Predicting Extinction Risk for Data Deficient Bats
Abstract
:1. Introduction
2. Materials and Methods
2.1. Trait Data Collection
2.2. Phylogenetic Tree Construction
2.3. Model Construction
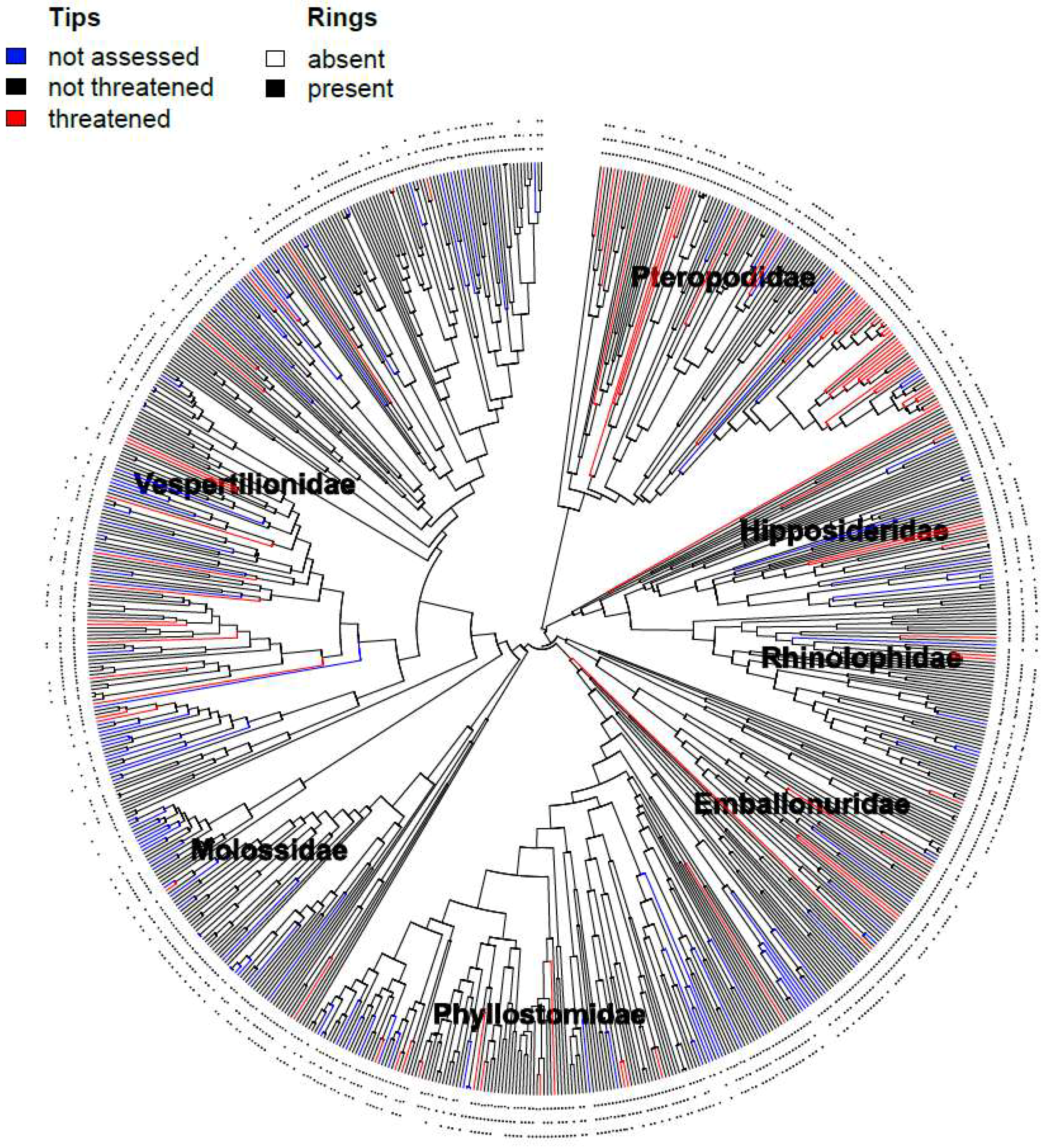
3. Results
3.1. Correlates of Extinction Risk
3.2. Models for Predicting Extinction Risk
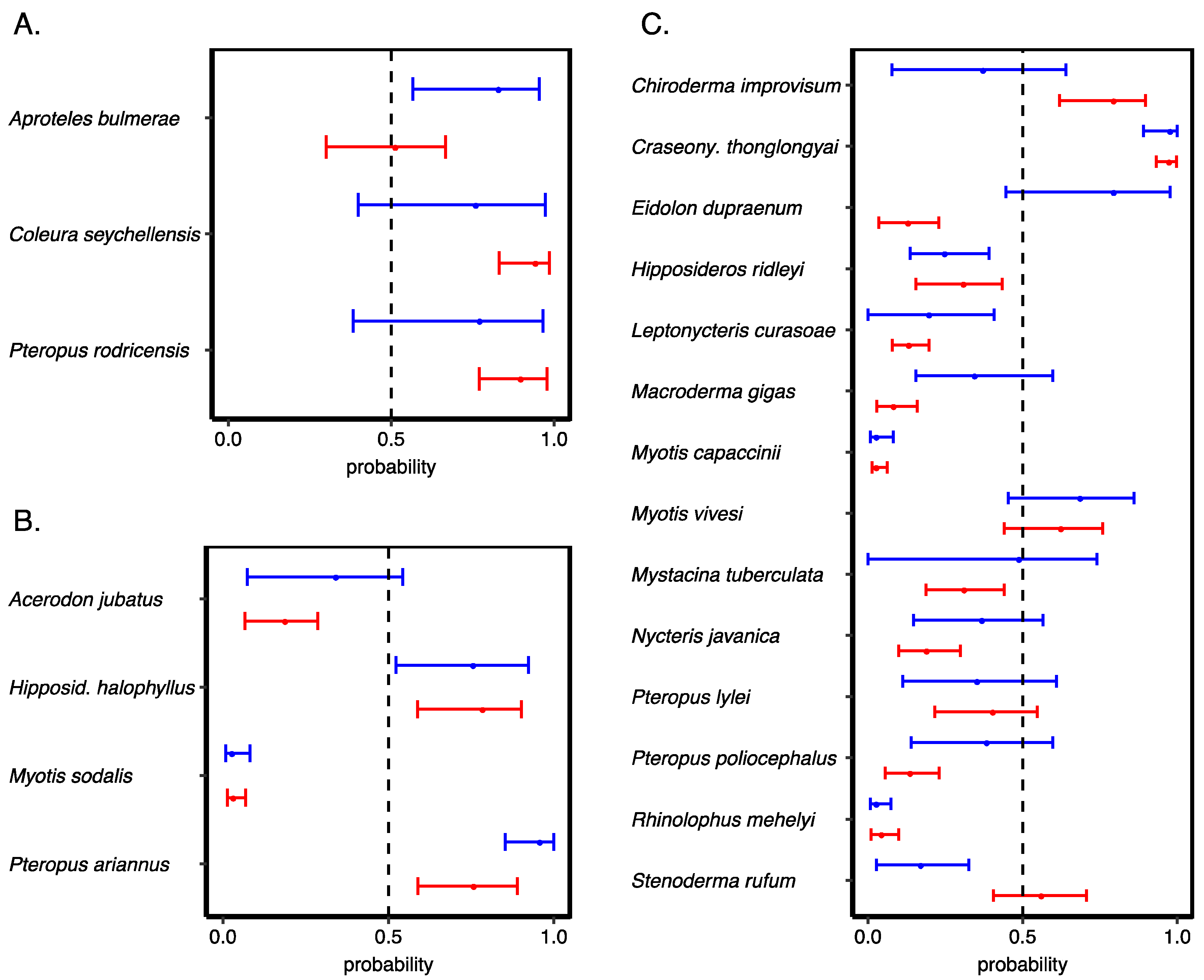
3.3. Validating Our Predictions of Extinction Risk
3.4. Species Most at Risk of Extinction
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- The IUCN Red List of Threatened Species, Version 2013. Available online: iucnredlist.org (accessed on 26 March 2013).
- Hutson, A.M.; Mickleburgh, S.P.; Racey, P.A. Microchiropteran Bats: Global Status Survey Conservation Action Plan; IUCN/SSC Chiroptera Specialist Group: Gland, Switzerland; Cambridge, UK, 2001; 258p. [Google Scholar]
- IUCN Standards Petitions Subcommittee. Guidelines for Using the IUCN Red List Categories and Criteria; Version 11; IUCN: Gland, Switzerland, 2014; 87p. [Google Scholar]
- Racey, P.A.; Entwistle, E.C. Conservation ecology of bats. In Bat Ecology; Kunz, T.H., Fenton, M.B., Eds.; University of Chicago Press: Chicago, IL, USA, 2005; pp. 680–743. [Google Scholar]
- Juffe-Bignoli, D.; Brooks, T.M.; Butchart, S.H.; Jenkins, R.B.; Boe, K.; Hoffmann, M.; Angulo, A.; Bachman, S.; Böhm, M.; Brummitt, N.; et al. Assessing the cost of global biodiversity and conservation knowledge. PLoS ONE 2016, 11, e0160640. [Google Scholar] [CrossRef] [PubMed]
- Campbell, L.M. Seeing red: Inside the science and politics of the IUCN Red List. Conserv. Soc. 2012, 10, 367. [Google Scholar] [CrossRef]
- Jetz, W.; Freckleton, R.P. Towards a general framework for predicting threat status of data-deficient species from phylogenetic, spatial and environmental information. Philos. Trans. R. Soc. B 2015, 370, 20140016. [Google Scholar] [CrossRef] [PubMed]
- Jeppsson, T.; Forslund, P. Species’ traits explain differences in Red list status and long-term population trends in longhorn beetles. Anim. Conserv. 2014, 17, 332–341. [Google Scholar] [CrossRef]
- Olah, G.; Butchart, S.H.; Symes, A.; Guzmán, I.M.; Cunningham, R.; Brightsmith, D.J.; Heinsohn, R. Ecological and socio-economic factors affecting extinction risk in parrots. Biodivers. Conserv. 2016, 25, 205–223. [Google Scholar] [CrossRef]
- Davidson, A.D.; Hamilton, M.J.; Boyer, A.G.; Brown, J.H.; Ceballos, G. Multiple ecological pathways to extinction in mammals. Proc. Natl. Acad. Sci. USA 2009, 106, 10702–10705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Böhm, M.; Williams, R.; Bramhall, H.R.; McMillan, K.M.; Davidson, A.D.; Garcia, A.; Bland, L.M.; Bielby, J.; Collen, B. Correlates of extinction risk in squamate reptiles: The relative importance of biology, geography, threat and range size. Glob. Ecol. Biogeogr. 2016, 25, 391–405. [Google Scholar] [CrossRef]
- Garland, T.J.; Ives, A.R. Using the past to predict the present: Confidence intervals for regression equations in phylogenetic comparative methods. Am. Nat. 2000, 155, 346–364. [Google Scholar] [CrossRef] [PubMed]
- Purvis, A.; Agapow, P.M.; Gittleman, J.L.; Mace, G.M. Nonrandom extinction and the loss of evolutionary history. Science 2000, 288, 328–330. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Phylogenies and the comparative method. Am. Nat. 1985, 125, 1–15. [Google Scholar] [CrossRef]
- Purvis, A.; Gittleman, J.L.; Cowlishaw, G.; Mace, G.M. Predicting extinction risk in declining species. Proc. R. Soc. B Biol. Sci. 2000, 267, 1947–1952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardillo, M.; Purvis, A.; Sechrest, W.; Gittleman, J.L.; Bielby, J.; Mace, G.M. Human population density and extinction risk in the world’s carnivores. PLoS Biol. 2004, 2, e197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardillo, M.; Mace, G.M.; Jones, K.E.; Bielby, J.; Bininda-Emonds, O.R.; Sechrest, W.; Orme, C.D.; Purvis, A. Multiple causes of high extinction risk in large mammal species. Science 2005, 309, 1239–1241. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.E.; Purvis, A.; Gittleman, J.L. Biological correlates of extinction risk in bats. Am. Nat. 2003, 161, 601–614. [Google Scholar] [CrossRef] [PubMed]
- Simmons, N.B. Order Chiroptera. In Mammal Species of the World, 3rd ed.; Wilson, D.E., Reeder, D.M., Eds.; The John Hopkins University Press: Baltimore, MD, USA, 2005; Volume 1, pp. 312–529. [Google Scholar]
- Benson, D.A.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2015, 43, D30–D35. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.E.; Bielby, J.; Cardillo, M.; Fritz, S.A.; O’Dell, J.; Orme, C.D.L.; Safi, K.; Sechrest, W.; Boakes, E.H.; Carbone, C.; et al. PanTHERIA: A species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology 2009, 90, 2648. [Google Scholar] [CrossRef]
- Jones, K.E.; Purvis, A.; Maclarnon, A.N.N.; Bininda-Emonds, O.R.; Simmons, N.B. A phylogenetic supertree of the bats (Mammalia: Chiroptera). Biol. Rev. 2002, 77, 223–259. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.A.; Beaulieu, J.M.; Donoghue, M.J. Mega-phylogeny approach for comparative biology: An alternative to supertree and supermatrix approaches. BMC Evolut. Biol. 2009, 9, 37. [Google Scholar] [CrossRef] [PubMed]
- Teeling, E.C.; Springer, M.S.; Madsen, O.; Bates, P.; O’Brien, S.J.; Murphy, W.J. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 2005, 307, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Almeida, F.C.; Giannini, N.P.; DeSalle, R.; Simmons, N.B. Evolutionary relationships of the old world fruit bats (Chiroptera, Pteropodidae): Another star phylogeny? BMC Evolut. Biol. 2011, 11, 281. [Google Scholar] [CrossRef] [PubMed]
- Ruedi, M.; Stadelmann, B.; Gager, Y.; Douzery, E.J.; Francis, C.M.; Lin, L.K.; Guillén-Servent, A.; Cibois, A. Molecular phylogenetic reconstructions identify East Asia as the cradle for the evolution of the cosmopolitan genus Myotis (Mammalia, Chiroptera). Mol. Phylogenet. Evol. 2013, 69, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Agnarsson, I.; Zambrana-Torrelio, C.M.; Flores-Saldana, N.P.; May-Collado, L.J. A time-calibrated species-level phylogeny of bats (Chiroptera, Mammalia). PLoS Curr. 2011, 3. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.A.; O’Meara, B.C. treePL: Divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 2012, 28, 2689–2690. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.J.; Rabosky, D.L. Speciation dynamics during the global radiation of extant bats. Evolution 2015, 69, 1528–1545. [Google Scholar] [CrossRef] [PubMed]
- Springer, M.S.; Teeling, E.C.; Madsen, O.; Stanhope, M.J.; de Jong, W.W. Integrated fossil and molecular data reconstruct bat echolocation. Proc. Natl. Acad. Sci. USA 2001, 98, 6241–6246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyles, J.G.; Storm, J.J. The perils of picky eating: Dietary breadth is related to extinction risk in insectivorous bats. PLoS ONE 2007, 2, e672. [Google Scholar] [CrossRef] [PubMed]
- Ives, A.R.; Garland, T.J. Phylogenetic logistic regression for binary dependent variables. Syst. Biol. 2009, 59, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Tung Ho, L.S.; Ané, C. A linear-time algorithm for Gaussian and non-Gaussian trait evolution models. Syst. Biol. 2014, 63, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Freckleton, R.P.; Harvey, P.H.; Pagel, M. Phylogenetic analysis and comparative data: A test and review of evidence. Am. Nat. 2002, 160, 712–726. [Google Scholar] [CrossRef] [PubMed]
- Blomberg, S.P.; Garland, T.J.; Ives, A.R. Testing for phylogenetic signal in comparative data: Behavioral traits are more labile. Evolution 2003, 57, 717–745. [Google Scholar] [CrossRef] [PubMed]
- Cooney, R. The Precautionary Principle in Biodiversity Conservation and Natural Resource Management: An Issues Paper for Policy-Makers, Researchers and Practitioners; IUCN Policy and Global Change Series No. 2; IUCN: Gland, Switzerland; Cambridge, UK, 2004; 51p. [Google Scholar]
- Ruedi, M.; Eger, J.L.; Lim, B.K.; Csorba, G. A new genus and species of vespertilionid bat from the Indomalayan Region. J. Mammal. 2017, 99, 209–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arroyo-Cabrales, J.; Álvarez-Castañeda, S.T.; Myotis, S. The IUCN Red List of Threatened Species; IUCN: Gland, Switzerland, 2008. [Google Scholar] [CrossRef]
- Fisher, D.O.; Owens, I.P. The comparative method in conservation biology. TREE 2004, 19, 391–398. [Google Scholar] [PubMed]
- Jones, K.E.; Mickleburgh, S.P.; Sechrest, W.; Walsh, A.L. Global overview of the conservation of island bats: Importance, challenges and opportunities. In Island Bats: Evolution, Ecology, and Conservation; Fleming, T.H., Racey, P.A., Eds.; University of Chicago Press: Chicago, IL, USA, 2009; pp. 496–530. [Google Scholar]
- Safi, K.; Kerth, G. A comparative analysis of specialization and extinction risk in temperate-zone bats. Conserv. Biol. 2004, 18, 1293–1303. [Google Scholar] [CrossRef] [Green Version]
- Norberg, U.M.; Rayner, J.M. Ecological morphology and flight in bats (Mammalia; Chiroptera): Wing adaptations, flight performance, foraging strategy and echolocation. Philos. Trans. R. Soc. B 1987, 316, 335–427. [Google Scholar] [CrossRef]

| Variable | Definition | Source |
|---|---|---|
| aspect ratio | continuous: wingspan squared divided by wing area | Jones et al., 2003 |
| endemism | binary: observed (endemic) only on islands, excluding Australia | IUCN Red List 3 |
| forearm length (mm) | continuous: adults, total length from elbow to wrist; measures of central tendency | PanTHERIA 1 |
| diet breadth | discrete: number of dietary categories eaten; measures of central tendency; categories defined as vertebrate, invertebrate, fruit, flowers/nectar/pollen, leaves/branches/bark, seeds, grass, and roots/tubers; maximum observed = 4 | PanTHERIA 1 |
| litter size | continuous: number of offspring born per littler per female; measures of central tendency | PanTHERIA 1 |
| litters per year | continuous: number of litters; central tendency | PanTHERIA 1 |
| mass (g) | continuous: adult body mass, excluding pregnant females; measures of central tendency | PanTHERIA 1 |
| nucleotide sequences | CytB, Rag2, Val, 12S, 16S | GenBank |
| range (km2) | continuous: total extent of a species geographic range area from Sechrest 2003 2 | PanTHERIA 1 |
| threat category | binary: 1 = Critically Endangered (CR), Endangered (EN), Vulnerable (VU), 0 = Near Threatened (NT), Least Concern (LC); NA = Data Deficient (DD), Not Evaluated (NE) | IUCN Red List 3 |
| trophic level | categorical: measures of central tendency; categories defined as 0 = herbivore, 1 = omnivore, 2 = carnivore | PanTHERIA 1 |
| wing loading | continuous: body mass times gravity acceleration divided by wing area | Jones et al., 2003 |
| Variables | All Bats | Yinpterochiroptera | Yangochiroptera | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| α | b | P(1) | R2 | α | b | P(1) | R2 | α | b | P(1) | R2 | |
| aspect ratio | 0.084 | −8.089 | 0.000 | 0.058 | 0.281 | −7.379 | 0.001 | 0.017 | 0.148 | −7.793 | 0.000 | 0.051 |
| diet breadth | 0.547 | 0.600 | 0.646 | 0.037 | 0.509 | 0.881 | 0.707 | 0.104 | 0.557 | 0.401 | 0.599 | 0.010 |
| endemism | 0.099 | 1.643 | 0.838 | 0.139 | 0.133 | 1.035 | 0.738 | 0.067 | 0.243 | 1.942 | 0.875 | 0.138 |
| forearm length | 0.160 | 3.902 | 0.980 | 0.054 | 0.246 | 4.695 | 0.991 | 0.095 | 0.678 | −2.353 | 0.087 | 0.005 |
| litter size | 0.082 | −23.657 | 0.000 | 0.050 | 0.592 | −10.664 | 0.000 | 0.010 | 0.404 | −4.755 | 0.009 | 0.052 |
| litters per year | 0.240 | −9.702 | 0.000 | 0.062 | 0.502 | −10.511 | 0.000 | 0.141 | 0.448 | −2.591 | 0.070 | 0.050 |
| mass | 0.266 | 1.574 | 0.828 | 0.074 | 0.517 | 1.461 | 0.812 | 0.153 | 0.345 | −0.011 | 0.497 | 0.000 |
| range | 0.020 | −1.462 | 0.188 | 0.468 | 0.138 | −1.284 | 0.217 | 0.405 | 0.028 | −1.774 | 0.145 | 0.527 |
| range abbr. | 0.021 | −1.109 | 0.248 | 0.264 | 0.032 | −0.903 | 0.288 | 0.221 | 0.039 | −1.270 | 0.219 | 0.261 |
| wing loading | 0.336 | −0.584 | 0.358 | 0.001 | 0.293 | −0.394 | 0.403 | −0.003 | 0.413 | −0.881 | 0.293 | 0.002 |
| range × endemism | 0.019 | 0.964 | 0.724 | 0.493 | 0.052 | 1.017 | 0.734 | 0.469 | 0.029 | 0.550 | 0.634 | 0.537 |
| range × mass | 0.020 | 0.129 | 0.532 | 0.373 | 0.031 | 0.085 | 0.521 | 0.396 | 0.033 | −0.759 | 0.319 | 0.340 |
| mass × endemism | 0.056 | −0.066 | 0.484 | 0.123 | 0.539 | 0.902 | 0.711 | 0.188 | 0.354 | 1.319 | 0.789 | 0.120 |
| Variable | b |
|---|---|
| range | −2.292 |
| mass | 0.546 |
| diet breadth | 1.499 |
| trophic level (omnivore) | −2.131 |
| trophic level (carnivore) | 0.817 |
| endemism | −7.516 |
| range × endemism | 1.196 |
| Variable | b |
|---|---|
| range | −1.875 |
| endemism | −5.463 |
| clade | 1.346 |
| range × endemism | 0.956 |
| range × clade | −0.345 |
| Rank | Family | Species | IUCN Status | Lower Estimate | Median Estimate | Upper Estimate |
|---|---|---|---|---|---|---|
| 1 | Craseonycteridae | Craseonycteris thonglongyai | VU | 0.891 | 0.981 | 1.000 |
| 2 | Pteropodidae | Pteropus mariannus | EN | 0.853 | 0.961 | 1.000 |
| 3 | Pteropodidae | Aproteles bulmerae | CR | 0.566 | 0.833 | 0.953 |
| 4 | Pteropodidae | Eidolon dupreanum | VU | 0.445 | 0.798 | 0.977 |
| 5 | Pteropodidae | Pteropus conspicillatus | LC | 0.539 | 0.795 | 0.950 |
| 6 | Pteropodidae | Pteropus rodricensis | CR | 0.383 | 0.775 | 0.965 |
| 7 | Emballonuridae | Coleura seychellensis | CR | 0.399 | 0.764 | 0.973 |
| 8 | Hipposideridae | Hipposideros halophyllus | EN | 0.523 | 0.760 | 0.922 |
| 9 | Vespertilionidae | Myotis vivesi | VU | 0.453 | 0.690 | 0.860 |
| 10 | Pteropodidae | Pteropus dasymallus | NT | 0.000 | 0.655 | 0.934 |
| Rank | Family | Species | IUCN Status | Lower Estimate | Median Estimate | Upper Estimate |
|---|---|---|---|---|---|---|
| 1 | Vespertilionidae | Eptesicus dimissus | DD | 0.996 | 1.000 | 1.000 |
| 2 | Molossidae | Otomops wroughtoni | DD | 0.992 | 1.000 | 1.000 |
| 3 | Vespertilionidae | Myotis annamiticus | DD | 0.990 | 0.998 | 1.000 |
| 4 | Phyllostomidae | Artibeus incomitatus | CR | 0.963 | 0.996 | 1.000 |
| 5 | Vespertilionidae | Myotis anjouanensis | DD | 0.969 | 0.994 | 1.000 |
| 6 | Pteropodidae | Latidens salimalii | EN | 0.949 | 0.986 | 1.000 |
| 7 | Phyllostomidae | Micronycteris matses | DD | 0.946 | 0.986 | 1.000 |
| 8 | Vespertilionidae | Arielulus cuprosus | DD | 0.914 | 0.981 | 0.998 |
| 9 | Craseonycteridae | Craseonycteris thonglongyai | VU | 0.932 | 0.977 | 0.998 |
| 10 | Pteropodidae | Pteropus voeltzkowi | VU | 0.932 | 0.977 | 0.998 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Welch, J.N.; Beaulieu, J.M. Predicting Extinction Risk for Data Deficient Bats. Diversity 2018, 10, 63. https://doi.org/10.3390/d10030063
Welch JN, Beaulieu JM. Predicting Extinction Risk for Data Deficient Bats. Diversity. 2018; 10(3):63. https://doi.org/10.3390/d10030063
Chicago/Turabian StyleWelch, Jessica Nicole, and Jeremy M. Beaulieu. 2018. "Predicting Extinction Risk for Data Deficient Bats" Diversity 10, no. 3: 63. https://doi.org/10.3390/d10030063
APA StyleWelch, J. N., & Beaulieu, J. M. (2018). Predicting Extinction Risk for Data Deficient Bats. Diversity, 10(3), 63. https://doi.org/10.3390/d10030063





