Abstract
We present thorough this review the developments in the field, point out their current limitations, and outline its timelines and unique potential. In order to do so we introduce the methods used in each of the advances in the application of deep learning (DL) to coral research that took place between the years: 2016–2018. DL has unique capability of streamlining the description, analysis, and monitoring of coral reefs, saving time, and obtaining higher reliability and accuracy compared with error-prone human performance. Coral reefs are the most diverse and complex of marine ecosystems, undergoing a severe decline worldwide resulting from the adverse synergistic influences of global climate change, ocean acidification, and seawater warming, exacerbated by anthropogenic eutrophication and pollution. DL is an extension of some of the concepts originating from machine learning that join several multilayered neural networks. Machine learning refers to algorithms that automatically detect patterns in data. In the case of corals these data are underwater photographic images. Based on “learned” patterns, such programs can recognize new images. The novelty of DL is in the use of state-of-art computerized image analyses technologies, and its fully automated methodology of dealing with large data sets of images. Automated Image recognition refers to technologies that identify and detect objects or attributes in a digital video or image automatically. Image recognition classifies data into selected categories out of many. We show that Neural Network methods are already reliable in distinguishing corals from other benthos and non-coral organisms. Automated recognition of live coral cover is a powerful indicator of reef response to slow and transient changes in the environment. Improving automated recognition of coral species, DL methods already recognize decline of coral diversity due to natural and anthropogenic stressors. Diversity indicators can document the effectiveness of reef bioremediation initiatives. We explored the current applications of deep learning for corals and benthic image classification by discussing the most recent studies conducted by researchers. We review the developments in the field, point out their current limitations, and outline their timelines and unique potential. We also discussed a few future research directions in the fields of deep learning. Future needs are the age detection of single species, in order to track trends in their population recruitment, decline, and recovery. Fine resolution, at the polyp level, is still to be developed, in order to allow separation of species with similar macroscopic features. That refinement of DL will allow such comparisons and their analyses. We conclude that the usefulness of future, more refined automatic identification will allow reef comparison, and tracking long term changes in species diversity. The hitherto unused addition of intraspecific coral color parameters, will add the inclusion of physiological coral responses to environmental conditions and change thereof. The core aim of this review was to underscore the strength and reliability of the DL approach for documenting coral reef features based on an evaluation of the currently available published uses of this method. We expect that this review will encourage researchers from computer vision and marine societies to collaborate on similar long-term joint ventures.
1. Introduction
Coral reefs are among the most diverse ecosystems in the world. Their area comprises less than one percent of the total area of the oceans and seas, nevertheless they are the home of about 25 percent of all marine species [1], thereby maintaining the high biodiversity of coastal tropical marine ecosystems. The total number of extant reef-building species worldwide is estimated at 3235 [2]. Corals are divided into hard and soft corals. Hard corals or Scleractinia are the most important hermatypic (reef-building) organisms [3] that play a key role in forming the framework of coral reefs and in providing food, substrate, and shelter for a wide variety of organisms [4]. Acute damage to these corals results in the collapse of the complex community of organisms that live in close association with them. Reef-building corals have hard, calcium carbonate skeletons that offer partial protection from predators, and form the bulk of the coral reef’s structure. Symbiotic algae living in the corals are called zooxanthellae by the first person to report them [5], these microalgae provide the corals with most of the energy requirements of the coral host through their photosynthesis [6,7]. Hence, hard corals are limited to shallow depths, where they can gain access to sufficient sunlight on which the zooxanthellae depend [8]. Different light and nutrient conditions affect the chlorophyll content of the algal symbionts and their density [6]. The potential value of easily deep learning (DL) recognizable color changes in the same species, adds a further promising venue of the automated inspection of corals.
Corals supplement the photosynthate translocated from the zooxanthellae with predation on zooplankton that provides them with essential nutrients, which are in short supply in tropical seas. In deep-water, dim-light corals satisfy their energy needs by preying on zooplankton (see review by [9]). By the CO2 uptake of the photosynthesizing zooxanthellae, the internal pH of the coral is raised; enhancing the skeletal deposition of calcium carbonate in a process termed light-enhanced calcification [10], a process recently challenged by Cohen et al. [11].
2. The Future of Coral Reefs
Coral reefs are under multiple threats from global climate changes, mainly oceanic acidification and warming [12], blast and cyanide fishing [13], coral collection for the marine aquaria trade [14], sunscreen use [15], light pollution [16], and excessive SCUBA diving pressure [17]. Wastewater and fertilizer discharge creates the phenomenon of anthropogenic eutrophication, or over-fertilization of the reef water with excess nutrients that can harm reefs by encouraging excess seaweed growth. This phenomenon of enrichment violates the equilibrium in the reef by causing algal blooms that take over the existing corals and prevent the settlement of new ones [18].
Sedimentation has been identified as a stressor for the existence and recovery of corals and their habitats [19]. In addition, urbanization is a widespread threat to coral reefs, which includes multiple aspects such as eutrophication and light pollution [20].
Global warming that adversely affects coral reefs also reveals that certain species have far greater tolerance to climate change and coral bleaching than others [21]. Such “bleaching-resistant” species could be a key factor in the bioremediation of reefs damaged by global climate change. Coral reefs suffer from bleaching due to an increase in water temperature, ocean acidification, anthropogenic eutrophication, and light pollution [22,23]. Loss of the algal symbionts (bleaching, reduced calcification, disruption of reproductive timing, and increased seaweed competition) endanger the survival of the already declining coral reefs.
Due to those hazards to the health and extended survival of coral reefs and declining state of coral reef worldwide, there is a crucial need for large scale, real time, reef monitoring/surveys. Using further developed and refined automated technologies and methods such as DL for the auto annotation of corals are the only way to accomplish the urgent task of monitoring changes in the composition, diversity, recruitment, bleaching, disease, and live cover of coral reefs. The resulting “big data” sets will allow charting trajectories and future trends in coral reefs, in attempt to assure their survival.
3. Application of CNN and DBN
A number of convolutional neural networks (CNN) CNN-based optical character and handwriting recognition systems were developed and deployed by Microsoft. In the early 1990s, CNNs were tested for object detection in natural images for face recognition purposes [24]. Speech recognition and document reading applications were already used at the early 1990s. The document-reading system used a convolutional net (ConvNets or CNNs). By the late 1990s, this system was reading over 10% of all the checks in the United States.
Since the early 2000s, CNNs have been used for the detection, segmentation, and recognition of objects and regions in images. These relatively abundant tasks used labeled data, such as traffic sign, text, recognition, and the segmentation of biological images, particularly for the detection of faces, pedestrians, and human bodies in natural images [24].
CNNs have achieved recognition accuracy exceeding that of humans in several visual recognition tasks [25], including recognizing traffic signs [26], faces [27,28], and handwritten digits [26,29].
Deep neural networks learn hierarchical feature representations [24] (Figure 1). CNNs apply convolutions or filters across an image. Each filter can be responsive to different features of the image. Filters at early layers of the network tend to select out low-level properties of the input image, including spatial frequency, edges, and color. As we continue to stack additional convolutional layers, more complicated filters begin to evolve. By examining the output of a CNN, we can understand what properties of an image contribute to its classification in a particular category. Face recognition is a series of several related steps, starting with analyzing the picture and finding all the faces in it, focusing on each face, and understanding that even if a face turned in a weird direction or was in bad lighting, it is still of the same person. CNNs start by identifying simple small features, such as lines and circles. They then build these simple features into more complex ones, such as eyes and noses. Finally, the CNN aggregates those features into complete faces.
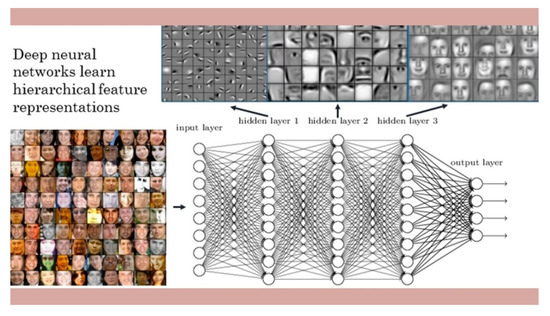
Figure 1.
Deep neural networks learn hierarchical feature representations. After (LeCun et al. (2015)) [24].
CNNs have been designed to recognize visual patterns directly from pixel images with minimal preprocessing. They also recognize patterns with extreme variability (such as handwritten characters), susceptible to distortions and simple geometric transformations. LeNet-5 by LeCun et al. (1998) [30] is such a convolutional network designed for handwritten and machine-printed character recognition (Figure 2).
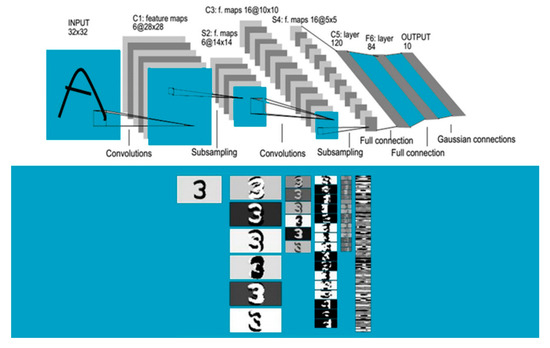
Figure 2.
LeNet-5 First back-propagation convolutional neural networks (CNN) for hand written digit recognition. After (LeCun et al. (1998)) [30].
CNN used for text detection [31] (Figure 3). Operation of detection and recognition modules. The detector decides whether a single 32-by-32 image patch contains text or not. The recognizer identifies the characters present in a text-containing input patch [32].
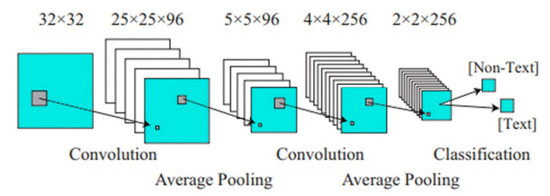
Figure 3.
CNN used for text detection. After (Wang et al. (2012)) [31].
The ECOUAN team in the LifeCLEF 2015 lab that is a part of the Conference and Labs of the Evaluation Forum: CLEF 2015, used a DL approach to pre-train a CNN for plant identification tasks using 1.8 million images. That project was designed to recognize the species of a plant based on observation of the plant’s features, using a set of images (1–5) from different angles. These points of view include scans of the entire plant, branches, leaves, fruit, flower and stem. The plant identification task was based on the Pl@ntView dataset. It included 1000 herb, tree and fern species centered on France and neighboring countries [33].
Mahmood et al. (2017) [34] summarizes the current uses of DL methods in the analysis of underwater images, especially for coral species classification and reef description. They show the challenges associated with the computerized analysis of marine data, and explore the applications of DL for automatic annotation of coral reef images.
Deep convolutional nets (DBN) have brought about breakthroughs in processing images, video, speech, and audio (Figure 4).
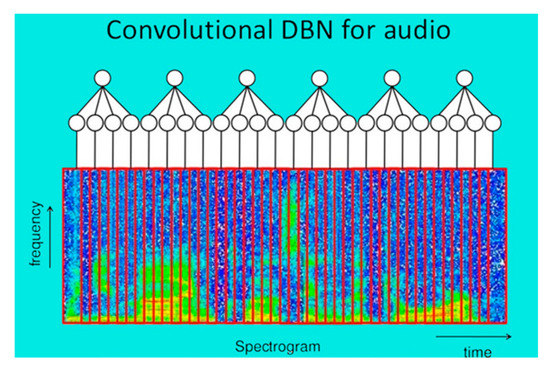
Figure 4.
Deep convolutional nets (DBN) for audio. After (Lee et al. (2009)) [35].
For the application of DBNs to audio data, the first step is to convert time-domain signals into spectrograms and then apply principal component analysis (PCA) Whitening. PCA is the general name for a technique which uses sophisticated underlying mathematical principles to transforms a number of possibly correlated variables into a smaller number of variables called principal components. Whitening is a useful preprocessing method that completely removes the second-order information of the data. “Second-order” means here correlations and to the spectrograms and create lower dimensional representations variances. It allows us to concentrate on properties that are not dependent on co-variances.
Christin et al. [36], present in their article a thorough review of DL applications used in ecology, and provide useful resources and recommendations for ecologists on how to use DL as a powerful tool in their research. The aim of their study was to evaluate the performance of state-of-the-art identification tools for biological data and to show that DL tools are choice methods to fulfil such an aim.
4. Applications to the Study of Corals
Any coral-reef classification has to follow three main steps:
- Underwater image photography, followed by de-noising (preprocessing step), due to different challenges to quality (motion blurring, color attenuation, refracted sunlight patterns, sky color variation, scattering effects, presence of particles, etc.). The obtained raw images must be enhanced in order to visualize the coral species in detail for the steps to follow [37].
- Feature extraction: for images of different coral species, one needs to find salient features in each species in order to identify and reliably distinguish it from other species [38], avoiding errors due to illumination, rotation, size, view angle, camera distance, etc.
- The extracted features are used as input for DL [39].
Efficient classification methods of coral species, based on the recent machine learning (ML) methodology of DL, have been developed. These consist of algorithms designed to reveal and extract high-level patterns and features in large photographic coral image datasets by using architecture composed of multiple nonlinear transformations. The two most popular algorithms applied to coral reef data are the CNN and the deep belief net (DBN). The first, CNN, is a neural network planned to recognize visual objects directly from images with minimal or without any preprocessing. The second, DBN, is a class of deep neural network which comprises of multiple layer of graphical model having both directed and undirected edges. One of the common features of a deep belief network is that although layers have connections between them, the network does not include connections between units in a single layer. A CNN consists of one or more convolutional layers, often with a subsampling layer, followed by one or more fully connected layer. The design of a CNN was motivated by the discovery of a visual mechanism, the visual cortex, in the brain. The convolution layer in a CNN performs the function of the cells in the visual cortex.
5. Published Coral Classifications Based on Machine Learning
Recent studies of coral species classification applied CNN (DL method) as a feature extraction and classification technique.
Coral classification using a semi-autonomous machine learning (ML) approach was used with a robotic underwater vehicle (AUV), conducted on Australia’s Great Barrier Reef in order to gather statistics about coral density and composition to the genus resolution. Some 10,000 full resolution images yielded 3000 segments which were hand labeled into the four classes and classified using SVM method. Distinction among these was based upon features of the coral [40]. As shown in Figure 5, the green sector with the branching pattern is the area with corals. The blue rectangles enclose identified areas with classified coral genera or rock. The black area is sand.
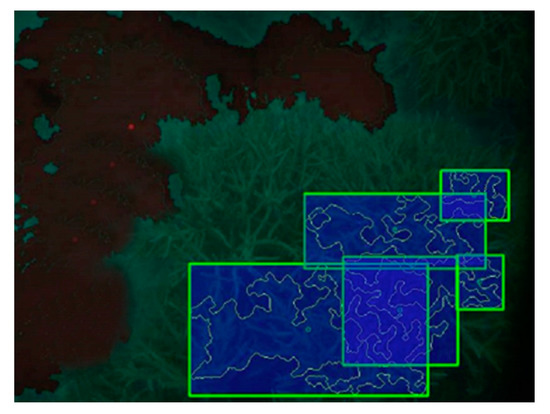
Figure 5.
Colors represent coral genus classification. The green is the area with coral reef not identified by the program. The rectangles enclose identified coral genera or rock. The black area is sand. After (Johnson-Roberson et al. (2007)) [40].
Pizarro et al. [38] analyzed benthic marine habitats including corals. They used a feature vector based on the normalized color coordinates NCC histogram, a Scale-Invariant Feature Transform (SIFT) that is a computer vision algorithm used to detect and describe local features in images. The algorithm was published by David Lowe (1999) [41]. SIFT descriptor is a 3-D spatial histogram of the image gradients characterizing the appearance of a key-point. The gradient at each pixel is regarded as a sample of a three-dimensional elementary feature vector, formed by the pixel location and the gradient orientation. Using a "bag of features" approach (BoF) based on random collections of quantized local image descriptors, they discard spatial information and are therefore conceptually and computationally simpler than many alternative methods [42]. The authors included those classes: (1) Coralline Rubble, (2) Hard Coral, (3) Hard Coral, Soft Coral, and Coralline Rubble, (4) Halimeda, Hard Coral, and Coralline Rubble, (5) Macroalgae, (6) Rhodolith, (7) Sponges, and (8) uncolonized substrate.
Stokes and Deane (2009) [43] used color space normalization (CSN) that is derived by a linear transformation of the RGB information, and a discrete cosine transform (DCT) that helps separate the image into parts (or spectral sub-bands) of differing importance (with respect to the image’s visual quality) to extract texture features [44]. They used one dataset of more than 3000 images provided by the National Oceanic and Atmospheric Administration (NOAA) of the U.S. Department of Commerce Ocean Explorer. They resolved discrimination metrics for four selected library images: (1) Pseudodiploria clivosa (a brain coral), (2) sand, (3) macroalgae (mainly Dictyota sp.), and (4) Montastraea annularis (a boulder star coral).
Beijbom et al. (2012) [45] used an SVM classifier with a Radial Basis Function (RBF) kernel for classification. They introduced the Moorea Labeled Corals (MLC) dataset, which is a subset of The Moorea Coral Reef Long Term Ecological Research (MCR-LTER) project that has been collecting image data from the island of Moorea (French Polynesia) since 2005.
The MLC dataset includes 2055 images collected over three years: 2008, 2009, and 2010.
It contains nine labels, four non coral labels: (1) Crustose Coralline Algae (CCA), (2) Turf algae, (3) Macroalgae and (4) Sand, and five coral genera: (5) Acropora, (6) Pavona, (7) Montipora, (8) Pocillopora, and (9) Porites.
Shihavuddin et al. (2013) [46] describes a method for classifying images of coral reefs. They used for classification, either k-nearest neighbor (KNN), neural network (NN), support vector machine (SVM), or probability density weighted mean distance (PDWMD). The main difference of this new method and the previous algorithms [45] is that:
Classification schemes are flexible, depending on the characteristics of each dataset (e.g., the size of the dataset, number of classes, resolution of the samples, color information availability, class types, etc.). In order to increase accuracy, different combinations of features and classifiers are tuned to each dataset prior to classification. Image classification methods were applied to a composite mosaic image that covers areas larger than a single image. They recognized in the EILAT dataset [46] eight classes: sand, urchin, branched type I coral, brain coral, favid coral, branched type II, dead coral and branched type III). The EILAT Red sea dataset contains 1123 image patches, each being 64 × 64 pixels in size. The accuracy they achieved was 83.7%. Other datasets used by the same author are: the Rosenstiel School of Marine and Atmospheric Sciences (RSMAS) dataset, consists of 766 image patches; each 256 × 256 pixels in size, of 14 different classes (1) Acropora cervicornis, (2) Acropora palmata, (3) Colpophyllia natans, (4) Diadema antillarum (sea urchin), (5) Pseudodiploria strigosa, (6) Gorgonians, (7) Millepora alcicornis (Fire coral), (8) Montastraea cavernosa, (9) Meandrina meandrites, (10) Montipora spp., (11) Palythoas palythoa, (12) Sponge fungus, (13) Siderastrea siderea, and (14) tunicates. A third dataset studied by Shihavuddin et al. [46] was the Moorea-Labeled Corals (MLC) dataset. The 2008 dataset was used 18,872 image patches of 312 × 312 pixels in size, centered on the annotated points were selected randomly. All nine classes were represented in the random image patches (Figure 6) and achieved the accuracy of 85.5%.
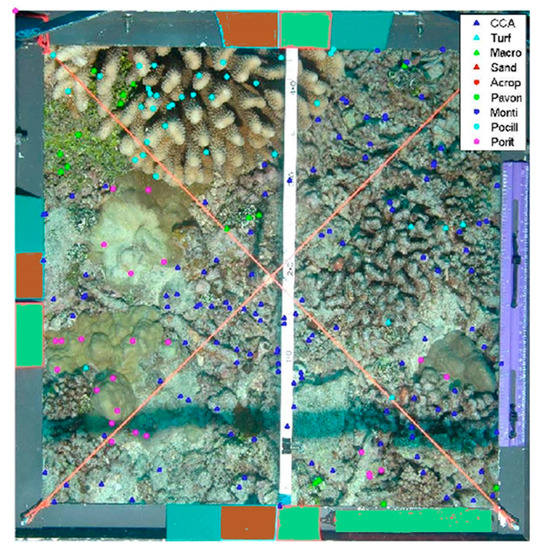
Figure 6.
A sample image from the Moorea Labeled Corals (MLC) dataset shows randomly labeled points. After (Beijbom et al. (2012)) [45].
Elawady (2015) [39] first enhanced the input raw images via color correction and smoothing filtering. Then, the trained LeNet-5 [30] based model, whose input layer consisted of three basic channels of color image plus extra channels for texture and shape descriptors. He also used the Moorea Labeled Corals (MLC) and chose nine classes as labels. These included five coral classes: (1) Acropora, (2) Pavona, (3) Montipora, (4) Pocillopora, and (5) Porites and four non-coral classes: (1) Crustose Coralline Algae, (2) Turf algae, (3) Macroalgae, and (4) Sand. The highest accuracy was achieved in identification rates for Acropora (coral) and for Sand (non-coral), and the lowest rates were for Pavona (coral) and Turf (non-coral). Misclassification of Pavona as Montipora/Macroalgae and of Turf as Macroalgae/Crustose Coralline Algae (CCA)/Sand was due to similarity in their shape properties.
Around 50 images were expertly annotated (200 labeled points per image) describing different, types of Lophelia coral habitats and the surrounding soft sediment Logachev mounds (Rockall Trough) Nine classes are classified: (1) Dead Coral, (2) Encrusting White Sponge, (3) Leiopathes Species, (4) Lophelia, and (5) Rubble Coral, (6) Boulder, (7) Darkness, (8) Gravel, and (9) Sand. Darkness had almost perfect classification rate due to its distinct nature. The black coral Leiopathes sp. also had an excellent classification rate due to its diagnostic color. According to the color diagnostic hypothesis, color should more strongly influence the recognition of high color diagnostic (HCD) objects (e.g., Lophelia (a coral)) than the recognition of low color diagnostic (LCD) objects (e.g., White Sponge) [47].
Mahmood et al. (2016) [48] combined CNN representations with hand-crafted features, using algorithms based on the information present in the image itself. These authors applied CNN representations extracted from VGGnet with a 2-layer MLP classifier (trained with the MLC, The Moorea Labeled Corals dataset). The accuracy they achieved was 77.9%.
We discuss in more detail the article by Gómez-Ríos et al. (2018) [49], since it is the most recent one, dealing with corals, and it resolves more corals than previous studies. In this study three CNNs were applied: Inception v3 [50], ResNet [51], and DenseNet [52] (Table 1). Two datasets were analyzed: EILAT and RSMAS both comprised of patches of coral images.

Table 1.
Accuracies obtained on two datasets. The accuracies obtained by Inception v3, ResNet-50, ResNet-152, DenseNet-121, DenseNet-161, and the classical state-of-the-art Shihavuddin model. Gómez-Ríos et al. (2018) [49].
ResNet is a short name for Residual Network. A residual network includes a number of residual blocks. Each block has a shortcut connection to the outputs of the next blocks (see Figure 7). Residual can be simply understood as subtraction of feature learned from input of that layer. ResNet does these using shortcut connections. Training this form of networks is easier than training simple deep CNN and the problem of degrading accuracy is resolved. Improved results for residual ResNet modifications are shown in Table 1. DenseNet differs from ResNet as it adds shortcuts among layers. Each subsequent layer receives all the outputs of previous layers and concatenates them in the depth dimension. In ResNet, a layer only receives outputs from the previous second or third layer, and the outputs are added together on the same depth. DenseNet adds a feature layer (convolutional layer) capturing low-level features from images, from several dense blocks and transition layers between adjacent dense blocks (see Figure 8).
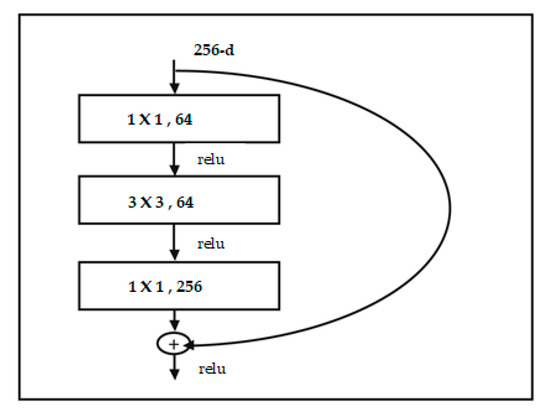
Figure 7.
ResNet building block from. After (He et al. 2016) [51].
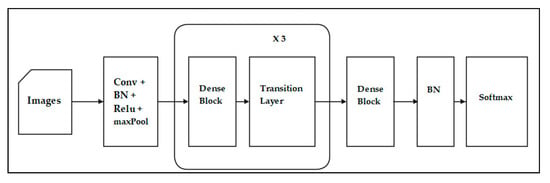
Figure 8.
The architecture of DenseNet.
CoralNet is a repository and a resource for benthic images analysis. The site https://coralnet.ucsd.edu/ deploys deep neural networks which allow fully or semi-automated annotation of images. It is used to upload and annotate coral reef images. It also serves as a convenient, user-friendly collaboration platform. CoralNet was conceived by Beijbom et al. [45,53]. In early 2019, Williams et al. [54] in a large study showed that the automated annotations for CoralNet Beta produced benthic cover estimates highly correlated with those derived from human annotation controls (see Table 2). The goal of the CoralNet, is that any user will be able to take advantage of their automated analysis, as stated by Hoegh-Guldberg “CoralNet will allow the world’s scientists to more quickly assess the health of coral reefs at scales never dreamed of before”, in https://blogs.nvidia.com/blog/2016/06/22/deep-learning-save-coral-reefs/. Using CoralNet as an automated analysis tool for a large-scale survey and monitoring program will encourage researchers to explore the coral reefs and find new solutions to save them.

Table 2.
Neural network (NN) reef studies from 2016–2018.
The above Table 2 reveals the already reliable power of neural networks in distinguishing corals from other benthos.
In summary, the findings of each study were as follows:
- Beijbom et al. (2012) [45] showed that their proposed method accurately estimates coral coverage across diverse reef sites and over multiple years, these offer timely and powerful potential for large scale coral reef analysis. The proposed algorithm accurately estimates coral coverage across locations and years, thereby taking a significant step towards reliable automated coral reef image annotation.
- Shihavuddin et al. (2013) [46] obtained better results by using a selective combination of various preprocessing steps, feature extraction methods, kernel mappings, priori designing a single method for all datasets. Using mosaic images, their method for large continuous areas resulted in 83.7% overall accuracy.
- Elawady, (2015) [39] presents the first application of DL techniques especially CNN in under-water image processing. He investigated CNN in handling noisy large-sized images, manipulating point-based multi-channel input data. Hybrid image patching procedure for multi-size scaling process across different square based windowing around labeled points.
- Mahmood et al. (2016) [48] proposed to use pre-trained CNN representations extracted from VGGnet with a 2-layer MLP classifier for coral classification. They also introduced a local-SPP approach to extract features at multiple scales to deal with the ambiguous class boundaries of corals. They then combined automated CNN based features with manually acquired hand-crafted features, while dealing effectively with the class imbalance problem.
- Mahmood et al. (2016a) [55] applied pre-trained CNN image representations extracted from VGGnet to a coral reef classification problem. They investigated the effectiveness of their trained classifier on unlabeled coral mosaics and analyzed the coral reef of the Abrolhos Islands in order to quantify the coral coverage and detect a decreasing trend in coral population automatically.
- Mahmood et al. (2017) [34] showed that the ResFeats data, extracted from the deeper layers of a ResNet performs better than the shallower ResFeats. They experimentally confirmed that their proposed approach consistently outperforms the CNN off-the-shelf features. Finally, they improved the state-of-the-art accuracy of the Caltech101, Caltech-256, and MLC datasets.
- Gómez-Ríos et al., (2018) [49] achieved state-of-the art accuracies using different variations of ResNet on the two small coral texture datasets, EILAT and RSMAS. They showed that using a simpler network, like ResNet-50, performs better than a more complex network, like DenseNet-121 or DenseNet-161 provided the datasets are small. When considering the impact of data augmentation, there is little benefit obtained from using such techniques.
- Williams et al. (2019) [54] used CoralNet to annotate points with an automated-analysis algorithm to achieve at least 90% certainty of a classification. CoralNet includes 822 data sets consisting of over 700,000 images. The site implements sophisticated computer vision algorithms, based on deep neural networks that allow researchers, agencies, and private partners to rapidly annotate benthic survey images. The site also serves as a repository and collaboration platform for the scientific community.
6. Discussion
The novelty of DL is the use of state-of-art computerized image analyses technologies, and its fully automated methodology of dealing with large data sets of images. We reviewed published applications DL to the marine benthos and corals. Early reef surveys were based on visual diver inspection of transects (e.g., [56,57]), followed by manual-visual expert study of photographic transects. For DL neither is required, as both are tedious, unsuited for large data sets and prone to human error. Hence, DL allows the process of identification to be done directly on a large number of photographs, minimizing human time, and errors, resulting in reliability, efficiency, and availability in terms of time and resources. These are essential for documenting and monitoring the worldwide processes that coral reef are undergoing.
Using CNN as a technique to classify coral assemblages and monitor their features is being increasingly used. This method will not always be the technique of choice for classifying corals underwater. The decision as to which technique to use depends, to a large extent, on the nature of the project and its goals. For example, it would be the method of choice when the objective of the project is to solve big data problems in ocean ecosystems generated by remotely operated vehicles (ROVs) and autonomous underwater vehicles (AUVs) and extensive video transects composed of thousands of images. All these provide repositories of big data that experts must process in order to assess policy-relevant metrics, such as species occurrence and mortality, providing the basis for operational conservation and remediation decisions. These methods will prove of special importance in detecting long-term change patterns and emerging trends in the structure of coral reefs due to global climate change effects, or conversely, in evaluating the effectiveness of coral-reef bioremediation enterprises. Image classification is time-consuming, costly to put into effect, and highly subject to differences in expert opinion. Using DL in a project whose goals are to classify and monitor the coral reef species during a long period of time based on a large amount of data, gives more accurate results in a shorter period of time compared to human classification. As pointed out, neural network methods are already reliable in distinguishing corals from other benthos and non-coral organisms [45], In general, corals were reliably separated from sand and turf [39], as well as from urchins, seaweeds, sand, and bare ground [46].
It is noteworthy that at present, the recognition of branching coral species is more trustworthy than that achieved among massive ones Shihavuddin et al. (2013) [46], For instance, the highest accuracy was achieved in identification rates for the branching coral Acropora and the lowest rates were for the coral Pavona The latter shortcoming needs more refinement of single polyp and calyx features, and less on colony parameters, due to their inter-specific similarities.
At its present state, the following difficulties are still hampering the full application of automated coral reef surveys: a) While video images have the clear advantage over single photographs, of generating vast larger data sets for comparable effort, their quality is inferior, limiting their usefulness. In addition, in video transects there are likely changes in camera angle, which introduce inevitable difficulties in comparing shots from a sequence. A similar difficulty in using video transects results from the camera movement that degrades image resolution. b) Photographic surveys are likely to miss cryptic coral species, hidden under large coral colonies, as well as small juvenile colonies visible to the human eye. c) Currently available automated coral survey applications are unable to distinguish bleached from diseased corals, as both are likely to show pigmentation loss, as compared to healthy colonies. d) Environmentally caused phenotypic plasticity of corals is well known [58]. These include large changes in colony architecture, size, and pigmentation. Such well documented variability results from factors like current velocity and direction [59], light intensity [9], temperature, and pH [60]. These changes are known to present a challenge to human specialists, leading to miss-identifications. That problem is even more difficult to overcome for AI methodology based, coral identification to the species level. Hence future refinements have to add a further classifying trait, the high resolution, close range, images of single coral polyps.
7. Conclusions
CNN may be especially useful when the primary goal is trend identification and outcome prediction while revealing hidden interactions or complex nonlinearities in a data set as these inherent in the coral reef itself. Further research will be required to determine, under which circumstances the advantages of CNN exceed their disadvantages in the classification and prediction of ecological outcomes.
The advantages of using DL are: reliability and efficiency in terms of time and resources.
The significance:
- Working with “big data” in order to address the urgent ecological need of classifying corals.
- So far, the classification of corals has been performed by using outdated painstaking, time consuming manual methods, which highly limit its performance and usefulness. As a result, current coral reef species censuses are usually limited to single reefs [61]. That does not allow comparison among different reefs and their responses to specific stressors. However, Cleary et al. [62,63,64] present evidence to the contrary. A further important goal is to refine discrimination acuity and reliability sufficiently to use photographic images of larger reef sectors. A complementary possibility will be to extract large data sets from continuous film transects, all the above will be achievable only by the combination between NN future capabilities with cloud large data set storage volume.Furthermore, the painstaking nature of coral censuses limits the possibility of non-specialized technicians to gather large amounts of temporarily sequential data sets that can track decline and/or recovery of reefs in response to different IPCC scenarios and bioremediation measures.The usefulness of future, more refined NN will allow such comparisons and their analyses.The impressive power of NN in advanced application to plants has already been demonstrated [33].
- Developing and applying the automatic methods of machine based DL neuronal networks for classifying corals and using future large dataset solutions, where programs are made freely available to allow researchers anywhere to use their own images to train the algorithms then run their own analysis will help monitoring the global reefs health all over the world. An additional, sorely needed, but achievable development would be the possibility of NN to identify different age colonies of the same species, in order to follow population level processes, crucial for recruitment and inter-specific competition outcomes.
The future vision of the application of DL to coral reef ecology should aim at the following capabilities.
- Using video transects, rather than stills, as the image source.
- Allow recognition of different ages of the same coral species in order to extract population dynamics features such as population age structure and recruitment or decline of species.
- Calculate species diversity parameters from the image sets.
- Construct trend lines from sequential photo surveys of the same reef providing future likely outcomes.
- Estimate reef health based on bleaching and mortality.
Author Contributions
A.R. wrote the manuscript with input from all authors. Z.D. shared his extensive experience and guided the authors with a deeper understanding of the coral reefs and marine ecosystems. D.I. actively participated throughout the research process. N.S.N. helped and advised with deep learning throughout the research process. throughout the research process. All authors have read and agreed to the published version of the manuscript.
Acknowledgments
The authors thank Irit Shoval from the Scientific Equipment Center of the Faculty of Life Sciences at Bar-Ilan University for helping with the data analysis and processing of the results. We also thank Jennifer Benichou Israel Cohen from the Statistics Unit at Bar-Ilan University for helping with presentation of the results and preparation for statistical analyses. We thank Sharon Victor for her help with formatting the manuscript. No funding was received for this research.
Conflicts of Interest
We hereby declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Field, M.E.; Cochran, S.A.; Evans, K.R. U.S. Coral Reefs; Imperiled National Treasures; 025-02; USGS U.S. Geological Survey Fact: Reston, VA, USA, 2002. Available online: https://pubs.usgs.gov/fs/2002/fs025-02/ (accessed on 12 November 2019).
- Roberts, C.M.; McClean, C.J.; Veron, J.E.N.; Hawkins, J.P.; Allen, G.R.; McAllister, D.E.; Mittermeier, C.G.; Schueler, F.W.; Spalding, M.; Wells, F.; et al. Marine biodiversity hotspots and conservation priorities for tropical reefs. Science 2002, 295, 1280–1284. [Google Scholar] [CrossRef]
- Schumacher, H.; Zibrowius, H. What is hermatypic? A redefinition of ecological groups in corals and other organisms. Coral Reefs 1985, 4, 1–9. [Google Scholar] [CrossRef]
- Jones, C.G.; Lawton, J.H.; Shachak, M. Organisms as Ecosystem Engineers. In Ecosystem Management; Springer: New York, NY, USA, 1994; pp. 130–147. [Google Scholar]
- Brandt, K. Uber die symbiose von algen und tieren. Arch. Anat. Physiol. 1883, 1, 445–454. [Google Scholar]
- Odum, H.T.; Odum, E.P. Trophic structure and productivity of a windward coral reef community on Eniwetok Atoll. Ecol. Monogr. 1955, 25, 291–320. [Google Scholar] [CrossRef]
- Iluz, D.; Fermani, S.; Ramot, M.; Reggi, M.; Caroselli, E.; Prada, F.; Dubinsky, Z.; Goffredo, S.; Falini, G. Calcifying response and recovery potential of the brown alga Padina pavonica under ocean acidification. ACS Earth Space Chem. 2017, 1, 316–323. [Google Scholar] [CrossRef]
- Burns, T.P. Hard-coral distribution and cold-water disturbances in South Florida: Variation with depth and location. Coral Reefs 1985, 4, 117–124. [Google Scholar] [CrossRef]
- Dubinsky, Z.; Iluz, D. Corals and Light: From Energy Source to Deadly Threat. In The Cnidaria, Past, Present and Future: The World of Medusa and Her Sisters; Goffredo, S., Dubinsky, Z., Eds.; Springer: Cham, Switzerland, 2016; pp. 469–487. [Google Scholar]
- Goreau, T.F. The physiology of skeleton formation in corals. I. A method for measuring the rate of calcium deposition by corals under different conditions. Biol. Bull. 1959, 116, 59–75. [Google Scholar] [CrossRef]
- Cohen, A.L.; McCorkle, D.C.; de Putron, S.; Gaetani, G.A.; Rose, K.A. Morphological and compositional changes in the skeletons of new coral recruits reared in acidified seawater: Insights into the bio mineralization response to ocean acidification. Geochem. Geophys. Geosyst. 2009, 10. [Google Scholar] [CrossRef]
- Anthony, K.R.N.; Kline, D.I.; Diaz-Pulido, G.; Dove, S.; Hoegh-Guldberg, O. Ocean acidification causes bleaching and productivity loss in coral reef builders. Proc. Natl. Acad. Sci. USA 2008, 105, 17442–17446. [Google Scholar] [CrossRef]
- Fox, H.E.; Pet, J.S.; Dahuri, R.; Caldwell, R.L. Recovery in rubble fields: Long-term impacts of blast fishing. Mar. Pollut. Bull. 2003, 46, 1024–1031. [Google Scholar] [CrossRef]
- Wood, E. Collection of Coral Reef Fish for Aquaria: Global Trade, Conservation Issues and Management Strategies; Marine Conservation Society: Ross-on-Wye, UK, 2001. [Google Scholar]
- Downs, C.A.; Kramarsky-Winter, E.; Segal, R.; Fauth, J.; Knutson, S.; Bronstein, O.; Pennington, P. Toxicopathological effects of the sunscreen UV filter, oxybenzone (benzophenone-3), on coral planulae and cultured primary cells and its environmental contamination in Hawaii and the US Virgin Islands. Arch. Environ. Contam. Toxicol. 2016, 70, 265–288. [Google Scholar] [CrossRef] [PubMed]
- Longcore, T.; Rich, C. Ecological light pollution. Front. Ecol. Environ. 2004, 2, 191–198. [Google Scholar] [CrossRef]
- Wielgus, J.; Balmford, A.; Lewis, T.B.; Mora, C.; Gerber, L.R. Coral reef quality and recreation fees in marine protected areas. Conserv. Lett. 2010, 3, 38–44. [Google Scholar] [CrossRef]
- Littler, M.M.; Littler, D.S.; Brooks, B.L. Harmful algae on tropical coral reefs: Bottom-up eutrophication and top-down herbivory. Harmful Algae 2006, 5, 565–585. [Google Scholar] [CrossRef]
- Fabricius, K.E. Effects of terrestrial runoff on the ecology of corals and coral reefs: Review and synthesis. Mar. Pollut. Bull. 2005, 50, 125–146. [Google Scholar] [CrossRef] [PubMed]
- Heery, E.C.; Hoeksema, B.W.; Browne, N.K.; Reimer, J.D.; Ang, P.O.; Huang, D.; Alsagoff, N. Urban coral reefs: Degradation and resilience of hard coral assemblages in coastal cities of East and Southeast Asia. Mar. Pollut. Bull. 2018, 135, 654–681. [Google Scholar] [CrossRef]
- West, J.M.; Salm, R.V. Resistance and resilience to coral bleaching: Implications for coral reef conservation and management. Conserv. Biol. 2003, 17, 956–967. [Google Scholar] [CrossRef]
- Gorbunov, M.Y.; Falkowski, P.G. Photoreceptors in the cnidarian hosts allow symbiotic corals to sense blue moonlight. Limnol. Oceanogr. 2002, 47, 309–315. [Google Scholar] [CrossRef]
- Tamir, R.; Lerner, A.; Haspel, C.; Dubinsky, Z.; Iluz, D. The spectral and spatial distribution of light pollution in the waters of the northern Gulf of Aqaba (Eilat). Sci. Rep. 2017, 7, 42329. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification. In Proceedings of the IEEE International Conference on Computer Vision, Copenhagen, Denmark, 6 February 2015; pp. 1026–1034. [Google Scholar]
- Ciregan, D.; Meier, U.; Schmidhuber, J. Multi-Column Deep Neural Networks for Image Classification. In Proceedings of the 2012 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Providence, RI, USA, 16–21 June 2012; pp. 3642–3649. [Google Scholar]
- Sun, Y.; Chen, Y.; Wang, X.; Tang, X. Deep Learning Face Representation by Joint Identification-Verification. In Proceedings of the Advances in Neural Information Processing Systems (1988–1996), Montreal, QC, Canada, 8–13 December 2014. [Google Scholar]
- Taigman, Y.; Yang, M.; Ranzato, M.A.; Wolf, L. DeepFace: Closing the Gap to Human-Level Performance in Face Verification. In Proceedings of the 2014 IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 1701–1708. [Google Scholar]
- Wan, L.; Zeiler, M.; Zhang, S.; LeCun, Y.; Fergus, R. Regularization of Neural Networks Using DropConnect. In Proceedings of the 30th International Conference on Machine Learning, Atlanta, GA, USA, 16–21 June 2013; pp. 1058–1066. [Google Scholar]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Wang, T.; Wu, D.J.; Coates, A.; Ng, A.Y. End-to-end Text Recognition with Convolutional Neutral Networks. In Proceedings of the 21st International Conference on Pattern Recognition (ICPR2012), Tsukuba, Japan, 11–15 November 2012; pp. 3304–3308. [Google Scholar]
- Coates, A.; Carpenter, B.; Case, C.; Satheesh, S.; Suresh, B.; Wang, T.; Ng, A.Y. Text Detection and Character Recognition in Scene Images with Unsupervised Feature Learning. ICDAR 2011, 11, 440–445. [Google Scholar] [CrossRef]
- Reyes, A.K.; Caicedo, J.C.; Camargo, J.E. Fine-Tuning Deep Convolutional Networks for Plant Recognition. In Proceedings of the Working Notes of CLEF 2015 Conference, Toulouse, France, 8–11 September 2015. [Google Scholar]
- Mahmood, A.; Bennamoun, M.; An, S.; Sohel, F. Resfeats: Residual Network Based Features for Image Classification. In Proceedings of the 2017 IEEE International Conference on Image Processing (ICIP), Beijing, China, 17–20 September 2017; pp. 1597–1601. [Google Scholar]
- Lee, H.; Pham, P.T.; Largman, Y.; Ng, A.Y. Unsupervised Feature Learning for Audio Classification Using Convolutional Deep Belief Networks. In Proceedings of the Advances in Neural Information Processing Systems 22 (NIPS 2009), Vancouver, BC, Canada, 10–12 December 2009; pp. 1096–1104. [Google Scholar]
- Christin, S.; Hervet, E.; Lecomte, N. Applications for deep learning in ecology. bioRxiv 2018, bioRxiv334854. [Google Scholar] [CrossRef]
- Ravikiran, C.; Prasad, V.; Rehna, V.J. Enhancing Underwater Gray Scale Images Using a Hybrid Approach of Filtering and Stretching Technique. Int. J. Adv. Eng. Manag. Sci. 2015, 1, 3. [Google Scholar]
- Pizarro, O.; Rigby, P.; Johnson-Roberson, M.; Williams, S.B.; Colquhoun, J. Towards Image-Based Marine Habitat Classification. In Proceedings of the OCEANS, Basel, Switzerland, 15–18 September 2008; pp. 1–7. [Google Scholar]
- Elawady, M. Sparse Coral Classification Using Deep Convolutional Neural Networks (VIBOT 2014). arXiv 2015, arXiv:1511.09067v1. [Google Scholar]
- Johnson-Roberson, M.; Kumar, S.; Williams, S. Segmentation and Classification of Coral for Oceanographic Surveys: A Semi-Supervised Machine Learning Approach. In Proceedings of the OCEANS—Asia Pacific, Singapore, 16–19 May 2006; pp. 1–6. [Google Scholar]
- Lowe, D.G. Object Recognition from Local Scale-Invariant Features. In Proceedings of the 7th IEEE International Conference on Computer Vision, Kerkyra, Greece, 20–27 September 1999; Volume 2, pp. 1150–1157. [Google Scholar]
- O’Hara, S.; Draper, B.A. Introduction to the Bag of Features paradigm for image classification and retrieval. arXiv 2011, arXiv:1101.3354. [Google Scholar]
- Stokes, M.D.; Deane, G.B. Automated processing of coral reef benthic images. Limnol. Oceanogr. Methods 2009, 7, 157–168. [Google Scholar] [CrossRef]
- Yang, J.; Liu, C.J.; Zhang, L. Color space normalization: Enhancing the discriminating power of color spaces for face recognition. Pattern Recognit. 2010, 43, 1454–1466. [Google Scholar] [CrossRef]
- Beijbom, O.; Edmunds, P.J.; Kline, D.I.; Mitchell, B.G.; Kriegman, D. Automated Annotation of Coral reef Survey Images. In Proceedings of the 2012 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Providence, RI, USA, 16–21 June 2012. [Google Scholar]
- Shihavuddin, A.S.M.; Gracias, N.; Garcia, R.; Gleason, A.C.; Gintert, B. Image-based coral reef classification and thematic mapping. Remote Sens. 2013, 5, 1809–1841. [Google Scholar] [CrossRef]
- Tanaka, J.W.; Presnell, L.M. Color diagnosticity in object recognition. Percept. Psychophys. 1999, 61, 1140–1153. [Google Scholar] [CrossRef]
- Mahmood, A.; Bennamoun, M.; An, S.; Sohel, F.; Boussaid, F.; Hovey, R.; Kendrick, G.; Fisher, R.B. Coral classification with hybrid feature representations. In Proceedings of the IEEE International Conference on Image Processing (ICIP), Phoenix, AZ, USA, 25–28 September 2016. [Google Scholar]
- Gómez-Ríos, A.; Tabik, S.; Luengo, J.; Shihavuddin, A.S.M.; Krawczyk, B.; Herrera, F. Towards highly accurate coral texture images classification using deep convolutional neural networks and data augmentation. arXiv 2018, arXiv:1804.00516. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Vegas, NV, USA, 26 June–1 July 2016; pp. 2818–2826. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity Mappings in Deep Residual Networks. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 11–14 October 2016; Springer: Cham, Switzerland, 2016; pp. 630–645. [Google Scholar]
- Huang, G.; Liu, S.; van der Maaten, L.; Weinberger, K.Q. CondenseNet: An Efficient DenseNet Using Learned Group Convulutions. Group 2017, 3, 11. [Google Scholar]
- Beijbom, O.; Edmunds, P.J.; Roelfsema, C.; Smith, J.; Kline, D.I.; Neal, B.P.; Chan, S. Towards automated annotation of benthic survey images: Variability of human experts and operational modes of automation. PLoS ONE 2015, 10, e0130312. [Google Scholar] [CrossRef] [PubMed]
- Williams, I.D.; Couch, C.S.; Beijbom, O.; Oliver, T.A.; Vargas-Angel, B.; Schumacher, B.D.; Brainard, R.E. Leveraging Automated Image Analysis Tools to Transform Our Capacity to Assess Status and Trends of Coral Reefs. Front. Mar. Sci. 2019. [Google Scholar] [CrossRef]
- Mahmood, A.; Bennamoun, M.; An, S.; Sohel, F.; Boussaid, F.; Hovey, R.; Kendrick, G.; Fisher, R. Automatic annotation of coral reefs using deep learning. In Proceedings of the OCEANS 2016 MTS/IEEE Monterey, Monterey, CA, USA, 19–23 September 2016; pp. 1–5. [Google Scholar] [CrossRef]
- Loya, Y. Community structure and species diversity of hermatypic corals at Eilat, Red Sea. Mar. Biol. 1972, 13, 100–123. [Google Scholar] [CrossRef]
- Samoilys, M.A.; Carlos, G. Determining methods of underwater visual census for estimating the abundance of coral reef fishes. Environ. Biol. Fishes 2000, 57, 289–304. [Google Scholar] [CrossRef]
- Shaish, L.; Abelson, A.; Rinkevich, B. How plastic can phenotypic plasticity be? The branching coral Stylophora pistillata as a model system. PLoS ONE 2007, 2, e644. [Google Scholar] [CrossRef]
- Kaandorp, J.A.; Koopman, E.A.; Sloot, P.M.; Bak, R.P.; Vermeij, M.J.; Lampmann, L.E. Simulation and analysis of flow patterns around the scleractinian coral Madracis mirabilis (Duchassaing and Michelotti). Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2003, 358, 1551–1557. [Google Scholar] [CrossRef]
- Goffredo, S.; Caroselli, E.; Mattioli, G.; Pignotti, E.; Dubinsky, Z.; Zaccantia, F. Inferred level of calcification decreases along an increasing temperature gradient in a Mediterranean endemic coral. Limnol. Oceanogr. 2009, 54, 930–937. [Google Scholar] [CrossRef]
- Hoey, A.S.; Bellwood, D.R. Limited functional redundancy in a high diversity system: Single species dominates key ecological process on coral reefs. Ecosystems 2009, 12, 1316–1328. [Google Scholar] [CrossRef]
- Cleary, D.F.; De Vantier, L.; Vail, L.; Manto, P.; de Voogd, N.J.; Rachello-Dolmen, P.G.; Hoeksema, B.W. Relating variation in species composition to environmental variables: A multi-taxon study in an Indonesian coral reef complex. Aquat. Sci. 2008, 70, 419–431. [Google Scholar] [CrossRef]
- Cleary, D.F.; Polónia, A.R.; Renema, W.; Hoeksema, B.W.; Wolstenholme, J.; Tuti, Y.; de Voogd, N.J. Coral reefs next to a major conurbation: A study of temporal change (1985–2011) in coral cover and composition in the reefs of Jakarta, Indonesia. Mar. Ecol. Prog. Ser. 2014, 501, 89–98. [Google Scholar] [CrossRef]
- Cleary, D.F.R.; Polónia, A.R.M.; Renema, W.; Hoeksema, B.W.; Rachello-Dolmen, P.G.; Moolenbeek, R.G.; Hariyanto, R. Variation in the composition of corals, fishes, sponges, echinoderms, ascidians, molluscs, foraminifera and macroalgae across a pronounced in-to-offshore environmental gradient in the Jakarta Bay–Thousand Islands coral reef complex. Mar. Pollut. Bull. 2016, 110, 701–717. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).