Abstract
Naturally occurring nucleoside-peptide antibiotics such as muraymycins or caprazamycins are of major interest for the development of novel antibacterial agents. However, the synthesis of new analogues of these natural products for structure–activity relationship (SAR) studies is challenging. In our synthetic efforts towards a muraymycin-derived nucleoside building block suitable for attachment to a solid support, we came across an interesting side product. This compound resulted from an undesired Fmoc deprotection with subsequent cyclization, thus furnishing a remarkable caprazamycin-like seven-membered diazepanone ring.
1. Introduction
Emerging bacterial strains that are resistant to antibiotics are a severe threat in healthcare [1,2,3]. To address this problem, novel antibacterial agents with previously unexploited modes of action are urgently needed [4]. A promising class that meets this requirement are uridine-derived nucleoside-peptide antibiotics that interfere with bacterial cell wall biosynthesis [5,6,7]. They inhibit MraY, an enzyme that catalyzes a membrane-associated step in the intracellular part of peptidoglycan formation [8,9,10,11,12,13]. Among this structurally diverse class, our research focusses mainly on the subclasses of muraymycins [14,15] and caprazamycins [16,17,18,19,20,21,22] (Figure 1). For muraymycins, a co-crystal structure in complex with MraY has been reported, enabling deeper insights into the inhibitor-target interaction [23,24,25]. However, experimental structure–activity relationship (SAR) studies are still crucial due to the pronounced conformational plasticity of MraY, and have therefore been carried out extensively as exemplified by the according work on muraymycins [26,27,28,29,30,31,32,33,34,35].
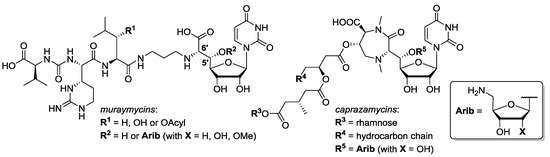
Figure 1.
Structures of the nucleoside-peptide antibiotics muraymycins and caprazamycins.
In contrast to previously reported solution-phase syntheses of muraymycin derivatives [28,30,31], we have recently described a solid phase-supported approach for the preparation of structurally simplified muraymycin analogues [36]. This novel strategy was based on the stepwise construction of the muraymycin peptide unit on the solid support. As part of our initial studies on this approach, we had also investigated a potential attachment of the muraymycin nucleoside unit (or an analogue thereof) to the solid phase. Hence, we had envisioned to prepare uridine-derived building block 5 (see Scheme 2) that could then be connected to the solid phase. The design of 5 was based on our previously reported strategy to simplify the muraymycin core structure by omission of the 5′-substituent (“5′-deoxy” analogues) [30,33,35,36,37,38]. Carboxylic acid 5 was intended to be attached to a trityl resin by formation of a trityl ester. In order to then apply standard solid-phase protocols for the assembly of the peptide unit, we chose Fmoc as protecting group for the terminal amino group of the linker moiety. For the preparation of 5, we intended to selectively cleave the tert-butyl ester in the 7′-position of precursor 4 (Scheme 1 and Scheme 2). Studies on this particular transformation have furnished the unexpected side reaction reported in this work.
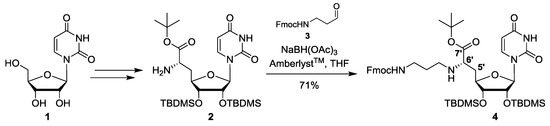
Scheme 1.
Synthesis of protected uridine-derived precursor building block 4.
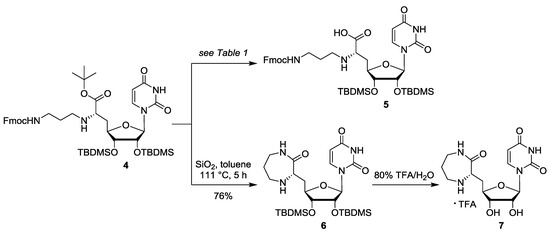
Scheme 2.
Attempted deprotection of 4 towards 5, yielding diazepanone 6 as major product instead.
2. Results
The synthesis of the fully protected uridine-derived precursor building block 4 involved protocols that were previously reported by our group. Thus, uridine 1 was transformed into the protected nucleosyl amino acid 2 using an established five-step route (Scheme 1) [30,37,38]. This was followed by reductive amination of 2 with Fmoc-protected aldehyde 3 [36]. The reductive amination step was carried out in a similar way as with other Cbz-protected aldehydes as previously reported [30] (i.e., using amberlyst as acidic activator and sodium triacetoxyborohydride as reducing agent). The resultant product 4 was obtained in 71% yield (Scheme 1).
For the envisioned selective deprotection of the 7′-carboxylate of 4, we needed a method that would cleave the tert-butyl ester but leave the tert-butyldimethylsilyl (TBDMS) groups intact. We therefore studied the mild acidic cleavage of the tert-butyl ester of 4—as previously reported by us for similar transformations [39]—in order to potentially obtain 5 (Scheme 2). In first attempts, the reaction was carried out by stirring a solution of 4 in toluene in the presence of silica at reflux temperature (i.e., 111 °C (Table 1, entries 1–3)). The conversion of 4 was monitored by thin layer chromatography (TLC). Surprisingly, a seven-membered diazepanone ring was formed to give main product 6 (without the Fmoc protecting group) in a yield of 41% (Table 1, entry 1). This was unexpected, as the Fmoc group usually requires basic conditions for its cleavage. We speculated that, under the applied reaction conditions (high temperature, extended reaction period of 5 days), the secondary amine at the 6′-position of 4 might have been sufficiently basic to mediate Fmoc deprotection. The resultant primary amine then probably attacked the 7′-tert-butyl ester of 4, thus leading to ring formation (proposed mechanism not shown). By significantly decreasing the reaction period, the yield of side product 6 even increased to 68% (after 1.5 days, Table 1, entry 2) and 76% (after 5 h, entry 3), respectively. This demonstrated that 6 was indeed the main product of this transformation at elevated temperatures. Variation of the temperature then revealed that the desired product 5 might have been formed at lower temperatures, but only in small amounts. Target compound 5 was obtained when the reaction mixture was stirred at 80 °C for 1 day and then at room temperature (rt) for further 2 days, furnishing a moderate yield of 48% of 5 (Table 1, entry 4). Further attempts to repeat this reaction towards 5 and possibly improve its yield only gave worse results (i.e., lower yields with significant amounts of unidentified impurities). We also explored other conditions to selectively cleave the tert-butyl ester, mainly the treatment of 4 with TFA (Table 1, entries 5–7) or with pig liver esterase (PLE, entry 8), but none of these experiments afforded satisfying conversions to the desired carboxylic acid 5.

Table 1.
Investigated conditions for the attempted reaction of 4 to 5 (see Scheme 2).
Since no reaction conditions were identified that gave the desired uridine-derived building block 5 in robust, reproducible and satisfying yields, this strategy for the solid phase-supported synthesis of muraymycin analogues was discarded, and our reported route (vide supra) was established instead [36]. The obtained diazepanone side product 6 represented an interesting structure as it strongly resembles the core structure (“caprazol”) of caprazamycin antibiotics (cf. Figure 1). We therefore intended to evaluate a deprotected derivative of 6 for its biological activity. Uridine-derived diazepanone 6 was globally deprotected using 80% aqueous TFA for acidic desilylation (Scheme 2). The resultant fully deprotected caprazol analogue 7 was purified by semipreparative HPLC. In the 1H NMR spectrum of the obtained product, two sets of signals for the H-5 and H-6 protons of the nucleobase were observed, hinting at a uracil-derived impurity that could not be separated from target compound 7 and that could not be identified. However, the amount of this impurity could be estimated from the 1H NMR to be ~30% based on the uracil-associated signals. We therefore decided to test this mixture of 7 and the unidentified impurity for its potential to inhibit the bacterial target protein MraY, using an established in vitro assay for MraY activity [32,33,40]. It was found that the IC50 value of the mixture containing caprazol analogue 7 was outside of the relevant range (IC50 > 50 μg/mL), thus demonstrating that 7 did not inhibit MraY to a relevant extent (also see Supplementary Materials, Figure S1, for measured data).
3. Discussion
The encountered side reaction from precursor 4 to diazepanone 6 is highly interesting as the construction of the diazepanone scaffold represents one of the main challenges in the total synthesis of caprazamycins and their analogues [18,19,21,22]. To the best of our knowledge, a simple thermally promoted nucleophilic attack of the linker unit to a 7′-carboxylic acid ester moiety has not been successfully explored for the diazepanone-forming step before. The good yields of 6 obtained from this reaction strongly suggest that it should be investigated for caprazamycin syntheses in a more systematic way in the future. Additionally, it has not escaped our notion that this transformation might even be considered to be “biomimetic” with respect to the proposed biosynthesis of caprazamycins [41,42]. However, it seems possible that a cyclization precursor decorated with more functional groups (i.e., an analogue of 4 closer resembling a non-simplified caprazamycin precursor) might undergo decomposition reactions at the elevated temperatures needed for diazepanone formation. Such hurdles might be overcome by the choice of more reactive (i.e., less bulky) ester units in the 7′-position, thus potentially allowing lower temperatures for the ring closure reaction after Fmoc deprotection.
The missing inhibitory potential of the deprotected uridine-derived diazepanone 7 towards MraY shows that a simplified 5′-defunctionalized (“5′-deoxy”) version of the caprazamycin core structure (“caprazol”) does not inhibit this bacterial target protein, at least not in the relevant concentration range. This supports the conclusion that at least some of the functionalized side chain structures found in caprazamycins (cf. Figure 1) are relevant for biological activity.
In summary, we report an interesting and unexpected formation of a caprazamycin-like seven-membered diazepanone ring in a supposedly facile ester cleavage en route to muraymycin analogues. It is particularly remarkable that this side product was obtained in yields much higher than those of the desired product. Additionally, the complete cleavage of the Fmoc group under the (supposedly slightly acidic) reaction conditions was surprising. The observed side reaction might pave the way for the development of more efficient strategies for diazepanone formation in the synthesis of caprazamycin analogues.
4. Materials and Methods
4.1. Synthesis
General methods: All chemicals were purchased from standard suppliers and used without further purification. Reactions involving oxygen and/or moisture sensitive reagents were carried out under an atmosphere of nitrogen using anhydrous solvents. Anhydrous solvents were obtained in the following manner: THF was dried with a solvent purification system (MBRAUN MB SPS 800, M. Braun, Garching, Germany). All other solvents were of technical quality and distilled prior to use, and deionized water was used throughout. Analytical TLC was performed on aluminum plates precoated with silica gel 60 F254 (VWR, Darmstadt, Germany). Visualization of the spots was carried out using UV light (254 nm) and/or staining under heating (H2SO4 staining solution: 4 g vanillin, 25 mL conc. H2SO4, 80 mL AcOH and 680 mL MeOH; KMnO4 staining solution: 1 g KMnO4, 6 g K2CO3 and 1.5 mL 1.25 m NaOH solution, all dissolved in 100 mL H2O; ninhydrin staining solution: 0.3 g ninhydrin, 3 mL AcOH and 100 mL 1-butanol). Preparative TLC was carried out on a ChromatotronTM 7924T by T-Squared Technology (San Bruno/CA, USA), using glass plates coated with silica gel 60 PF254 containing a fluorescent indicator (VWR, Darmstadt, Germany; thickness depending on the amount of crude material to be separated, for 50–500 mg: 1 mm layer). Column chromatography was carried out on silica gel 60 (0.040–0.063 mm, 230–400 mesh ASTM, VWR, Darmstadt, Germany) under flash conditions. Semipreparative HPLC was performed on an Agilent Technologies 1200 Series system equipped with an MWD detector (254/280) (Agilent Trechnologies, Waldbronn, Germany) and a LiChroCartTM column (10 × 250 mm) containing reversed phase silica gel PurospherTM RP18e (5 μm, VWR, Darmstadt, Germany). Method: eluent A water, eluent B MeCN; 0–35 min gradient of B (10%–100%), 35–39 min 100% B, 39–40 min gradient of B (100%–10%), 40–45 min 10% B; flow 3 mL/min. 500 MHz-1H, 126 MHz-13C, and 376 MHz-19F NMR spectra were recorded on Bruker AVANCE-500 spectrometers (Bruker, Bremen, Germany). All 13C and 19F NMR spectra were 1H-decoupled. All spectra were recorded at room temperature and were referenced internally to solvent reference frequencies wherever possible. Chemical shifts (δ) are given in ppm and coupling constants (J) are reported in Hz. Assignment of signals was carried out using 1H,1H-COSY, HSQC and HMBC spectra. High-resolution ESI mass spectra were measured on a Dionex UltiMate 3000 HPLC system and on a Bruker time-of-flight (TOF) maXis (Bruker, Bremen, Germany). Infrared spectra (IR) were measured on a Bruker Vertex 70 spectrometer equipped with an integrated ATR unit (PlatinumATRTM, Bruker, Bremen, Germany). Wavenumbers (ῦ) are quoted in cm–1. UV spectra were measured on an Agilent Cary 100 spectrophotometer (Agilent Trechnologies, Waldbronn, Germany). Wavelengths of maximum absorption (λmax) are reported in nm.
N-Fmoc-protected aminoalkylated uridinyl amino acid tert-butyl ester (4): Protected nucleosyl amino acid 2 [30,37,38] (98.0 mg, 0.167 mmol) was dissolved in THF (6 mL) over molecular sieves (4 Å). N-Fmoc-protected aldehyde 3 [36] (75.0 mg, 0.254 mmol) was added and the mixture was stirred at rt for 21 h. Amberlyst™ (7.9 mg, 37 μmol) and sodium triacetoxyborohydride (75.7 mg, 0.357 mmol) were added and the solution was stirred at rt for further 24 h. The reaction mixture was then filtered, and the molecular sieves were washed with EtOAc. The combined filtrates were washed with saturated (sat.) Na2CO3 solution (50 mL) and the aqueous layer was extracted with EtOAc (50 mL). The organic layer was dried over Na2SO4 and the solvent was evaporated under reduced pressure. The resultant crude product was purified by column chromatography (100:0 → 99:1 → 98:2, CH2Cl2–MeOH) to give 4 as a colorless foam (102 mg, 71%). 1H NMR (500 MHz, CDCl3): δ [ppm] = 0.07 (s, 3H, SiCH3), 0.07 (s, 3H, SiCH3), 0.07 (s, 3H, SiCH3), 0.09 (s, 3H, SiCH3), 0.88 (s, 9H, SiC(CH3)3), 0.90 (s, 9H, SiC(CH3)3), 1.47 (s, 9H, OC(CH3)3), 1.65–1.71 (m, 2H, 2″-H), 1.83–1.89 (m, 1H, 5′-Ha), 1.98–2.02 (m, 1H, 5′-Hb), 2.54–2.59 (m, 1H, 1″-Ha), 2.70–2.75 (m, 1H, 1″-Hb), 3.24–3.35 (m, 3H, 3″-H, 6′-H), 3.66–3.68 (m, 1H, 3′-H), 4.10–4.14 (m, 1H, 4′-H), 4.20–4.25 (m, 2H, 2′-H, Fmoc-9-H), 4.33–4.42 (m, 2H, Fmoc-CH2), 5.41–5.44 (m, 1H, 3″-NH), 5.59 (s, 1H, 1′-H), 5.71 (d, J = 8.2 Hz, 1H, 5-H), 7.29 (t, J = 7.4 Hz, 2H, Fmoc-3-H, Fmoc-6-H), 7.34 (d, J = 8.2 Hz, 1H, 6-H), 7.38 (t, J = 7.4 Hz, 2H, Fmoc-2-H, Fmoc-7-H), 7.59 (d, J = 7.4 Hz, 2H, Fmoc-4-H, Fmoc-5-H), 7.75 (d, J = 7.4 Hz, 2H, Fmoc-1-H, Fmoc-8-H). 13C NMR (126 MHz, CDCl3): δ [ppm] = −4.74 (SiCH3), −4.67 (SiCH3), −4.44 (SiCH3), −4.02 (SiCH3), 18.08 (SiC(CH3)3), 18.18 (SiC(CH3)3), 25.88 (SiC(CH3)3), 25.95 (SiC(CH3)3), 28.21 (OC(CH3)3), 29.90 (C-2″), 37.36 (C-5′), 39.30 (C-3″), 45.67 (C-1″), 47.38 (Fmoc-C-9), 60.03 (C-6′), 66.72 (Fmoc-CH2), 74.87 (C-2′), 75.39 (C-3′), 81.08 (C-4′), 81.93 (OC(CH3)3), 92.41 (C-1′), 102.32 (C-5), 120.07 (Fmoc-C-1, Fmoc-C-8), 125.21 (Fmoc-C-4, Fmoc-C-5), 127.11 (Fmoc-C-3, Fmoc-C-6), 127.75 (Fmoc-C-2, Fmoc-C-7), 140.53 (C-6), 141.40 (Fmoc-C-1a, Fmoc-C-8a), 144.12 (Fmoc-C-4a, Fmoc-C-5a), 150.07 (C-2), 156.61 (urea-C=O), 163.22 (C-4), 173.96 (C-7′). HRMS (ESI): calcd. for C45H69N4O9Si2: 865.4598 [M + H]+, found 865.4608. IR (ATR): ῦ = 2929, 2856, 1687, 1450, 1251, 1151, 836, 776, 739. UV (CHCl3): λmax = 266, 301. optical rotation: = +90.0 (c = 1.0, CHCl3). TLC: Rf = 0.20 (19:1, CH2Cl2–MeOH).
N-Fmoc-protected aminoalkylated uridinyl amino acid (5): N-Fmoc-protected aminoalkylated uridinyl amino acid tert-butyl ester 4 (54.4 mg, 62.9 μmol) was dissolved in toluene (10 mL). Silica (409 mg) was added, the reaction mixture was heated to 80 °C and stirred at this temperature for 1 day. Then, the mixture was allowed to cool to rt and stirred at rt for further 2 days. The mixture was filtered, the silica was washed with a mixture of CH2Cl2 and MeOH (1:1), and the solvent of the combined filtrates was evaporated under reduced pressure. The resultant crude product was purified by preparative TLC (chromatotron, 9:1 → 4:1, CH2Cl2–MeOH) to give 5 as a yellowish solid (24.6 mg, 48%). 1H NMR (500 MHz, CDCl3): δ [ppm] = −0.13 (s, 3H, SiCH3), −0.01 (s, 3H, SiCH3), 0.07 (s, 6H, SiCH3), 0.81 (s, 9H, SiC(CH3)3), 0.89 (s, 9H, SiC(CH3)3), 1.78–1.94 (m, 2H, 2″-H), 2.03–2.33 (m, 2H, 5′-H), 3.02–4.53 (m, 11H, 1″-H, 3″-H, 6′-H, 2′-H, 3′-H, 4′-H, Fmoc-9-H, Fmoc-CH2), 5.02–5.19 (m, 2H, 1′-H, 3″-NH), 5.55–5.71 (m, 1H, 5-H), 7.26–7.29 (m, 2H, Fmoc-3-H, Fmoc-6-H), 7.33–7.37 (m, 3H, 6-H, Fmoc-2-H, Fmoc-7-H), 7.51–7.62 (m, 2H, Fmoc-4-H, Fmoc-5-H), 7.72 (d, J = 7.7 Hz, 2H, Fmoc-1-H, Fmoc-8-H). MS (ESI): m/z = 809.51 [M + H]+. HRMS (ESI): calcd. for C41H61N4O9Si2: 809.3972 [M + H]+, found 809.3949. TLC: Rf = 0.18 (9:1, CH2Cl2–MeOH).
Protected uridine-derived diazepanone (6): N-Fmoc-protected aminoalkylated uridinyl amino acid tert-butyl ester 4 (63.1 mg, 49.8 μmol) was dissolved in toluene (8 mL). Silica (208 mg) was added and the reaction mixture was stirred under reflux for 1.5 days. Then, the mixture was allowed to cool to rt and the solvent was evaporated under reduced pressure. The resultant crude product was purified by column chromatography (95:5 → 9:1 → 4:1, CH2Cl2–MeOH) to give 6 as a colorless oil (19.3 mg, 68%). 1H NMR (500 MHz, CD3OD): δ [ppm] = 0.04 (s, 3H, SiCH3), 0.09 (s, 3H, SiCH3), 0.13 (s, 3H, SiCH3), 0.16 (s, 3H, SiCH3), 0.89 (s, 9H, SiC(CH3)3), 0.94 (s, 9H, SiC(CH3)3), 1.57–1.66 (m, 1H, 2″-Ha), 1.71–1.77 (m, 2H, 2″-Hb, 5′-Ha), 2.32–2.37 (m, 1H, 5′-Hb), 2.90–2.95 (m, 1H, 1″-Ha), 3.23–3.27 (m, 2H, 1″-Hb, 3″-Ha), 3.35–3.42 (m, 1H, 3″-Hb), 3.55 (dd, J = 8.2, 5.3 Hz, 1H, 6′-H), 3.99 (dd, J = 4.4, 3.3 Hz, 1H, 3′-H), 4.13 (ddd, J = 10.6, 3.3, 3.3 Hz, 1H, 4′-H), 4.40 (dd, J = 5.9, 4.4 Hz, 1H, 2′-H), 5.77 (d, J = 8.1 Hz, 1H, 5-H), 5.84 (d, J = 5.9 Hz, 1H, 1′-H), 7.67 (d, J = 8.1 Hz, 1 H, 6-H). 13C NMR (126 MHz, CD3OD): δ [ppm] = −4.64 (SiCH3), −4.38 (SiCH3), −4.35 (SiCH3), −4.17 (SiCH3), 18.87 (SiC(CH3)3), 18.97 (SiC(CH3)3), 26.34 (SiC(CH3)3), 26.42 (SiC(CH3)3), 31.43 (C-2″), 36.99 (C-5′), 41.99 (C-3″), 51.58 (C-1″), 58.68 (C-6′), 75.35 (C-2′), 77.09 (C-3′), 84.67 (C-4′), 91.31 (C-1′), 103.23 (C-5), 143.12 (C-6), 152.28 (C-2), 165.97 (C-4), 178.69 (C-7′). MS (ESI): m/z = 569.33 [M + H]+. HRMS (ESI): calcd. for C26H49N4O6Si2: 569.3185 [M + H]+, found 569.3177. IR (ATR): ῦ = 2920, 2851, 1738, 1457, 1374, 1229, 1070, 797. UV (CHCl3): λmax = 224. TLC: Rf = 0.24 (9:1, CH2Cl2–MeOH).
Uridine-derived diazepanone (7): Protected uridine-derived diazepanone 6 (6.1 mg, 10.7 μmol) was dissolved in TFA (80% in water, 3 mL). The reaction mixture was stirred at rt for 24 h and the solvent was evaporated under reduced pressure. The resultant crude product was purified by semipreparative HPLC to give 7 (TFA salt) as a colorless solid (2.0 mg containing ~30% of an unknown uracil-derived impurity; 100% yield: 4.9 mg). 1H NMR (500 MHz, D2O): δ [ppm] = 1.77–1.85 (m, 1H, 2″-Ha), 1.94–2.05 (m, 2H, 5′-Ha, 2″-Hb), 2.50–2.55 (m, 1H, 5′-Hb), 3.22–3.27 (m, 1H, 3″-Ha), 3.35–3.37 (m, 1H, 1″-Ha), 3.47–3.52 (m, 2H, 1″-Hb, 3″-Hb), 4.07–4.11 (m, 2H, 3′-H, 4′-H), 4.14–4.17 (m, 1H, 6′-H), 4.43 (dd, J = 5.0, 3.9 Hz, 1H, 2′-H), 5.71 (d, J = 3.9 Hz, 1H, 1′-H), 5.76 (d, J = 7.7 Hz, 1H, unknown, probably 5-H), 5.86 (d, J = 8.1 Hz, 1 H, 5-H), 7.50 (d, J = 7.6 Hz, 1 H, unknown, probably 6-H), 7.63 (d, J = 8.1 Hz, 1H, 6-H). 13C NMR (126 MHz, D2O): δ [ppm] = 26.70 (C-2″), 32.73 (C-5′), 39.77 (C-3″), 48.83 (C-1″), 56.78 (C-6′), 72.66 (C-2′), 73.15 (C-3′), 80.67 (C-4′), 91.97 (C-1′), 101.10 (unknown, probably C-5), 102.34 (C-5), 142.95 (C-6), 143.47 (unknown, probably C-6), 151.52 (C-2), 166.31 (C-4), 173.19 (C-7′). 19F NMR (376 MHz, D2O): −75.69 (TFA-CF3). MS (ESI): m/z = 341.11 [M + H]+. HRMS (ESI): calcd. for C14H21N4O6: 341.1456 [M + H]+, found 341.1448. UV (H2O): λmax = 202, 260. HPLC: tR = 8.4 min.
4.2. Overexpression of MraY from S. aureus
The overexpression of MraY was performed as described before [32,33,40].
4.3. Fluorescence-Based MraY Assay
The MraY assay was performed as described before [32,33,40]. Measured data are shown in the Supplementary Materials, Figure S1.
Supplementary Materials
The following are available online, data from the MraY assays, copies of NMR and IR spectra.
Author Contributions
Conceptualization, C.D.; formal analysis, K.L., S.K. and C.D.; funding acquisition, C.D.; investigation, K.L., S.K.; project administration, C.D.; supervision, C.D.; writing—original draft preparation, K.L.; writing—review and editing, K.L., S.K. and C.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Deutsche Forschungsgemeinschaft (DFG), grant DU 1095/5-1. The APC was funded by Saarland University.
Acknowledgments
We thank Jannine Ludwig (Saarland University) for technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Taubes, G. The bacteria fight back. Science 2008, 321, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Cooper, M.A.; Shlaes, D. Fix the antibiotics pipeline. Nature 2011, 472, 32. [Google Scholar] [CrossRef] [PubMed]
- Von Nussbaum, F.; Brands, M.; Hinzen, B.; Weigand, S.; Häbich, D. Antibacterial Natural Products in Medicinal Chemistry - Exodus or Revival? Angew. Chem. Int. Ed. 2006, 45, 5072–5129. [Google Scholar] [CrossRef]
- Walsh, C. Where will new antibiotics come from? Nat. Rev. Microbiol. 2003, 1, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Winn, M.; Goss, R.J.M.; Kimura, K.; Bugg, T.D.H. Antimicrobial nucleoside antibiotics targeting cell wall assembly: Recent advances in structure–function studies and nucleoside biosynthesis. Nat. Prod. Rep. 2010, 27, 279–304. [Google Scholar] [CrossRef]
- Wiegmann, D.; Koppermann, S.; Wirth, M.; Niro, G.; Leyerer, K.; Ducho, C. Muraymycin nucleoside-peptide antibiotics: Uridine-derived natural products as lead structures for the development of novel antibacterial agents. Beilstein J. Org. Chem. 2016, 12, 769–795. [Google Scholar] [CrossRef]
- Ichikawa, S.; Yamaguchi, M.; Matsuda, A. Antibacterial Nucleoside Natural Products Inhibiting Phospho-MurNAc-Pentapeptide Translocase; Chemistry and Structure–activity Relationship. Curr. Med. Chem. 2015, 22, 3951–3979. [Google Scholar] [CrossRef]
- Struve, W.G.; Neuhaus, F.C. Evidence for an Initial Acceptor of UDP-NAc-Muramyl-Pentapeptide in the Synthesis of Bacterial Mucopeptide. Biochem. Biophys. Res. Commun. 1965, 18, 6–12. [Google Scholar] [CrossRef]
- Anderson, J.S.; Matsuhashi, M.; Haskin, M.A.; Strominger, J.L. Lipid-Phosphoacetylmuramyl-pentapeptide and Lipid-Phosphodisaccharide-pentapeptide: Presumed Membrane Transport Intermediates in Cell Wall Synthesis. Proc. Natl. Acad. Sci. USA 1965, 53, 881–889. [Google Scholar] [CrossRef]
- Heydanek, M.G., Jr.; Struve, W.G.; Neuhaus, F.C. Initial state in peptidoglycan synthesis. III. Kinetics and uncoupling of phospho-N-acetylmuramyl-pentapeptide translocase. Biochemistry 1969, 8, 1214–1221. [Google Scholar] [CrossRef]
- Ikeda, M.; Wachi, M.; Jung, H.K.; Ishino, F.; Matsuhashi, M. The Escherichia coli mraY gene encoding UDP-N-acetylmuramoyl-pentapeptide: Undecaprenyl-phosphate phospho-N-acetylmuramoyl-pentapeptide transferase. J. Bacteriol. 1991, 173, 1021–1026. [Google Scholar] [CrossRef] [PubMed]
- Boyle, D.S.; Donachie, W.D. mraY is an essential gene for cell growth in Escherichia coli. J. Bacteriol. 1998, 180, 6429–6432. [Google Scholar] [CrossRef] [PubMed]
- Bouhss, A.; Mengin-Lecreulx, D.; Le Beller, D.; Van Heijenoort, J. Topological analysis of the MraY protein catalysing the first membrane step of peptidoglycan synthesis. Mol. Microbiol. 1999, 34, 576–585. [Google Scholar] [CrossRef] [PubMed]
- McDonald, L.A.; Barbieri, L.R.; Carter, G.T.; Lenoy, E.; Lotvin, J.; Petersen, P.J.; Siegel, M.M.; Singh, G.; Williamson, R.T. Structures of the Muraymycins, Novel Peptidoglycan Biosynthesis Inhibitors. J. Am. Chem. Soc. 2002, 124, 10260–10261. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Wang, X.; Koppermann, S.; Thorson, J.S.; Ducho, C.; Van Lanen, S.G. Antibacterial Muraymycins from Mutant Strains of Streptomyces sp. NRRL 30471. J. Nat. Prod. 2018, 81, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, M.; Nakagawa, N.; Doi, N.; Hattori, S.; Naganawa, H.; Hamada, M. Caprazamycin B, a Novel Anti-tuberculosis Antibiotic, from Streptomyces sp. J. Antibiot. 2003, 56, 580–583. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, M.; Takahashi, Y.; Shitara, T.; Nakamura, H.; Naganawa, H.; Miyake, T.; Akamatsu, Y. Caprazamycins, Novel Lipo-nucleoside Antibiotics, from Streptomyces sp. J. Antibiot. 2005, 58, 327–337. [Google Scholar] [CrossRef]
- Nakamura, H.; Tsukano, C.; Yasui, M.; Yokouchi, S.; Igarashi, M.; Takemoto, Y. Total Synthesis of (−)-Caprazamycin A. Angew. Chem. Int. Ed. 2015, 54, 3136–3139. [Google Scholar] [CrossRef]
- Abe, H.; Gopinath, P.; Ravi, G.; Wang, L.; Watanabe, T.; Shibasaki, M. Synthesis of caprazamycin B. Tetrahedron Lett. 2015, 56, 3782–3785. [Google Scholar] [CrossRef]
- Hirano, S.; Ichikawa, S.; Matsuda, A. Synthesis of Caprazamycin Analogues and Their Structure−Activity Relationship for Antibacterial Activity. J. Org. Chem. 2008, 73, 569–577. [Google Scholar] [CrossRef]
- Linder, R.; Ducho, C. Unified Synthesis of Densely Functionalized Amino Acid Building Blocks for the Preparation of Caprazamycin Nucleoside Antibiotics. Eur. J. Org. Chem. 2019, 2019, 1523–1534. [Google Scholar] [CrossRef]
- Sarabia, F.; Vivar-García, C.; García-Ruiz, C.; Martín-Ortiz, L.; Romero-Carrasco, A. Exploring the Chemistry of Epoxy Amides for the Synthesis of the 2′′-epi-Diazepanone Core of Liposidomycins and Caprazamycins. J. Org. Chem. 2012, 77, 1328–1339. [Google Scholar] [CrossRef] [PubMed]
- Chung, B.C.; Zhao, J.; Gillespie, R.A.; Kwon, D.-Y.; Guan, Z.; Hong, J.; Zhou, P.; Lee, S.-Y. Crystal Structure of MraY, an Essential Membrane Enzyme for Bacterial Cell Wall Synthesis. Science 2013, 341, 1012–1016. [Google Scholar] [CrossRef] [PubMed]
- Chung, B.C.; Mashalidis, E.H.; Tanino, T.; Kim, M.; Matsuda, A.; Hong, J.; Ichikawa, S.; Lee, S.-Y. Structural insights into inhibition of lipid I production in bacterial cell wall synthesis. Nature 2016, 533, 557–560. [Google Scholar] [CrossRef]
- Koppermann, S.; Ducho, C. Natural Products at Work: Structural Insights into Inhibition of the Bacterial Membrane Protein MraY. Angew. Chem. Int. Ed. 2016, 55, 11722–11724. [Google Scholar] [CrossRef]
- Yamashita, A.; Norton, E.; Petersen, P.J.; Rasmussen, B.A.; Singh, G.; Yang, Y.; Mansour, T.S.; Ho, D.M. Muraymycins, novel peptidoglycan biosynthesis inhibitors: Synthesis and SAR of their analogues. Bioorg. Med. Chem. Lett. 2003, 13, 3345–3350. [Google Scholar] [CrossRef]
- Tanino, T.; Ichikawa, S.; Al-Dabbagh, B.; Bouhss, A.; Oyama, H.; Matsuda, A. Synthesis and Biological Evaluation of Muraymycin Analogues Active against Anti-Drug-Resistant Bacteria. ACS Med. Chem. Lett. 2010, 1, 258–262. [Google Scholar] [CrossRef] [PubMed]
- Tanino, T.; Al-Dabbagh, B.; Mengin-Lecreulx, D.; Bouhss, A.; Oyama, H.; Ichikawa, S.; Matsuda, A. Mechanistic Analysis of Muraymycin Analogues: A Guide to the Design of MraY Inhibitors. J. Med. Chem. 2011, 54, 8421–8439. [Google Scholar] [CrossRef]
- Takeoka, Y.; Tanino, T.; Sekiguchi, M.; Yonezawa, S.; Sakagami, M.; Takahashi, F.; Togame, H.; Tanaka, Y.; Takemoto, H.; Ichikawa, S.; et al. Expansion of Antibacterial Spectrum of Muraymycins toward Pseudomonas aeruginosa. ACS Med. Chem. Lett. 2014, 5, 556–560. [Google Scholar] [CrossRef]
- Spork, A.P.; Büschleb, M.; Ries, O.; Wiegmann, D.; Boettcher, S.; Mihalyi, A.; Bugg, T.D.; Ducho, C. Lead Structures for New Antibacterials: Stereocontrolled Synthesis of a Bioactive Muraymycin Analogue. Chem.- Eur. J. 2014, 20, 15292–15297. [Google Scholar] [CrossRef]
- Mitachi, K.; Aleiwi, B.A.; Schneider, C.M.; Siricilla, S.; Kurosu, M. Stereocontrolled Total Synthesis of Muraymycin D1 Having a Dual Mode of Action against Mycobacterium tuberculosis. J. Am. Chem. Soc. 2016, 138, 12975–12980. [Google Scholar] [CrossRef] [PubMed]
- Koppermann, S.; Cui, Z.; Fischer, P.D.; Wang, X.; Ludwig, J.; Thorson, J.S.; Van Lanen, S.G.; Ducho, C. Insights into the Target Interaction of Naturally Occurring Muraymycin Nucleoside Antibiotics. ChemMedChem 2018, 13, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Spork, A.P.; Koppermann, S.; Schier (née Wohnig), S.; Linder, R.; Ducho, C. Analogues of Muraymycin Nucleoside Antibiotics with Epimeric Uridine-Derived Core Structures. Molecules 2018, 23, 2868. [Google Scholar] [CrossRef] [PubMed]
- Wiegmann, D.; Koppermann, S.; Ducho, C. Aminoribosylated Analogues of Muraymycin Nucleoside Antibiotics. Molecules 2018, 23, 3085. [Google Scholar] [CrossRef] [PubMed]
- Heib, A.; Niro, G.; Weck, S.C.; Koppermann, S.; Ducho, C. Muraymycin Nucleoside Antibiotics: Structure–activity Relationship for Variations in the Nucleoside Unit. Molecules 2020, 25, 22. [Google Scholar] [CrossRef]
- Leyerer, K.; Koppermann, S.; Ducho, C. Solid Phase-Supported Synthesis of Muraymycin Analogues. Eur. J. Org. Chem. 2019, 45, 7420–7431. [Google Scholar] [CrossRef]
- Spork, A.P.; Ducho, C. Novel 5′-deoxy nucleosyl amino acid scaffolds for the synthesis of muraymycin analogues. Org. Biomol. Chem. 2010, 8, 2323–2326. [Google Scholar] [CrossRef]
- Spork, A.P.; Wiegmann, D.; Granitzka, M.; Stalke, D.; Ducho, C. Stereoselective Synthesis of Uridine-Derived Nucleosyl Amino Acids. J. Org. Chem. 2011, 76, 10083–10098. [Google Scholar] [CrossRef]
- Schmidtgall, B.; Höbartner, C.; Ducho, C. NAA-modified DNA oligonucleotides with zwitterionic backbones: Stereoselective synthesis of A–T phosphoramidite building blocks. Beilstein J. Org. Chem. 2015, 11, 50–60. [Google Scholar] [CrossRef]
- Wohnig, S.; Spork, A.P.; Koppermann, S.; Mieskes, G.; Gisch, N.; Jahn, R.; Ducho, C. Total Synthesis of Dansylated Park’s Nucleotide for High-Throughput MraY Assays. Chem. – Eur. J. 2016, 22, 17813–17819. [Google Scholar] [CrossRef]
- Kaysser, L.; Lutsch, L.; Siebenberg, S.; Wemakor, E.; Kammerer, B.; Gust, B. Identification and Manipulation of the Caprazamycin Gene Cluster Lead to New Simplified Liponucleoside Antibiotics and Give Insights into the Biosynthetic Pathway. J. Biol. Chem. 2009, 284, 14987–14996. [Google Scholar] [CrossRef] [PubMed]
- Wiker, F.; Hauck, N.; Grond, S.; Gust, B. Caprazamycins: Biosynthesis and structure activity relationship studies. Int. J. Med. Microbiol. 2019, 309, 319–324. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).