Abstract
(E)-1-(4-Ethoxycarbonylphenyl)-5-(3,4-dimethoxyphenyl)-3-(3,4-dimethoxystyryl)-2-pyrazoline was synthesized via the cyclization reaction between the monocarbonyl curcuminoid (2E,6E)-2,6-bis(3,4-dimethoxybenzylidene)acetone and ethyl hydrazinobenzoate in high yield and purity (>95% by High-performance liquid chromatography (HPLC)). The compound has been fully characterized by 1H, 13C NMR, FTIR, UV-Vis and HRMS and its activity was evaluated in terms of its potential interaction with DNA as well as its cytotoxicity against resistant and non-resistant tumor cells. Both DNA thermal denaturation and DNA viscosity measurements revealed that a significant intercalation binding takes place upon treatment of the DNA with the synthesized pyrazoline, causing an increase in melting temperature by 3.53 ± 0.11 °C and considerable DNA lengthening and viscosity increase. However, neither re-sensitisation of Doxorubicin (DO X)-resistant breast cancer and multidrug resistance (MDR) reversal nor synergistic activity with DOX by potentially increasing the DOX cell killing ability was observed.
1. Introduction
Nitrogen-containing heterocycles are important core structures found in many natural products [1] and synthetic compounds exhibiting a broad range of biological activities. Recent analysis of a Food and Drug Administration-approved drug database revealed that 59% of unique small-molecule drugs contain at least one nitrogen heterocycle [2]. Pyrazolines in particular, constitute a class of five-membered heterocycles incorporating a nitrogen-nitrogen (N-N) bond [3], with remarkable pharmacological applications [4] as anticancer [5,6,7,8,9], anti-inflammatory [10,11,12], antimicrobial [13,14], and antimalarial drugs [15].
Representative examples of synthetic bioactive pyrazolines are depicted in Figure 1. Thiazolone containing pyrazoline 1 has been found to be selectively active against colon cancer cell lines, especially on HT-29 [6]. Benzimidazole pyrazoline 2 has been reported by Shaharyar et al. as the most active antitumor compound among a library of selected similar derivatives [7], whereas benzenesulfonamide pyrazoline 3 was shown by Rathish et al. in in vitro and in vivo studies to be a more potent anti-inflammatory agent than celecoxib [10]. Manna et al. investigated 1-acetyl-3,5-diaryl-pyrazolines 4a and 4b for their anticancer activity and binding affinity to P-glycoprotein [16]. On the other hand, 3,5-divinylpyrazole 5 (Figure 1) which is a curcumin derivative, has been reported by Kolotova et al. as a potent inhibitor of P-glycoprotein, which is associated with the induction of multidrug resistance in cancer chemotherapy [17,18]. Such molecules have consequently caused resensitisation of the resistant cells and reversal of the multidrug resistance phenomenon [18]. Taking into account the aforementioned promising findings, we synthesized compound 6 (Figure 2) which is the pyrazoline analogue of the curcuminoid pyrazole derivative 5, and we studied its interaction with DNA as well as its cytotoxicity against chemo-resistant and non-resistant tumor cells.
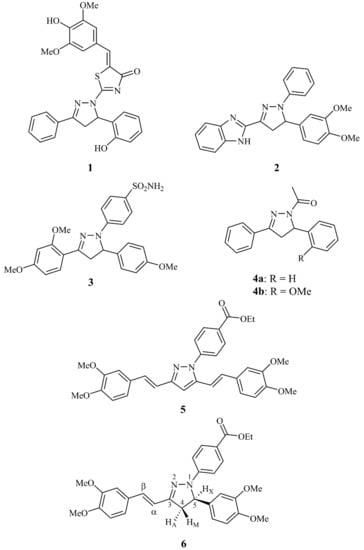
Figure 1.
Structures of known biologically active pyrazolines 1–4, pyrazole 5 and title pyrazoline 6. The numbering of compound 6 skeleton is according to common pyrazoline nomenclature. The product 6 is racemic, however the relative configuration is shown for clarity and simplicity purposes to describe the AMX system and the assignments.
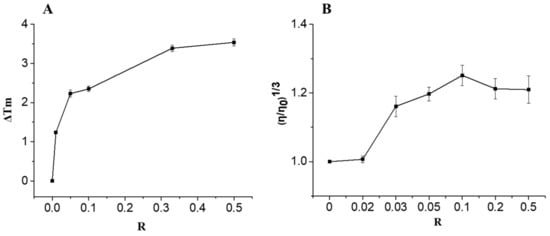
Figure 2.
Thermal denaturation data of CT-DNA (A) and Effect of increasing amounts on the relative viscosity of CT-DNA (B) upon addition of compound 6 with ratios R = [compound 6]/[DNA] ranging from 0 to 0.5.
2. Results and Discussion
2.1. Synthesis and Characterization
The synthetic route towards the preparation of the title compound 6 is outlined in Scheme 1 and Scheme 2. In the first step, the intermediate symmetrical monocarbonyl curcuminoid 7 was synthesized from acetone and veratraldehyde via base-catalyzed Claisen–Schmidt condensation reaction and used after recrystallization from ethanol (Scheme 1).
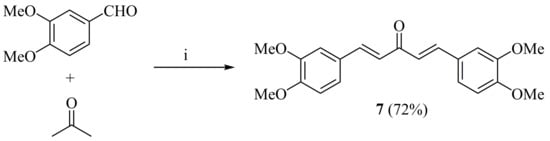
Scheme 1.
Synthesis of monocarbonyl curcuminoid intermediate 7. Reagents and conditions: (i) NaOH, EtOH, 2 h, r.t.
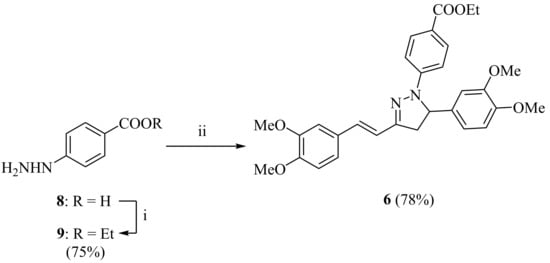
Scheme 2.
Synthesis of (E)-1-(4-ethoxycarbonylphenyl)-5-(3,4-dimethoxyphenyl)-3-(3,4-dimetho- xystyryl)-2-pyrazoline 6. Reagents and conditions: (i) H2SO4, EtOH, reflux, 5 h; (ii) 7, glacial AcOH, reflux, 5 h.
In the second step, 7 reacted with ethyl hydrazinobenzoate 9, prepared from the esterification of the corresponding carboxylic acid 8 with ethanol and concentrated sulfuric acid. The cyclization reaction between curcuminoid 7 and the hydrazine 9 in refluxing glacial acetic acid afforded the desired pyrazoline 6 in high yield and purity without the need for further purification. However, for analytical or biological purposes, even higher purity was achieved by ethanol precipitation (≥ 95%), as determined by high performance liquid chromatography (HPLC, see Supplementary Materials).
The structure of 6 has been confirmed by NMR spectroscopy and HRMS spectrometry. The characteristic signals assigned to the protons on C-4 (HM, HA) and C-5 (HX) of the pyrazoline ring (Figure 1) can be described as a typical AMX spin system. Three doublets of doublets appear at 3.03 (HA), 3.78 (HM) and 5.51 (HX) ppm. The coupling constants conform to this abovementioned pattern having the following J values: JAM = 17.2 Hz the JAX = 4.6 Hz for the cis- configuration and JMX = 12.2 Hz for the trans. Moreover, the large J value (16.4 Hz) of the vinylic Hα and Hβ protons, appearing at 6.81 and 7.21 ppm respectively, is in accordance with the expected E-configuration of the double bond. The rest of the 1H signals, along with the 13C signals, were assigned by means of 2D NMR (COSY, HSQC and HMBC—see Supporting material).
The FTIR spectrum of the prepared compound further supports the formation of a 2-pyrazoline. The characteristic C=N and C-N bonds of the molecule appear at 1600 and 1250 cm–1 respectively as strong bands, while the medium intensity band at 1700 cm–1 is assigned to the C=O of the aromatic ester.
2.2. Biological Evaluation
2.2.1. DNA Binding Studies
The potential of compound 6 to interact with CT-DNA was studied by means of the biophysical methods of DNA melting and DNA viscosity changes. Firstly, in thermal denaturation experiments for CT-DNA alone, under our experimental conditions, a Tm of 69.02 ± 0.31 °C was recorded, a value that is in good agreement with reported literature values [19]. Upon co-incubation of increasing concentrations of compound 6 with CT-DNA (5 × 10–5 M), a concentration-dependent rise of Tm values was recorded reaching a ΔTm value of 3.53 ± 0.11 °C at the highest [compound 6]/[DNA] ratio of R = 0.5 (Figure 2A). As in the literature an increase in melting temperature with ΔTm values ranging from +3 °C to +14 °C is associated with intercalative binding [20], the observed ΔΤm value suggests that compound 6 may interact with DNA through intercalation. To further investigate the nature of this interaction with CT-DNA, viscosity measurements were carried out. In general, interaction of a compound with DNA results in changes in the length of DNA [21]. When classical intercalation occurs, DNA lengthening is usually observed, due to the separation of base pairs at the intercalation site, which produces a concomitant increase in the relative specific viscosity of such solutions. As it is shown by the viscosity data obtained (Figure 2B), with increasing concentration of compound 6, the relative viscosity of CT-DNA increases continuously, providing additional evidence of intercalation mode of binding. Curcumin and curcumin derivatives are known to interact with DNA via groove binding [22,23,24,25], as demonstrated by UV-visible, competitive fluorescence studies, CD spectroscopy, DNA viscosity and melting studies. The interaction of the pyrazoline curcuminoid 6, with DNA via the intercalative mode, demonstrates the fact that structural modifications of bioactive molecules modulate dramatically their mechanism, mode and specificity of action.
2.2.2. In Vitro Cytotoxicity Studies Against Doxorubicin-Resistant Breast Cancer Cells
To assess the relative toxicity of compound 6, Doxorubicin (DOX) and their combination, the MTT colorimetric method was employed against MCF-7 breast cancer cell line and the corresponding DOX-resistant (MCF-7/DOXR) cell line. Compound 6 exhibited moderate toxicity against both MCF-7 DOX-sensitive and resistant cells, with IC50 values of 53.09 μM and 85.11 μM respectively, following a 48 h treatment (Table 1). Furthermore, the IC50 value obtained for the cytotoxity of DOX in MCF-7 was in good agreement with literature data [26], and as expected, in the MCF-7/DOXR cells DOX was found to be less toxic compared to corresponding non-resistant cells (14-fold lower). In order to evaluate the potential ability of compound 6 to re-sensitize the DOX-resistant cells and re-establish a low IC50 value for DOX, or investigate the potential synergistic activity of compound 6 with DOX in the parental cell line, both the MCF-7 and the MCF-7/DOXR cells were pre-treated with a small amount of compound 6 before the DOX co-incubation. As presented in Table 1, compound 6 did not alter IC50 values of DOX in either cell lines and no significant reduction was observed. Our data reveal that even though compound 6 has a close structural similarity with 5 it does not seem to reverse drug resistance in DOX-resistant MCF-7/DOXR and re-sensitize them as well as no synergistic activity was exhibited with DOX against the parental MCF-7 cells.

Table 1.
Effect of curcuminoid pyrazoline 6 on DOX sensitivity in MCF-7 and MCF-7/DOXR.
3. Materials and Methods
3.1. General
All reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA) and Alfa Aesar (Lancaster, UK) unless otherwise stated and used without further purification. All the media/agents for the cultures of cells were purchased from Thermo Fisher Scientific (Cleveland, OH, USA). Melting points were determined with a Gallenkamp MFB-595 melting point apparatus (Weiss Gallenkamp, London, UK). NMR spectra were recorded with a Bruker Avance 500 MHz spectrometer (Bruker, Rheinstetten, Germany) operating at 500 MHz (1H) and 125 (13C). Chemical shifts are reported in ppm relative to DMSO-d6 (1H: δ = 2.50 ppm, 13C: δ = 39.52 ± 0.06 ppm) or CDCl3 (1H: δ = 7.26 ppm). UV-Vis spectra were recorded with a Hitachi U-3010 spectrophotometer (Hitachi, Tokyo, Japan). IR spectra were recorded on a Perkin-Elmer Spectrum 100-IR spectrophotometer (Perkin-Elmer, San Francisco, CA, USA). HRMS spectra were recorded on UHPLC LC-MSn Orbitrap Velos-Thermo instrument (Thermo Scientific; Bremen, Germany) in the Institute of Biology, Medicinal Chemistry and Biotechnology of the National Hellenic Research Foundation. HPLC analysis was performed with a Waters 600E Chromatography System coupled with a Waters 486 UV detector (at 254 nm) (Waters, Manchester, UK). Separation was achieved on a C-18 reverse phase column (250 × 4 mm, 5μm) eluted with a binary gradient system at 1 mL/min flow rate. Mobile phase A was methanol containing 0.1% trifluoroacetic acid, while mobile phase B was water containing 0.1% trifluoroacetic acid. The elution gradient was 0–1 min 5% A (95% B), followed by a linear gradient to 70% A (30% B) in 5 min; this composition was held for another 3 min; 85% A (15% B) for 17 min. After a column wash with 95% A (5% B) for 10 min, the column was re-equilibrated by applying the initial conditions 5% A (95% B) for 10 min prior to the next injection.
3.2. (1E,4E)-1,5-Bis(3,4-dimethoxyphenyl)penta-1,4-dien-3-one (7)
To a solution of veratraldehyde (3.32 g, 20 mmol) and acetone (0.98 g, 10 mmol) in ethanol (10 mL) was added a suspension of NaOH (5 g, 125 mmol) in ethanol (40 mL). The resulting suspension was stirred for 2 h at room temperature. After 10 min, a yellow solid started to precipitate. The mixture was filtered, washed with water (×3) and dried. The resulting solid was recrystallized from ethanol to afford the product as yellow needles (2.84 g, 7.20 mmol, 72%). Mp: 79–80 °C; 1H NMR (500 MHz, DMSO-d6): δ 3.82 (s, 6H, OMe), 3.84 (s, 6H, OMe), 7.03 (d, J = 8.3 Hz, 2H, Ph), 7.23 (d, J = 15.9 Hz, 2H, -CH=CH-Ph), 7.33 (d, J = 8.3 Hz, 2H, Ph), 7.40 (s, 2H, Ph), 7.70 (d, J = 15.9 Hz, 2H, -CH=CH-Ph) [27,28,29].
3.3. Ethyl 4-hydrazinobenzoate (9)
To a solution of 4-hydrazinobenzoic acid (1.52 g, 10 mmol) in absolute ethanol (20 mL) was added dropwise concentrated H2SO4 (1 mL). After stirring under reflux for 8 h, the mixture was cooled to room temperature and then concentrated under vacuum. The residue was suspended in AcOEt (40 mL) and washed with a saturated K2CO3 solution. The latter was extracted two times with AcOEt (20 mL). The combined organic solutions were dried over Na2SO4 and the solvent was evaporated to afford the product as off-white solid (1.35 g, 7.5 mmol, 75%). Mp: 106–107 °C; 1H NMR (500MHz, CDCl3): δ 1.37 (t, J = 7.0 Hz, 3H, CH3CH2), 3.31 (br, 2H, NHNH2), 4.33 (q, J = 7.0 Hz, 2H, CH3CH2), 5.52 (br, 1H, NHNH2), 7.40 (d, J = 7.4 Hz, 2H, Ph), 7.92 (d, J = 7.4 Hz, 2H, Ph) [30].
3.4. (E)-1-(4-Ethoxycarbonylphenyl)-5-(3,4-dimethoxyphenyl)-3-(3,4-dimethoxystyryl)-2-pyrazoline (6)
A mixture of the curcuminoid 7 (177 mg, 0.50 mmol) and ethyl 4-hydrazinobenzoate 9 (270 mg, 1.50 mmol) was refluxed in glacial acetic acid (6 mL) for 5 h. The solution was cooled to 50 °C and then poured into ice-cold water (50 mL). The completion of the reaction was monitored by TLC (n-hexane/AcOEt = 7:3) after mini work-up. The precipitate was filtered and washed thoroughly with water (×3). The product (199 mg, 0.39 mmol, 78%) was dried in a desiccator over CaCl2 overnight. For analytical purposes, it was precipitated from hot ethanol. Yellow solid. Mp: 76-77 °C; UV-Vis (EtOH) λmax (log ε): 388 nm (4.65); FTIR (KBr, cm–1): 1700, 1600, 1510, 1265, 1170, 1105, 1025, 765; 1H NMR (500 MHz, DMSO-d6): δ 1.26 (t, J = 6.8 Hz, 3H, CH3CH2O-), 3.03 (dd, JMX = 4.6 Hz, JAM = 17.1 Hz, 1H, CHAHM), 3.69 (s, 3H, MeO), 3.71 (3H, s, MeO), 3.74 (dd, JAX = 11.8 Hz, JAM = 17.1 Hz, 1H, CHAHM), 3.77 (s, 3H, MeO), 3.81 (s, 3H, MeO), 4.21 (q, J = 6.8 Hz, 2H, CH3CH2O-), 5.51 (dd, JMX = 4.6 Hz, 1H, JAX = 11.8 Hz, CHX), 6.65 (d, J = 7.8 Hz, 1H, Ph), 6.81 (d, J = 16.2 Hz, 1H, Hβ), 6.87 (1H, s, Ph), 6.88 (d, J = 7.8 Hz, 1H, Ph), 6.94 (d, J = 8.4 Hz, 1H, Ph), 6.99 (d, J = 8.6 Hz, 2H, Ph), 7.08 (d, J = 8.4 Hz, 1H, Ph), 7.21 (d, J = 16.2 Hz, 1H, Hα), 7.26 (s, 1H, Ph), 7.74 (d, J = 8.6 Hz, 2H, Ph); 13C NMR (125 MHz, DMSO-d6): δ 14.3 (CH3CH2), 42.0 (C-4), 55.46 (2C, CH3O-), 55.48 (2C, CH3O-), 59.8 (CH3CH2), 61.8 (C-5), 109.2 (CH-arom), 109.5 (CH-arom), 111.7 (CH-arom), 112.0 (2C, CH-arom), 112.2 (CH-arom), 117.3 (CH-arom), 118.8 (Cα), 120.8 (CH-arom), 129.2 (2C, Cq-arom), 130.6 (2C, CH-arom), 134.0 (Cq-arom), 135.1 (Cβ), 146.9 (Cq-arom), 148.1 (Cq-arom), 149.0 (Cq-arom), 149.1 (Cq-arom), 149.4 (Cq-arom), 151.9 (C-3), 165.6 (-COOEt); HRMS: calcd for C30H33N2O6 (M+ + H) 517.2333; found 517.2328; HPLC: tR = 16.5 min.
3.5. Preparation of DNA Samples
All DNA experiments were performed in phosphate buffer (0.05 M, pH = 7.2) consisting of Na2HPO4 and KH2PO4. The CT-DNA solution was sufficiently free of protein as evidenced by the ratio of its UV absorbance at 260 and 280 nm that was approx. 1.9:1 [20]. The DNA concentration per nucleotide was determined spectrophotometrically by using the molar absorption coefficient of ε = 6600 M–1·cm–1 per nucleotide at 260 nm. Before each measurement, fresh stock solutions of CT-DNA and compound 6 (in DMSO at a concentration of 10–3 M) were prepared and the corresponding mixtures were incubated for 24 h at 25 °C to reach the equilibrium state. Each reported measurement value is the average of three independent experiments.
3.6. Thermal Denaturation Studies
DNA melting temperature measurements were performed using a Varian Cary 300 spectrophotometer (Agilent Technologies, Inc., Santa Clara, CA, USA) equipped with a heating multiple cell block apparatus. Temperatures were maintained under computer control and were increased at a 0.5 °C/min rate. DNA melting experiments were carried out by monitoring the absorbance of DNA at 258 nm in the temperature range from 25.0 °C to 95.0 °C. Solutions were allowed to equilibrate for 1 min at each temperature. Cuvettes (1.0 cm capped quartz) were mounted in a thermal block and the solution temperatures were monitored by a thermistor in the reference cuvette. The melting temperature (Tm) of DNA was determined as the midpoint of the optically detected transition and the measurement was repeated three times for each sample. The melting experiments were performed in phosphate buffer solution (final percentage of DMSO = 2%) by keeping the DNA concentration constant (5 × 10–5 M) while varying the concentration of compound 6 (0 – 2.5 × 10-5 M) to achieve ratios R = [compound 6]/[DNA] of 0, 0.01, 0.05, 0.1, 0.33, 0.5.
3.7. Viscosity Studies
Viscosity measurements were carried out using Ostwald’s viscometer (Schott geräte, type 531 01/0a, thermostat Schott geräte GT 1150, meter Schott geräte AVS 300) at 25 °C (Schott –Instruments GmbH, Germany). The viscometer was thermostated at 25.0 ± 0.1 °C in a constant temperature bath. Flow time of solutions was measured with a digital stopwatch and each sample was measured three times and an average flow time was calculated. The values did not differ by more than 0.2 s from each other. All solutions were filtered through 0.22 μm filter (Millipore, Billerica, MA) prior to the measurements. Experiments were carried out in phosphate buffer solution (final percentage of DMSO = 2%) by keeping the DNA concentration constant (5 × 10–5 M) while varying the concentration of compound 6 (0–2.5 × 10-5 M) to achieve ratios R = [compound 6]/[DNA] of 0, 0.02, 0.033, 0.05, 0.1, 0.2, 0.5. The intrinsic viscosity [n] was calculated according to the relation [n] = (t - tb)/tb, where tb is the flow time for the buffer and t is the observed flow time for DNA in the presence or absence of the compound 6. Data were presented as (n/n0)1/3 versus R where n and n0 are the intrinsic viscosities in the presence or absence of the compound 6. For the low DNA concentrations used in these experiments, the intrinsic viscosity [n] is proportional to the difference in the flow time for the buffer with and without DNA, resulting in the following Equation (1) [31]:
where L and L0 are the DNA lengths and [n] and [n0] are the intrinsic viscosities with and without the compound. The tb, t0 and t are the flow times of the buffer, the plain DNA and the DNA/compound solution, respectively.
3.8. Cell Culture
Human breast adenocarcinoma MCF-7 cells (American Type Culture Collection, Manassas, VA, USA), were grown in DMEM growth medium of pH 7.4 supplemented with 10% FBS, 100 U/mL of penicillin, 2 mM glutamine and 100 μg/mL of streptomycin. Cell cultures were maintained in flasks and were grown at 37 °C in a humidified atmosphere of 5% CO2 in air. Subconfluent cells were detached using a 0.25% (w/v) trypsin—0.03% (w/v) EDTA solution and the split ratio was 1:3–1:5. Doxorubicin-resistance was established by stepwise exposure of MCF-7 cells to increased concentrations of DOX ranging between 0.01 and 1 μg/mL. Finally, the MCF-7/DOXR cells were able to grow continually in medium with 0.1 μg/mL DOX. The stock solutions of compound 6 and DOX at a concentration of 10 mM dissolved in DMSO were diluted to different concentrations as required.
3.9. In Vitro Cytotoxicity Assay
In vitro cytotoxicity of compound 6, DOX and the combination of them against the MCF-7 and MCF-7/DOXR cell lines was determined by the MTT colorimetric assay. Cells were seeded in 96-well plates (104 cells/well) and grown overnight at 37 °C in a 5% CO2 incubator. Exponentially growing cells were incubated for 48 h with various concentrations ranging between 10–3–10–8 M of compound 6 and DOX and the final DMSO concentration never exceeded 0.2%. For the combination experiments, MCF-7/DOXR cells were pre-incubated for 1 h with compound 6 (10–5 M) and then DOX was added in various concentrations (10–3–10–8 M) for another 47 h. The medium was then removed and replaced with 100 μL of MTT solution (Sigma-Aldrich, Darmstadt, Germany, 1 mg mL–1). After a 4 h incubation, the solution was aspirated, formazan crystals were solubilized in 100 μL of dimethyl sulfoxide (DMSO) and absorbance was recorded at 540 nm (Tecan well plate reader, Tecan, Grödig, Austria). The results were expressed as % cell viability = (mean optical density (OD) of treated cells/mean OD of untreated cells) × 100. Data were plotted against the corresponding compound concentration in a semi-log chart and the values of IC50 (the concentration of test compound required to reduce the fraction of live cells to 50% of the control) were calculated from the dose–response curves using GraphPad Prism 5.0 software (GraphPad Software Inc., CA, USA).
4. Conclusions
(E)-1-(4-Ethoxycarbonylphenyl)-5-(3,4-dimethoxyphenyl)-3-(3,4-dimethoxystyryl)-2-pyrazoline was successfully synthesized for the first time from the corresponding monocarbonyl acetone-derived curcuminoid and ethyl hydrazinobenzoate in high yield and purity. The compound exhibited significant intercalation binding activity with CT-DNA, which renders it a suitable lead compound for further derivatisation aiming to improve its DNA binding capacity. However, despite its structural similarity with the corresponding curcumin pyrazoline derivative which has shown MDR reversal activity no re-sensitisation of DOX-resistant breast cancer was observed and there was no synergistic activity with DOX against non-resistant cell lines. Further investigation is underway to identify whether conversion of the pyrazoline to pyrazole moiety may improve the MDR reversal activity of such monocarbonyl curcumin derivatives.
Supplementary Materials
The following are available online. All the spectroscopic data of the title compound 6 namely 1H and 13C NMR, 1H-1H COSY NMR, HSQC and HMBC NMR, FT-IR, HRMS, UV-Vis spectra, and HPLC are available online.
Author Contributions
Conceptualization, M.S.; methodology and data analysis, D.M and B.M.; 2D NMR analysis A.P. and M.P.; writing—original draft preparation, D.M., B.M., M.S.; writing—review and editing and funding acquisition, M.P.; supervision, M.S., C.M. and M.P. All authors have read and agreed to the published version of the manuscript.
Funding
The authors acknowledge financial support of this work by the project “Target Identification and Development of Novel Approaches for Health and Environmental Applications” (MIS 5002514) which is implemented under the Action for the Strategic Development on the Research and Technological Sectors, funded by the Operational Programme “Competitiveness, Entrepreneurship and Innovation” (NSRF 2014-2020) and co-financed by Greece and the European Union (European Regional Development Fund). B. Mavroidi gratefully acknowledges financial support by Stavros Niarchos Foundation (SNF) through implementation of the program of Industrial Fellowships at NCSR “Demokritos”.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Natural Products in Medicinal Chemistry; Hanessian, S., Ed.; Wiley-VCH:: Weinheim, Germany, 2014. [Google Scholar]
- Vitaku, E.; Smith, D.T.; Njardarson, J.T. Analysis of the structural diversity, substitution patterns, and frequency of nitrogen heterocycles among U.S. FDA approved pharmaceuticals. J. Med. Chem. 2014, 57, 10257–10274. [Google Scholar] [CrossRef] [PubMed]
- Varghese, B.; Al-Busafi, S.N.; Suliman, F.O.; Al-Kindy, M.Z. Unveiling a versatile heterocycle: Pyrazoline—A review. RSC Adv. 2017, 7, 46999–47016. [Google Scholar] [CrossRef]
- Shaaban, M.R.; Mayhoub, A.S.; Farag, A.M. Recent advances in the therapeutic applications of pyrazolines. Expert Opim. Ther. Patents 2012, 22, 253–291. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, J.; Khan, A.A.; Ali, Z.; Haider, R.; Yar, M.S. Structure-activity relationship (SAR) study and design strategies of nitrogen-containing heterocyclic moieties for their anticancer activities. Eur. J. Med. Chem. 2017, 125, 143–189. [Google Scholar] [CrossRef] [PubMed]
- Havrylyuk, D.; Zimenkovsky, B.; Vasylenko, O.; Zaprutko, L.; Gzella, A.; Lesyk, R. Synthesis of novel thiazolone-based compounds containing pyrazoline moiety and evaluation of their anticancer activity. Eur. J. Med. Chem. 2009, 44, 1396–1404. [Google Scholar] [CrossRef] [PubMed]
- Shaharyar, M.; Abdullah, M.M.; Bakht, M.A.; Majeed, J. Pyrazoline bearing benzimidazoles: Search for anticancer agent. Eur. J. Med. Chem. 2010, 45, 114–119. [Google Scholar] [CrossRef]
- Insuasty, B.; Montoya, A.; Becerra, D.; Quiroga, J.; Abonia, R.; Robledo, S.; Vélez, I.D.; Upegui, Y.; Nogueras, M.; Cobo, J. Synthesis of novel analogs of 2-pyrazoline obtained from [(7-chloroquinolin-4-yl)amino]chalcones and hydrazine as potential antitumor and antimalarial agents. Eur. J. Med. Chem. 2013, 67, 252–262. [Google Scholar] [CrossRef]
- Shin, S.Y.; Yoon, H.; Hwang, D.; Ahn, S.; Kim, D.-W.; Koh, D.; Lee, Y.H.; Lim, Y. Benzochalcones bearing pyrazoline moieties show anti-colorectal cancer activities and selective inhibitory effects on aurora kinases. Bioorg. Med. Chem. 2013, 21, 7018–7024. [Google Scholar] [CrossRef]
- Rathish, I.G.; Javed, K.; Ahmad, S.; Bano, S.; Alam, M.S.; Pillai, K.K.; Singh, S.; Bagchi, V. Synthesis and antiinflammatory activity of some new 1,3,5-trisubstituted pyrazolines bearing benzene sulfonamide. Bioorg. Med. Chem. Lett. 2009, 19, 255–258. [Google Scholar] [CrossRef]
- Bansal, E.; Srivastava, V.K.; Kumar, A. Synthesis and anti-inflammatory activity of 1-acetyl-5-substitute daryl-3-(b-aminonaphthyl)-2-pyrazolines and b-(substituted aminoethyl) amidonaphthalenes. Eur. J. Med. Chem. 2001, 36, 81–92. [Google Scholar] [CrossRef]
- Lokeshwari, D.M.; Achutha, D.K.; Srinivasan, B.; Shivalingegowda, N.; Krishnappagowda, L.N.; Kariyappa, A.K. Synthesis of novel 2-pyrazoline analogues with potent anti-inflammatory effect mediated by inhibition of phospholipase A2: Crystallographic, in silico docking and QSAR analysis. Bioorg. Med. Chem. Lett. 2017, 27, 3806–3811. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, M.S.; Holla, B.S.; Kumari, N.S. Synthesis and antimicrobial studies on novel chloro-fluorine containing hydroxy pyrazolines. Eur. J. Med. Chem. 2007, 42, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, Z.N.; Musthafa, T.N.M.; Ahmad, A.; Khan, A.U. Thermal solvent-free synthesis of novel pyrazolyl chalcones and pyrazolines as potential antimicrobial agents. Bioorg. Med. Chem. Lett. 2011, 21, 2860–2865. [Google Scholar] [CrossRef] [PubMed]
- Mishra, V.K.; Mishra, M.; Kashaw, V.; Kashaw, S.K. Synthesis of 1,3,5-trisubstituted pyrazolines as potential antimalarial and antimicrobial agents. Bioorg. Med. Chem. 2017, 25, 1949–1962. [Google Scholar] [CrossRef] [PubMed]
- Manna, F.; Chimenti, F.; Fioravanti, R.; Bolasco, A.; Secci, D.; Chimenti, P.; Ferlini, C.; Scambia, G. Synthesis of some pyrazole derivatives and preliminary investigation of their affinity binding to P-glycoprotein. Bioorg. Med. Chem. Lett. 2005, 15, 4632–4635. [Google Scholar] [CrossRef]
- Kolotova, E.S.; Shtil, A.A.; Novikov, F.N.; Chilov, G.G.; Stroganov, O.V.; Stroilov, V.S.; Zeifman, A.A.; Titov, I.Y.; Sagnou, M.; Alexiou, P. Novel Derivatives of 3,5-Divinyl-pyrazole for Medical Application. WO 2016/190770 A1, 1 December 2016. [Google Scholar]
- Gottesman, M.M.; Pastan, I.; Ambudkar, S.V. P-Glycoprotein and multidrug resistance. Curr. Opin. Gen. Dev. 1996, 6, 610–617. [Google Scholar] [CrossRef]
- Halevas, E.; Mavroidi, B.; Swanson, C.H.; Smith, G.C.; Moschona, A.; Hadjispyrou, S.; Salifoglou, A.; Pantazaki, A.A.; Pelecanou, M.; Litsardakis, G. Magnetic cationic liposomal nanocarriers for the efficient drug delivery of a curcumin-based vanadium complex with anticancer potential. J. Inorg. Biochem. 2019, 199, 110778. [Google Scholar] [CrossRef]
- Mavroidi, B.; Sagnou, M.; Stamatakis, K.; Paravatou-Petsotas, M.; Pelecanou, M.; Methenitis, G. Palladium(II) and platinum(II) complexes of derivatives of 2-(4′-aminophenyl)benzothiazole as potential anticancer agents. Inorg Chim. Acta 2016, 444, 63–75. [Google Scholar] [CrossRef]
- Suh, D.; Chaires, J.B. Criteria for the mode of binding of DNA binding agents. Bioorg. Med. Chem. 1995, 3, 723–728. [Google Scholar] [CrossRef]
- Zsila, F.; Bikádi, Z.; Simonyi, M. Circular dichroism spectroscopic studies reveal pH dependent binding of curcumin in the minor groove of natural and synthetic nucleic acids. Org. Biomol. Chem. 2004, 2, 2902–2910. [Google Scholar]
- Kunwar, A.; Simon, E.; Singh, U.; Chittela, R.; Sharma, D.; Sandur, S.K.; Priyadarsini, I.K. Interaction of a curcumin analogue dimethoxycurcumin with DNA. Chem. Biol. Drug Des. 2011, 77, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, B.K.; Ghosh, K.S.; Bera, R.; Dasgupta, S. Studies on the interaction of diacetylcurcumin with calf thymus-DNA. Chem. Phys. 2008, 351, 163–169. [Google Scholar] [CrossRef]
- Bera, R.; Sahoo, B.K.; Ghosh, K.S.; Dasgupta, S. Studies on the interaction of isoxazolcurcumin with calf thymus DNA. Int. J. Biol. Macromol. 2008, 42, 14–21. [Google Scholar]
- Arif, I.S.; Hooper, C.L.; Greco, F.; Williams, A.C.; Boateng, S.Y. Increasing doxorubicin activity against breast cancer cells using PPARγ-ligands and by exploiting circadian rhythms. Br. J. Pharmacol. 2013, 169, 1178–1188. [Google Scholar] [CrossRef]
- Li, Q.; Chen, J.; Luo, S.; Xu, J.; Huang, Q.; Liu, T. Synthesis and assessment of the antioxidant and antitumor properties of asymmetric curcumin analogues. Eur. J. Med. Chem. 2015, 93, 461–469. [Google Scholar] [CrossRef]
- Butcher, R.J.; Jasinski, J.P.; Yathirajan, H.S.; Bindya, S.; Narayana, B.; Sarojini, B.K. 1,5-Bis(3,4-dimethoxyphenyl)penta-1,4-dien-3-one. Acta Cryst. 2007, E63, o3115. [Google Scholar] [CrossRef]
- Du, Z.-Y.; Bao, Y.-D.; Liu, Z.; Qiao, W.; Ma, L.; Huang, Z.-S.; Gu, L.-Q.; Chan, A.S.C. Curcumin analogs as potent aldose reductase inhibitors. Arch. Pharm. Chem. Life Sci. 2006, 339, 123–128. [Google Scholar] [CrossRef]
- Bourrie, B.; Casellas, P.; Ciapetti, P.; Derocq, J.-M.; Jegham, S.; Muneaux, Y.; Wermuth, C.-G. Pyridoindolone Derivatives Substituted in the 3-Position by a Phenyl, Their Preparation and Their Application in Therapeutics. US 7,390,818 B2, 24 June 2008. [Google Scholar]
- Cohen, G. Eisenber, Viscosity and sedimentation study of sonicated DNA–proflavine complexes. Biopolymers 1969, 8, 45. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
