Abstract
(S)-1-Methyl-2-oxoimidazolidine-4-carboxylic acid 1 is an analog of (S)-pyroglutamic acid, a key component of naturally occurring peptide hormones and synthetic pharmaceutical candidates. The reaction of (S)-2-amino-3-(methylamino)propionic acid with COCl2 and aqueous NaHCO3 followed by ion exchange afforded 1, which was recrystallized from acetonitrile and then characterized by IR, 1H NMR, 13C NMR, polarimetry, elemental microanalysis, high-resolution mass spectrometry and single-crystal X-ray diffraction. The acid 1 crystallized in the orthorhombic chiral space group P212121 with cell constants a = 6.2275(4) Å, b = 8.3963(5) Å, c = 24.9490(14) Å. The X-ray crystal structure revealed that two distinct conformers of 1 occur at alternating positions within helices which are supported by hydrogen bonds. Each molecule of 1 is linked to its two neighbors in the helix by a total of three hydrogen bonds, and four molecules of 1 are contained within each turn of the helix. The pattern of hydrogen bonds illustrates a preference for the carboxylic acid group to act as a hydrogen bond donor and for the urea unit to be a hydrogen bond acceptor.
1. Introduction
The heterocyclic amino acid derivative (S)-1-methyl-2-oxoimidazolidine-4-carboxylic acid 1 (Figure 1) is a structural analogue of naturally occurring (S)-pyroglutamic acid 2, which forms the N-termini of several biologically active peptides, including thyrotropin-releasing hormone (TRH) and luteinizing hormone releasing hormone (LH-RH) [1,2]. 1 is both a precursor to, and a metabolite of, the angiotensin converting enzyme (ACE) inhibitor imidapril 3, which is used for the treatment of hypertension [3,4,5]. Furthermore, the incorporation of 1 into synthetic drug candidates has been reported in recent patents that are related to a range of conditions, including pain [6], cancer [7] and hepatitis C [8].
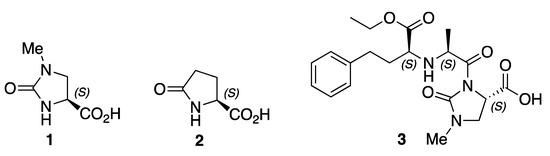
Figure 1.
Structural formulae of (S)-1-Methyl-2-oxoimidazolidine-4-carboxylic acid 1, l-pyroglutamic acid 2, and imidapril 3.
Previously, 1 has been prepared by the deprotection of the corresponding alkyl esters [8], which can be made in several steps from asparagine derivatives, using Hofmann degradation chemistry to modify the amide side chain [3]. Racemic 1 has also been formed in situ via a reaction of rac-2-amino-3-(methylamino)propionic acid hydrochloride 4 with phosgene under alkaline conditions but was further transformed into its methyl ester without being isolated [9]. Only limited characterization data (e.g., 1H NMR and low-resolution mass spectra) are available for 1. In particular, the crystal structure of 1 appears not to have been determined, despite the interesting possibilities for hydrogen bonding and polymorphism that exist in similar compounds, as exemplified by the reversible thermosalience (jumping when placed on a heated surface) of (S)-pyroglutamic acid 2 crystals [10].
Here, we report the preparation, isolation, characterization, and X-ray structure determination of (S)-1 formed in one synthetic step (Scheme 1), starting with (S)-2-amino-3-(methylamino)propionic acid (BMAA) hydrochloride 4, an amino acid salt that is commercially available from Merck and other suppliers of fine chemicals.
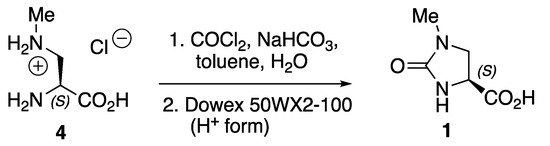
Scheme 1.
Preparation of (S)-1-methyl-2-oxoimidazolidine-4-carboxylic acid 1 from (S)-2-amino-3-(methylamino)propionic acid hydrochloride 4.
2. Results
(S)-1-Methyl-2-oxoimidazolidine-4-carboxylic acid 1 was prepared via the reaction of amino acid 4 with phosgene in the presence of excess sodium hydrogencarbonate (Scheme 1). The cyclized product 1 was water-soluble, so the aqueous solution was passed through a column of Dowex 50WX2-100 strongly acidic ion-exchange resin in the H+ form to convert the sodium salt into the free carboxylic acid species, which was recrystallized from hot acetonitrile after lyophilization to remove water. The acid 1 was characterized by elemental analysis, infrared, 1H NMR (Supplementary Materials, Figure S1) and 13C NMR data (Figure S2) and high-resolution mass spectrometry (Figure S3), which are consistent with the free acid obtained with >99% purity. The melting point and NMR data for acid 1 matched those of a sample that we prepared from N-benzyloxycarbonyl-L-asparagine in four steps according to the method of Lemieux et al. [7]. Crystals of 1 were heated on a hot-stage microscope from an ambient temperature to their melting point of 183 °C; thermosalience was not observed under these conditions, whereas it did occur for (S)-2.
A single-crystal X-ray diffraction study (Figure 2) confirmed the structure of 1, in which all molecules have the same enantiomeric form and occupy the chiral space group P212121; a CIF report for this crystal structure is available in the Supplementary Materials associated with this paper.
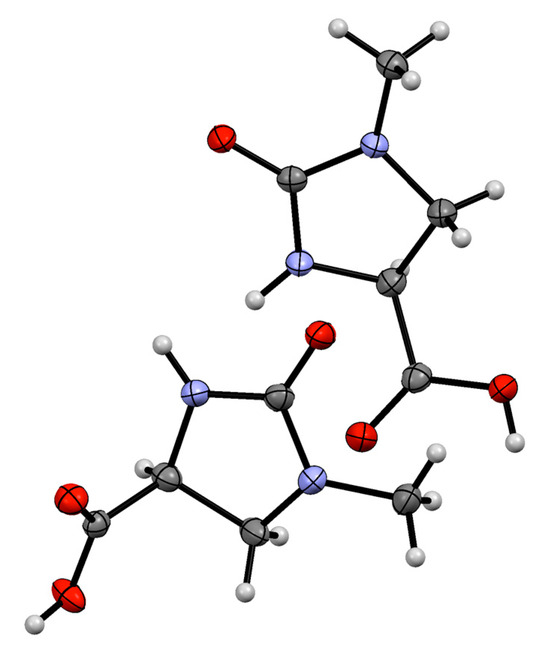
Figure 2.
ORTEP representation of the X-ray crystal structure of the acid 1. Key: C dark grey, H pale grey, O red, N blue. Thermal ellipsoids are shown at the 50% probability level. The unit cell contains two crystallographically independent conformers of (S)-1.
Two crystallographically independent conformers of (S)-1 were present within the unit cell of 1. Both conformers adopt flattened half-chair conformations to accommodate trigonal planar geometry at the urea carbonyl carbon atoms; in one of the conformers, the carboxylic acid substituent is pseudo-equatorial with respect to the heterocyclic ring, whereas in the other conformer, the carboxylic acid is pseudo-axial. These differences are illustrated by the observed Curea–N–C–Ccarboxyl torsion angles of 145.9° and 109.0° in the two conformers. The molecules of 1 each possess two potential hydrogen bond donor groups (urea N-H and carboxylic acid O–H) and two potential hydrogen bond acceptors (urea C=O and carboxylic acid C=O). The crystal structure of 1 contains helical assemblies of 1’s molecules linked by hydrogen bonds in which the two conformers alternate, with each turn of the helix comprising four molecules of 1 (Figure 3). Each molecule of 1 participates in a total of three hydrogen bonds that connect it to two neighbors within a helical chain. Every carboxylic acid O–H forms a hydrogen bond to the urea C=O of its neighbor (O–H···O distances 2.51 and 2.53 Å, angles 167.3° and 165.2°, respectively, depending on the role of each conformer). Additional hydrogen bonds are formed by using the urea N-H of one conformer to create a link with the acid C=O of the other conformer (N–H···O distance = 2.88 Å, angle = 151.6°). Half of the acid C=O groups and half of the urea N-H groups are thus not used for hydrogen bonding, showing a preference for the carboxylic acid groups to act as hydrogen bond donors and for the urea units to act as hydrogen bond acceptors, in accordance with the greater electronegativity of oxygen compared with nitrogen.
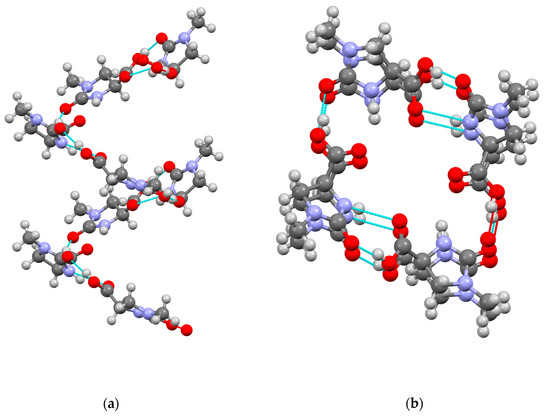
Figure 3.
Ball and stick representations of helical assemblies formed by hydrogen bonding (cyan and red) within the crystal structure of acid 1. Key: C dark grey, H pale grey, O red, N blue. Viewed (a) perpendicular to helix axis and (b) approximately parallel to helix axis.
3. Experimental
3.1. General Experimental Details
Unless otherwise stated, all commercially available solvents and reagents were used without further purification. Melting points were measured using a hot-stage microscope (Reichert). IR spectra were obtained by attenuated total reflection (ATR) using a Perkin-Elmer Spectrum 65 FT-IR spectrometer. NMR spectra were recorded using a Bruker AVIII 400 spectrometer. Elemental microanalysis was performed by Medac Ltd., Chobham, Surrey, UK. Mass spectra were obtained by the EPSRC NMSF, Swansea, UK.
3.2. Synthesis of (S)-1-Methyl-2-oxoimidazolidine-4-carboxylic Acid 1
A stirred solution of 4 (154.6 mg, 1.00 mmoL) in water (10 mL) was cooled in an ice bath and treated with NaHCO3 (840 mg, 10.0 mmoL), followed by 20% (w/w) phosgene in toluene (1.6 mL, 3.5 mmoL). The bath was allowed to attain room temperature; after 22 h, the aqueous phase of the reaction mixture was passed through a column of Dowex 50WX2-100 ion exchange resin (H+ form) and eluted with water. The combined filtrate and washings were lyophilized and recrystallized from hot acetonitrile to present title compound 1 (100.7 mg, 70%) as colorless needles, mp 183–185 °C. Found: C, 41.83; H, 5.61; N, 19.62. C5H8N2O3 requires C, 41.67; H, 5.59; N, 19.43%. [α]24D –9.4 (c 1.02 in H2O); IR νmax/cm–1 (ATR) 3317, 1708, 1626, 1516, 1452, 1242; 1H NMR δH (400 MHz, D2O) 2.63 (3H, s, NMe), 3.50 (1 H, dd, J = 9.7, 5.2 Hz, H-5), 3.72 (1 H, apparent t, J = 10.0 Hz, H-5), 4.27 (1 H, dd, J = 10.3, 5.2 Hz, H-4); 13C NMR δC (100.6 MHz, D2O) 29.5 (NMe), 49.9 (C-5), 51.2 (C-4), 163.5 (C-2), 175.6 (CO2H); high-resolution mass spectrum m/z (ESI–) found: 143.0458; C5H7N2O3– ([M–H]–) requires 143.0462.
3.3. X-ray Structure Determination of 1
Single-crystal X-ray diffraction was carried out at the Queen Mary University of London using the KAPPA APEX ii DUO diffractometer (Bruker UK Ltd., Coventry, UK), with MoKα radiation (λ = 0.71073 Å). X-ray crystal structures were solved and refined using the Bruker SHELXTL version 2018/2 software package.
A translucent colorless needle-like specimen of C5H8N2O3, approximate dimensions 0.080 mm × 0.090 mm × 0.300 mm, was used for the X-ray crystallographic analysis. The X-ray intensity data were measured. A total of 5961 frames were collected. The total exposure time was 16.56 h. The frames were integrated with the Bruker SAINT V8.18C software package using a narrow-frame algorithm. The integration of the data using an orthorhombic unit cell yielded a total of 9965 reflections to a maximum θ angle of 63.73° (0.86 Å resolution), of which 2073 were independent (average redundancy 4.807, completeness = 99.5%, Rint = 2.70%, Rsig = 1.89%) and 2055 (99.13%) were greater than 2σ(F2). The final cell constants of a = 6.2275(4) Å, b = 8.3963(5) Å, c = 24.9490(14) Å, and volume = 1304.53(14) Å3 are based upon the refinement of the XYZ-centroids of 9096 reflections above 20 σ(I) with 7.086° < 2θ < 127.4°. Data were corrected for absorption effects using the multi-scan method (SADABS). The ratio of minimum to maximum apparent transmission was 0.884. The calculated minimum and maximum transmission coefficients (based on crystal size) were 0.7430 and 0.9210, respectively.
The final anisotropic full-matrix least-squares refinement on F2 with 185 variables converged at R1 = 2.44%, for the observed data and wR2 = 6.58% for all data. The goodness of fit was 1.087. The largest peak in the final difference electron density synthesis was 0.156 e−/Å3, and the largest hole was −0.186 e−/Å3, with an RMS deviation of 0.037 e−/Å3. On the basis of the final model, the calculated density was 1.468 g/cm3 and F(000), 608 e−.
Supplementary Materials
CIF report for the crystal structure of 1, Figure S1: 1H NMR spectrum of 1 (400 MHz, D2O); Figure S2: 13C NMR spectrum of 1 (100.6 MHz, D2O); Figure S3: ESI mass spectrum of 1.
Author Contributions
Conceptualization, P.B.W.; methodology, A.L.D., P.B.W., M.M. and I.A.; investigation, A.L.D., P.B.W., M.M. and I.A.; writing—original draft preparation, P.B.W.; writing—review and editing, A.L.D., P.B.W. and I.A.; visualization, P.B.W., M.M. and I.A.; supervision, P.B.W.; project administration, P.B.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Crystallographic data (excluding structure factors) for the acid 1 are available in the Supplementary Materials of this paper and were deposited at the Cambridge Crystallographic Data Centre as supplementary publication no. CCDC 1549794. Copies of the data can be obtained, free of charge, upon application to CCDC, 12 Union Road, Cambridge CB2 1EZ, UK (fax: +44-(0)1223-336033 or e-mail: deposit@ccdc.cam.ac.uk).
Acknowledgments
We are grateful to the EPSRC NMSF at Swansea University for mass spectrometry and to Paul Cox, Brain Chemistry Laboratories, Jackson, Wyoming for a gift of amino acid 4.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bøler, J.; Enzmann, F.; Folkers, K.; Bowers, C.Y.; Schally, A.V. The identity of chemical and hormonal properties of the thyrotropin releasing hormone and pyroglutamyl-histidyl-proline amide. Biochem. Biophys. Res. Commun. 1969, 37, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Seeburg, P.H.; Adelman, J.P. Characterization of cDNA for precursor of human luteinizing hormone releasing hormone. Nature 1984, 311, 666–668. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Nunami, K.; Kato, J.; Yoneda, N.; Kubo, M.; Ochiai, T.; Ishida, R. Studies of Angiotensin Converting Enzyme Inhibitors. 4. Synthesis and Angiotensin Converting Inhibitory Activities of 3-Acyl-1-alkyl-2-oxoimidazolidine-4-carboxylic Acid Derivatives. J. Med. Chem. 1989, 32, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, M.; Horimoto, S.; Mabuchi, M.; Banno, K. Determination of three metabolites of a new angiotensin-converting enzyme inhibitor, imidapril, in plasma and urine by gas chromatography-mass spectrometry using multiple ion detection. J. Chromatograph. 1992, 581, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.M.; Curran, M.P.; Lyseng-Williamson, K.A. Imidapril A Review of its Use in Essential Hypertension, Type 1 Diabetic Nephropathy and Chronic Heart Failure. Drugs 2007, 67, 1359–1378. [Google Scholar] [CrossRef] [PubMed]
- Beswick, P.J.; Dean, D.K.; Gleave, R.J.; Moses, A.P. Walter DS. Imidazolidine Carboxamide Derivatives as P2X7 Modulators. World Patent WO2008119825A2, 9 October 2008. [Google Scholar]
- Lemieux, R.M.; Popovici-Muller, J.; Travins, J.M.; Cai, Z.; Cui, D.; Zhou, D. Therapeutically Active Compounds and Their Methods of Use. World Patent WO2015010297A1, 29 January 2015. [Google Scholar]
- Gai, Y.; Niu, D.; Or, Y.S.; Wang, Z. Cyclic P3 Tripeptide Hepatitis C Serine Protease Inhibitors. US Patent 2008/0267916 A1, 30 October 2008. [Google Scholar]
- Saijo, S.; Wada, M.; Himizu, J.-I.; Ishida, A. Heterocyclic Prostaglandins. VI. Synthesis of 11-Deoxy-8,10-diazaprostaglandin E1 and its 10-Methyl Derivative. Chem. Pharm. Bull. 1980, 28, 1459–1467. [Google Scholar] [CrossRef]
- Panda, M.K.; Runčevski Husain, A.; Dinnebier, R.E.; Naumov, P. Perpetually Self-Propelling Chiral Single Crystals. J. Am. Chem. Soc. 2015, 137, 1895–1902. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).