods 2013, 10, 999–1002. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glockner, F.O. The SILVA and "All-species Living Tree Project (LTP)" taxonomic frameworks. Nucleic. Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef]
- Leray, M.; Knowlton, N. Censusing marine eukaryotic diversity in the twenty-first century. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016, 371. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Jansson, J.K.; Knight, R. The Earth Microbiome project: Successes and aspirations. BMC Biol. 2014, 12, 69. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Xiong, J.; Yu, Y. Taxonomic resolutions based on 18S rRNA genes: A case study of subclass Copepoda. PLoS ONE 2015, 10, e0131498. [Google Scholar] [CrossRef] [PubMed]
- De Vargas, C.; Audic, S.; Henry, N.; Decelle, J.; Mahe, F.; Logares, R.; Lara, E.; Berney, C.; Le Bescot, N.; Probert, I.; et al. Ocean plankton. Eukaryotic plankton diversity in the sunlit ocean. Science 2015, 348, 1261605. [Google Scholar] [CrossRef] [PubMed]
- Degma, P.; Katina, S.; Sabatovičová, L. Horizontal distribution of moisture and Tardigrada in a single moss cushion. J. Zool. Syst. Evol. Res. 2011, 49, 71–77. [Google Scholar] [CrossRef]
- Rao, S.; Chan, Y.; Lacap, D.C.; Hyde, K.D.; Pointing, S.B.; Farrell, R.L. Low-diversity fungal assemblage in an Antarctic Dry Valleys soil. Polar Biol. 2012, 35, 567–574. [Google Scholar] [CrossRef]
- Meyer, H.A. Small-scale spatial distribution variability in terrestrial tardigrade populations. Hydrobiologia 2006, 558, 133–139. [Google Scholar] [CrossRef]
- Janssen, P.H. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 2006, 72, 1719–1728. [Google Scholar] [CrossRef]
- Rousk, K.; Jones, D.L.; Deluca, T.H. Moss-cyanobacteria associations as biogenic sources of nitrogen in boreal forest ecosystems. Front. Microbiol. 2013, 4, 150. [Google Scholar] [CrossRef]
- Billi, D.; Potts, M. Life and death of dried prokaryotes. Res. Microbiol. 2002, 153, 7–12. [Google Scholar] [CrossRef]
- Singh, H. Desiccation and radiation stress tolerance in cyanobacteria. J. Basic Microbiol. 2018, 58, 813–826. [Google Scholar] [CrossRef]
- Suzuki, A.C. Life history of Milnesium tardigradum Doyère (tardigrada) under a rearing environment. Zool. Sci. 2003, 20, 49–57. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).