Abstract
We herein report the synthesis of a derivative of the natural compound celastrol linked to the antimalarial drug primaquine through an amide obtained by the activation of the carboxylic acid with HOBt/EDC. The chemical structure of the new molecule was fully characterized by proton nuclear magnetic resonance (1H-NMR), carbon-13 nuclear magnetic resonance (13C-NMR), heteronuclear single quantum coherence (HSQC), correlation spectroscopy (1H-1H-COSY), distortionless enhancement by polarization transfer (DEPT), mass spectrometry, Fourier-transform infrared (FTIR), and ultraviolet (UV) spectroscopies. Computational studies were enrolled to predict the interaction of the synthesized compound with sarco-endoplasmic reticulum (SR) Ca2+ transport ATPase (SERCA), a target of relevance for developing new therapeutics against arthritis. The drug-likeness of the compound was also investigated by predicting its pharmacokinetic properties.
1. Introduction
Celastrol (1) is a naturally occurring compound that has gained significant attention in recent years for its potential therapeutic properties. Derived from the roots of Tripterygium wilfordii, a traditional Chinese medicinal plant, 1 has been used for centuries in traditional Chinese medicine to treat various diseases. However, it is only in the past few decades that scientists have begun to unravel its remarkable pharmacological effects and the involved molecular mechanisms.
In fact, it exhibits a wide range of beneficial properties, including anti-inflammatory [1], antioxidant [2], cardioprotective [3], anti-osteoarthritis, and anticancer activity [4], and it has been shown to combat neuronal degeneration [5] and obesity [6]. Thus, these properties have sparked intense research efforts to explore the therapeutic potential of celastrol in conditions such as cancer, neurodegenerative diseases, obesity, and metabolic disorders.
Research has shown that celastrol can inhibit the growth and proliferation of various cancer cells by targeting multiple signaling pathways involved in tumor development and progression. Indeed, one of the most intriguing aspects of 1 is its ability to modulate cellular signaling pathways that play a critical role in disease progression. More in-detail, studies have shown that celastrol can inhibit the activation of nuclear factor-kappa B (NF-κB) [7], Hsp90-Cdc37 [8] and sarco-endoplasmic reticulum (SR) Ca2+ transport ATPase (SERCA) [9].
SERCA is the only pump that moves calcium ions into the SR from the cytoplasm. It has been demonstrated that SERCA inhibitors cause cytosolic and mitochondrial Ca2+ buildup with ATP depletion, which can activate many signaling pathways leading to the death of cancer cells. Moreover, SERCA is a target of interest for the treatment of arthritis [9,10].
In addition to its antiproliferative and anti-inflammatory properties, 1 has also demonstrated potent antioxidant effects. The antioxidant activity of 1 helps by neutralizing harmful free radicals and protecting cells from oxidative damage, thereby contributing to its therapeutic potential. Although celastrol shows promising potential, further research is needed to fully understand its mechanisms of action and side effects. Additionally, the development of efficient delivery systems and formulation strategies to improve its bioavailability remains an active area of investigation.
This study describes the synthesis and computational studies of a celastrol derivative in which the scaffold has been linked to primaquine (3, Figure 1). Primaquine (3) is an important antimalarial drug which causes hemolytic anemia in patients with glucose-6-phosphate dehydrogenase (G6PDH) deficiency, probably due to the generation of oxidant species by its metabolites [11]. Bearing in mind that quinolines and some antimalarials were previously shown to target SERCA [12,13], the aim of this study was to design and synthesize a chimeric celastrol–primaquine derivative and to study its interaction with this the macromolecular target.
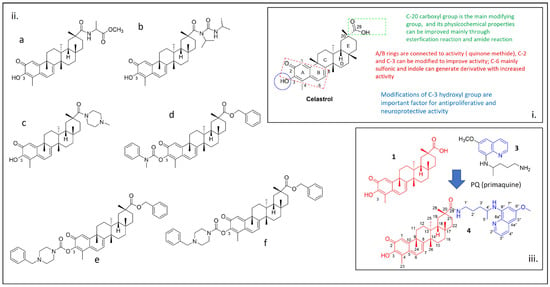
Figure 1.
(i) Functional groups of celastrol (1) that are important for its biological activity; (ii) example of celastrol derivatives reported in the literature (a) [14], (b) [15], (c) [5] and (d–f) [16]; (iii) chemical structures of primaquine (3) and compound 4 are reported in the inset of the figure.
2. Results and Discussion
The synthetic route for (2R,4aS,6aS,12bR,14aS,14bR)10-hydroxy-N-(4-((6-methoxyquinolin-8-yl)amino)pentyl)-2,4a,6a,9,12b,14a-hexamethyl-11-oxo-1,2,3,4,4a,5,6,6a,11,12b,13,14,14a,14b-tetradecahydropicene-2-carboxamide started from the preparation of the precursor primaquine (3, Scheme 1a).
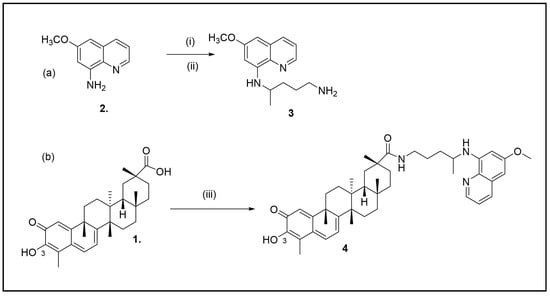
Scheme 1.
(a) Synthesis of N-4-(6-Methoxyquinolin-8-yl)Pentane-1,4-Diamine (i) EtOH, N2; (ii) phthalimidobromopentane (b) Synthesis of (2R,4aS,6aS,12bR,14aS,14bR)10-hydroxy-N-(4-((6-methoxyquinolin-8-yl)amino)pentyl)-2,4a,6a,9,12b,14a-hexamethyl-11-oxo- 1,2,3,4,4a,5,6,6a,11,12b,13,14,14a,14b-tetradecahydropicene-2-carboxamide 4:; (iii) EDC hydrochloride (1.5 equiv.), HOBt (1.5 equiv.), CH2Cl2.
Following the protocol reported in the literature [17], primaquine 3 was prepared by the reaction of 8-amino-6-methoxyquinoline (1.74 g, 0.01 mol) 2 and phthalimidobromopentane (1.46 g, 0.005 mol) refluxed in EtOH (20 mL) for 48 h (Scheme 1a).
The crude product was crystallized from MeOH (70 mL) at 10 °C in 24 h to afford 3 in 83% yield.
Primaquine (3) was used as a starting compound by a coupling reaction using HOBt/EDC/TEA as the activation conditions using a modified protocol reported by Pang et al. [14].
To a CH2Cl2 solution of celastrol, HOBt and EDC hydrochloride were added together with 3 equivalents of primaquine (Scheme 1b). After stirring at 65◦C for 24 h, compound 4 was isolated by column chromatography in a high yield (70%).
The chemical structure of compound 4 was confirmed by NMR, IR, UV, and mass spectrometry analysis.
The 1H-NMR spectrum showed a singlet at 3.88 ppm associated with the C6″- methoxyl group of the quinoline moiety (Figure S3), thus supporting the formation of the expected product. Regarding the 13C-NMR signals, the disappearance of peculiar peaks of carboxylic acid at 182 ppm, and the appearance of the peak at 179 ppm of the amide and many signals in the aromatic region were probably the most relevant features to verify the reaction with the primaquine moiety (Figure S4).
Heteronuclear single quantum coherence spectroscopy (HSQC), correlation spectroscopy (1H-1H-COSY), and distortionless enhancement by polarization transfer (DEPT-135, DEPT-90 and DEPT-45) were also used to assign 13C signals of compound 4, as shown in Table S1 (see Supplementary Materials for 2D spectra, Figures S5–S9). The 13C-NMR spectrum of 4 exhibited 44 carbon signals, classified by DEPT experiments as seven methyl groups, ten methylene, five methines, one methoxy, five aromatic, and sixteen quaternary carbons.
The IR spectrum of compound 4 showed characteristic N–H stretching at 3448 cm−1 and C=O stretching at 1577 cm−1 (Supplementary Materials, Figure S10b). Other vibrational peaks at 1411 and 1337 cm−1 were attributed to the N–H bending in the amide.
The UV spectrum of 4 was also recorded for further characterization (Supplementary Materials, Figure S11), showing an absorption peak at 237 nm and another lower absorption peak at 420 nm (n→π transition) (Supplementary Materials, Figure S11).
In order to investigate whether compound 4 could be a good candidate as a SERCA inhibitor, we first performed molecular modeling studies to assess its binding mode with the protein (Figure 2).
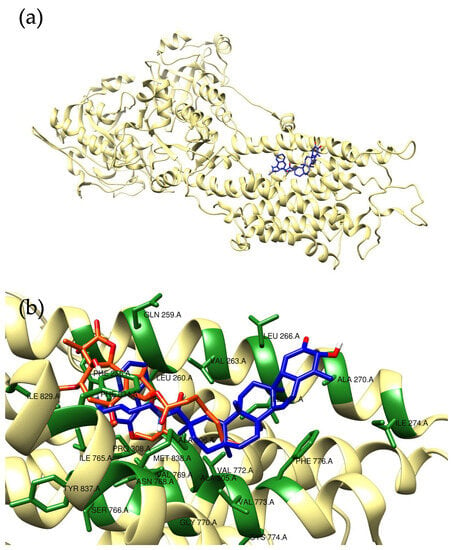
Figure 2.
Predicted binding motif for compound 4 (blue) with SERCA (a). A detailed view of the interaction pattern, in comparison to that of thapsigargin (orange), is reported in panel (b). The interacting residues of the protein have been highlighted in green and labeled.
In particular, molecular docking was used to preliminarily investigate the interaction motif of the molecule with this target. The crystal structure of SERCA (PDB ID 1IWO) was used to perform site-specific docking. For compound 4, a calculated binding energy value of −10.4 kcal/mol was computed (Figure 2a), which outperforms the one obtained for the thapsigargin, the molecule used as positive control (−8.4 kcal/mol). Compound 4 fitted within the identified binding pocket only partially sharing a similar interaction motif with respect to thapsigargin (Figure 2b). In fact, it must be noted that the two compounds show different chemical features and scaffolds, but a co-localization of hydrophobic parts of the molecules can be observed in the predicted poses. The residues within interaction distance (<5 Å) have been labelled in Figure 2b; as can be noted from the artwork, the binding pocked is indeed mainly constituted by hydrophobic residues (Ile, Leu, Val, Phe).
In view of potential pharmacological applications, compound 4 was also evaluated in terms of predicted physicochemical properties and oral bioavailability by using ligand-based predictive modeling [18]. The results of these calculations (see Supplementary Materials, Table S2) indicated that compound 4 violates two of Lipinski’s rules, suggesting that further optimization would be needed to fully satisfy drug-likeness criteria. Thus, even if the compound showed promising calculated binding energy values towards SERCA, it may be characterized by poor bioavailability. As a consequence, parameters such as molecular weight and LogP should be improved before further studies.
3. Materials and Methods
3.1. Chemistry
Silica gel (FCP 230–400 mesh) was used for column chromatography. Thin-layer chromatography was carried out on E. Merck precoated silica gel 60 F254 plates and visualized with phosphomolybdic acid, iodine, or a UV-visible lamp.
All chemicals were purchased from Bide Pharmatech., Ltd. (Shanghai, China) and J & K scientific (Hong Kong, China). 1H-NMR and 13C-NMR spectra were collected in CDCl3 and DMSO at 25 °C on a Bruker Ascend®−600 NMR spectrometer (600 MHz for 1H and 150 MHz for 13C). All chemical shifts were reported in the standard δ notation of parts per million using the peak of residual proton signals of CDCl3 or DMSO-d6 as an internal reference (CDCl3, δC 77.2 ppm, δH 7.26 ppm; DMSO-d6, δC 39.5 ppm, δH 2.50 ppm). High resolution mass spectra (HRMS) were measured using electrospray ionization (ESI). The measurements were performed in a positive ion mode (interface capillary voltage 4500 V); the mass-to-charge ratio was set from m/z 50 to 3000 Da; external/internal calibration was carried out with electrospray calibration solution. HRMS analyses were performed by an Agilent 6230 ESI time-of-flight (TOF) mass spectrometer with Agilent C18 column (4.6 mm × 150 mm, 3.5 μm). The mobile phase was isocratic (water + 0.01% TFA; CH3CN) at a flow rate of 0.35 mL/min. The peaks were determined at 254 nm by using a UV detector.
UV analysis was performed on a Shimadzu UV—2600 with 1 cm quartz cell and a slit width of 2.0 nm. The analysis was carried out using a wavelength in the range of 200–500 nm.
IR analysis (KBr) was performed on a Shimadzu IRAffinity-1S with a frequency range of 4000–500 cm−1.
3.1.1. Synthesis of N-4-(6-Methoxyquinolin-8-yl)pentane-1,4-diamine (3)
A mixture of 8-amino-6-methoxyquinoline (1.74 g, 0.01 mol) and phthalimidobromopentane (1.46 g, 0.005 mol) was refluxed for 48 h in EtOH (20 mL). The reaction mixture was concentrated under reduced pressure and the residue was extracted with EtOAc (3 × 30 mL). The combined EtOAc extract was washed with H2O (2 × 20 mL), dried over Na2SO4 and concentrated under reduced pressure. The residue was extracted with hot hexane and this extract was concentrated to give 6-methoxy-8-(1-methyl-4-phthalimidobutylamino) quinolone which was crystallized from MeOH (70 mL) at 10 °C in 24 h. Yield 83%. ESI-MS m/z 260.17 [M + H]+ (calcd. for C15H22N3O+, m/z 260.17).
The spectral characteristics are consistent with those of 3 in the literature [17].
3.1.2. Synthesis of (2R,4aS,6aS,12bR,14aS,14bR)10-Hydroxy-N-(4-((6-methoxyquinolin-8-yl)amino)pentyl)-2,4a,6a,9,12b,14a-hexamethyl-11-oxo-1,2,3,4,4a,5,6,6a,11,12b,13,14,14a,14b-tetradecahydropicene-2-carboxamide (4)
To a CH2Cl2 (6 mL) solution of celastrol (45 mg, 0.10 mmol), HOBT (41 mg, 0.30 mmol), and EDC-HCl (57 mg, 0.30 mmol) were added under the conditions of an ice/salt-bath (5 °C). Then, Et3N (70 µL) was added after 20 min, and the solution was stirred at 5 °C for 1 h. Then, 3 was added, the mixture was stirred at room temperature for 12–18 h. The mixture was washed thrice with water and the organic phase was collected, dried over anhydrous Na2SO4, filtered, and concentrated in vacuo to give the crude product. The purification was implemented by chromatography on a silica gel column (eluents: from CH2Cl2 to 5% MeOH). Yield 70%, δH (600 MHz, CDCl3) 0.60 (3H, s, CH3), 0.91–0.98 (1H, m, H-22), 1.09 (3H, s, CH3), 1.10 (3H, s, CH3), 1.23 (3H, s, CH3), 1.28 (3H, s, CH3), 1.42 (3H, s, CH3)), 1.51–1.62 (7H, m), 1.76–2.00 (6H, m), 2.07–2.11 (1H, m), 2.20 (3H, s, CH3), 2.40 (1H, t, H–19), 3.14 (2H, q), 3.60 (1H, m, H-4′), 3.88 (3H, s, OCH3), 5.70 (1H, m, NH), 5.99 (1H, m, NH), 6.25–6.27 (2H, m, H-7″ and H-7), 6.34 (1H, t, H-5″), 6.51 (1H, dd, H-1), 6.97–7.00 (2H, m, H-6), 7.30 (1H, m, H-3″), 7.92 (1H, m, H-4″), 8.53 (1H, m, H-2″) ppm; δC (150 MHz, CDCl3) 10.9 (CH3), 18.9 (CH3), 21.2 (CH3), 22.4 (CH3), 26.50, 26.57, 29.3, 30.0, 31.4, 31.6, 32.2, 34.1, 34.4, 34.4, 34.5, 35.6, 37.0, 38.7, 40.0, 40.2, 40.9, 43.6, 45.0, 45.7, 48.4, 55.8, 92.4, 97.5, 117.6, 118.6, 120.2, 122.5, 128.0, 130.6, 134.6, 135.5, 136.0, 145.0, 145.5, 146.6, 160.1, 165.4, 170.8, 178.3, 179.0 ppm;); ESI-MS m/z 714.44 [M + Na]+ (calcd. for C44H57N3O4Na+, m/z 714.42); UV (CH2Cl2) peaks at 237, 273, and 420 nm; IR (KBr) at 3448, 3136, 2962, 2924, 1577, 1411, and 1337 cm−1.
3.2. Computational Studies
The 3D X-ray crystal structure of SR Ca2+-ATPase was retrieved from the RCSB Protein Data Bank (www.rcsb.org, accessed on 12 June 2023, PDB ID 1IWO, resolution 3.10 Å) [19].
Prior to site-specific docking studies, the protein was prepared and selected chains were isolated and considered. Ligands were built and optimized using Avogadro [20]. The site-specific molecular docking study was performed using AutoDock Vina [21,22]. The search grid was set according to the following parameters: x = −7.8959, y = −28.092, z = 12.276; size: 30.000 × 30.000 × 30.000 Å. Docking simulation was carried out with default Vina parameters, the number of generated docking poses was set to 8 and the docking energy conformation value was expressed in -kcal/mol. The best scoring pose was considered for further analyses. The UCSF Chimera molecular viewer was used to produce the artworks [23].
4. Conclusions
The preparation of a celastrol derivative was presented in this work. The synthesized compound was characterized by using NMR, UV, and IR spectrometry. The drug-likeness of the compound was also determined by computational studies. Although the predicted physico-chemical properties suggest that the compound is not drug-like, the binding energy of the compound against the SERCA target outperformed that of the positive control. Thus, compound 4 represents a promising starting point for further optimization.
Supplementary Materials
The following supporting information are available online, Figure S1: 1H NMR compound 3, Figure S2: 13C NMR compound3 2, Figure S3: 1H NMR compound 4, Figure S4: 13C NMR compound 4, Figure S5a,b: HSQC NMR compound 4, Figure S6a,b: 1H-COSY NMR compound 4, Figures S7–S9: DEPT spectrum of 4, Figure S10: IR spectrum compound 1 and 4, Figure S11: UV spectrum of 4; Figure S12: mass spectra of 4; Table S1: 1H and 13C-nuclear magnetic spectroscopy (NMR) chemical shifts 4, Table S2: physicochemical properties of 4 calculated by SwissADME.
Author Contributions
Conceptualization, P.C.; methodology, Y.X., H.K. and Q.W.; software, H.K. and G.R.; validation, P.C.; formal analysis, Y.X.; investigation, Y.X.; data curation, Y.X.; writing—original draft preparation, Y.X. and H.K.; writing—review and editing, A.G., G.R. and P.C.; supervision, P.C.; project administration, P.C.; funding acquisition, P.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Faculty Grants from the Macao University of Science and Technology to PC (project code: FRG-22-077-SP).
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kannaiyan, R.; Shanmugam, M.K.; Sethi, G. Molecular targets of celastrol derived from Thunder of God Vine: Potential role in the treatment of inflammatory disorders and cancer. Cancer Lett. 2011, 303, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Allison, A.C.; Cacabelos, R.; Lombardi, V.R.M.; Álvarez, X.A.; Vigo, C. Central Nervous System Effects of Celastrol, a Potent Antioxidant and Antiinflammatory Agent. CNS Drug Rev. 2000, 6, 45–62. [Google Scholar] [CrossRef]
- Der Sarkissian, S.; Cailhier, J.; Borie, M.; Mansour, S.; Hamet, P.; Stevens, L.; Noiseux, N. Celastrol as a Novel Cardioprotective Drug. Can. J. Cardiol. 2013, 29, S331. [Google Scholar] [CrossRef]
- Jang, S.Y.; Jang, S.-W.; Ko, J. Celastrol inhibits the growth of estrogen positive human breast cancer cells through modulation of estrogen receptor α. Cancer Lett. 2011, 300, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Xu, L.; Yu, P.; Jiang, J.; Zhang, G.; Wang, Y. Synthesis and preliminary evaluation of neuroprotection of celastrol analogues in PC12 cells. Bioorg. Med. Chem. Lett. 2010, 20, 3844–3847. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lee, J.; Salazar Hernandez, M.A.; Mazitschek, R.; Ozcan, U. Treatment of Obesity with Celastrol. Cell 2015, 161, 999–1011. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.Z.; Hwang, B.Y.; Kim, H.S.; Lee, J.H.; Kim, Y.H.; Lee, J.J. Antiinflammatory Constituents of Celastrus orbiculatus Inhibit the NF-κB Activation and NO Production. J. Nat. Prod. 2002, 65, 89–91. [Google Scholar] [CrossRef]
- Zhang, T.; Li, Y.; Yu, Y.; Zou, P.; Jiang, Y.; Sun, D. Characterization of Celastrol to Inhibit Hsp90 and Cdc37 Interaction*. J. Biol. Chem. 2009, 284, 35381–35389. [Google Scholar] [CrossRef]
- Wong, V.K.W.; Qiu, C.; Xu, S.-W.; Law, B.Y.K.; Zeng, W.; Wang, H.; Michelangeli, F.; Dias, I.R.D.S.R.; Qu, Y.Q.; Chan, T.W.; et al. Ca2+ signalling plays a role in celastrol-mediated suppression of synovial fibroblasts of rheumatoid arthritis patients and experimental arthritis in rats. Br. J. Pharmacol. 2019, 176, 2922–2944. [Google Scholar] [CrossRef]
- Xu, S.-W.; Law, B.Y.K.; Qu, S.L.Q.; Hamdoun, S.; Chen, J.; Zhang, W.; Guo, J.-R.; Wu, A.-G.; Mok, S.W.F.; Zhang, D.W.; et al. SERCA and P-glycoprotein inhibition and ATP depletion are necessary for celastrol-induced autophagic cell death and collateral sensitivity in multidrug-resistant tumor cells. Pharmacol. Res. 2020, 153, 104660. [Google Scholar] [CrossRef]
- Hong, Y.L.; Pan, H.Z.; Scott, M.D.; Meshnick, S.R. Activated oxygen generation by a primaquine metabolite: Inhibition by antioxidants derived from Chinese herbal remedies. Free Radic. Biol. Med. 1992, 12, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Eckstein-Ludwig, U.; Webb, R.J.; Van Goethem, I.D.; East, J.M.; Lee, A.G.; Kimura, M.; O’Neill, P.M.; Bray, P.G.; Ward, S.A.; Krishna, S. Artemisinins target the SERCA of Plasmodium falciparum. Nature 2003, 424, 957–961. [Google Scholar] [CrossRef] [PubMed]
- Britzolaki, A.; Cronin, C.C.; Flaherty, P.R.; Rufo, R.L.; Pitychoutis, P.M. Chronic but not acute pharmacological activation of SERCA induces behavioral and neurochemical effects in male and female mice. Behav. Brain Res. 2021, 399, 112984. [Google Scholar] [CrossRef] [PubMed]
- Pang, C.; Luo, J.; Liu, C.; Wu, X.; Wang, D. Synthesis and Biological Evaluation of a Series of Novel Celastrol Derivatives with Amino Acid Chain. Chem. Biodivers. 2018, 15, e1800059. [Google Scholar] [CrossRef] [PubMed]
- Klaić, L.; Morimoto, R.I.; Silverman, R.B. Celastrol Analogues as Inducers of the Heat Shock Response. Design and Synthesis of Affinity Probes for the Identification of Protein Targets. ACS Chem. Biol. 2012, 7, 928–937. [Google Scholar] [CrossRef]
- Shan, W.-G.; Wang, H.-G.; Chen, Y.; Wu, R.; Wen, Y.-T.; Zhang, L.-W.; Ying, Y.-M.; Wang, J.-W.; Zhan, Z.-J. Synthesis of 3- and 29-substituted celastrol derivatives and structure-activity relationship studies of their cytotoxic activities. Bioorg. Med. Chem. Lett. 2017, 27, 3450–3453. [Google Scholar] [CrossRef]
- Azad, C.S.; Saxena, M.; Siddiqui, A.J.; Bhardwaj, J.; Puri, S.K.; Dutta, G.P.; Anand, N.; Saxena, A.K. Synthesis of primaquine glyco-conjugates as potential tissue schizontocidal antimalarial agents. Chem. Biol. Drug Des. 2017, 90, 254–261. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Toyoshima, C.; Nomura, H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature 2002, 418, 605–611. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).