3-{[(2,3-Dichlorophenyl)amino]methyl}-5-(furan-2-ylmethylidene)-1,3-thiazolidine-2,4-dione
Abstract
:1. Introduction
2. Results
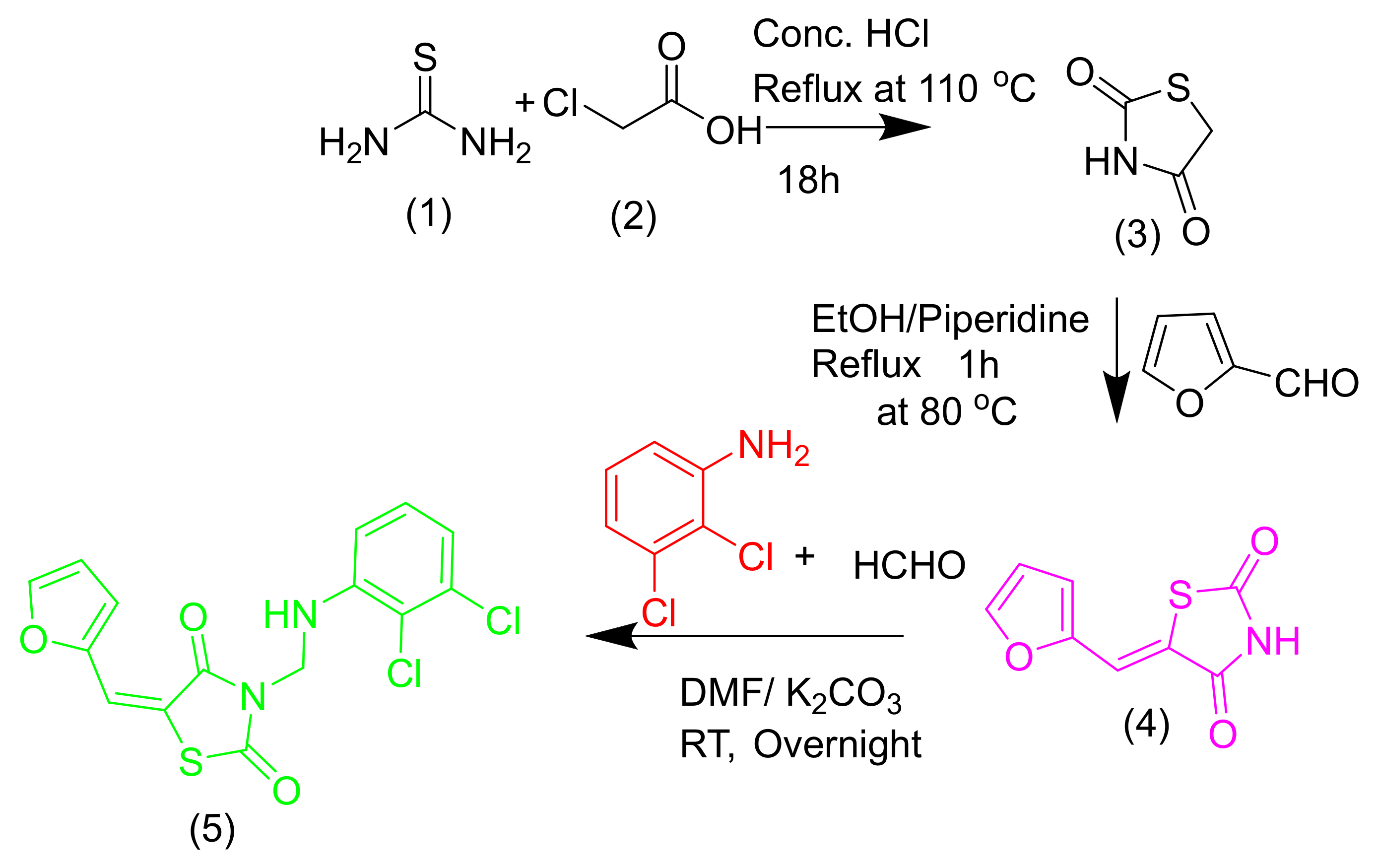
2.1. Chemistry
2.2. Biology
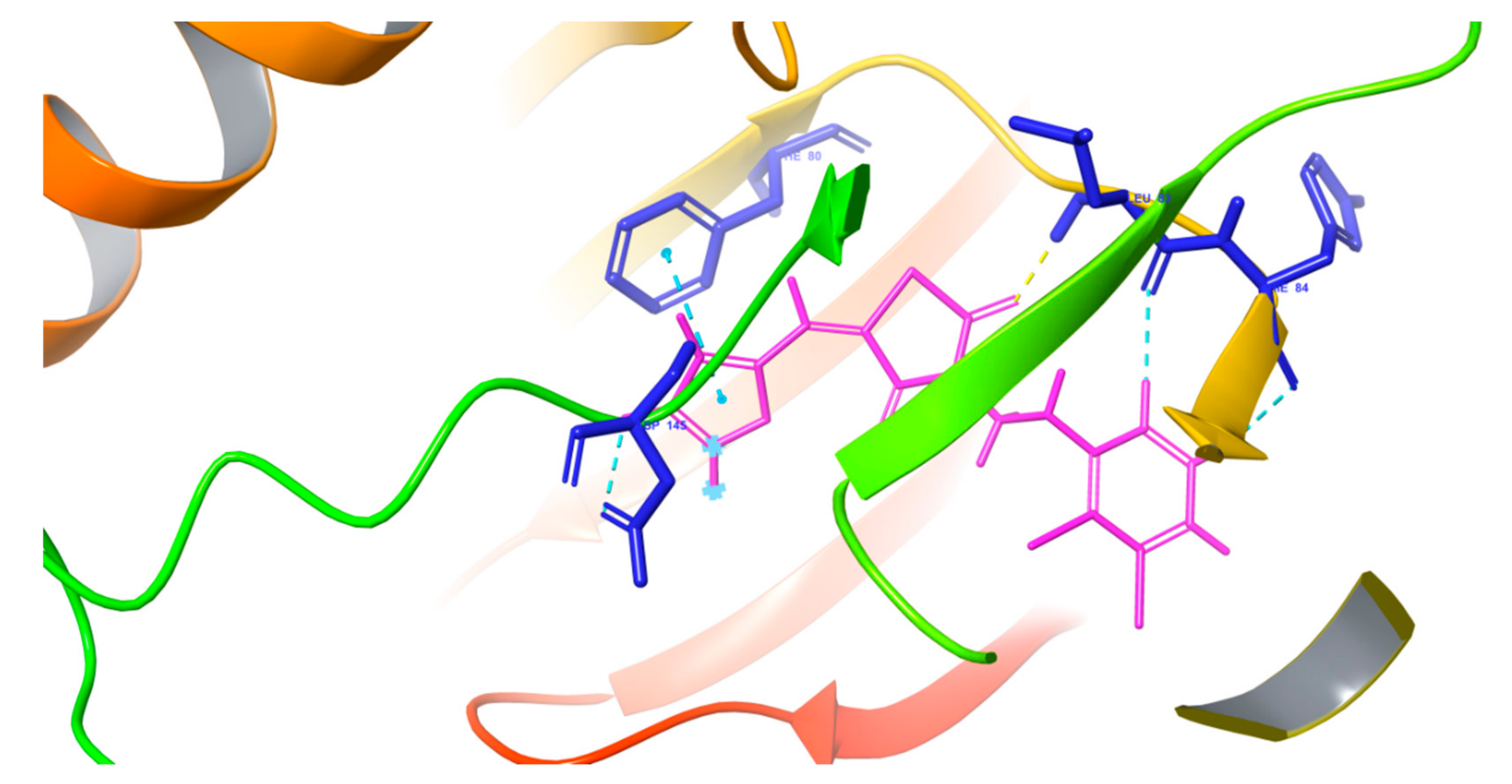
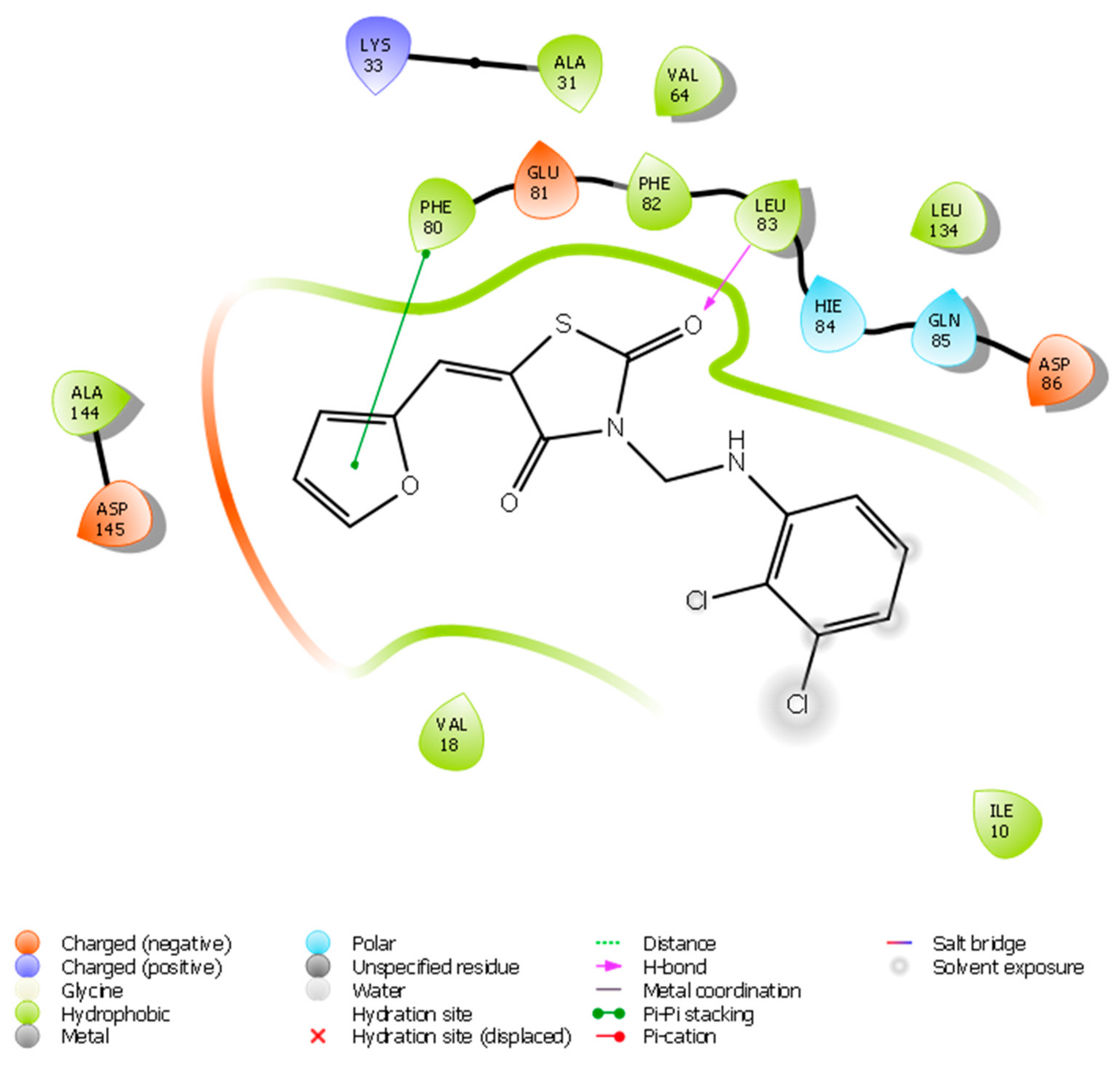
2.2.1. Molecular Docking
2.2.2. Anticancer Activity
2.2.3. Anti-Inflammatory Activity
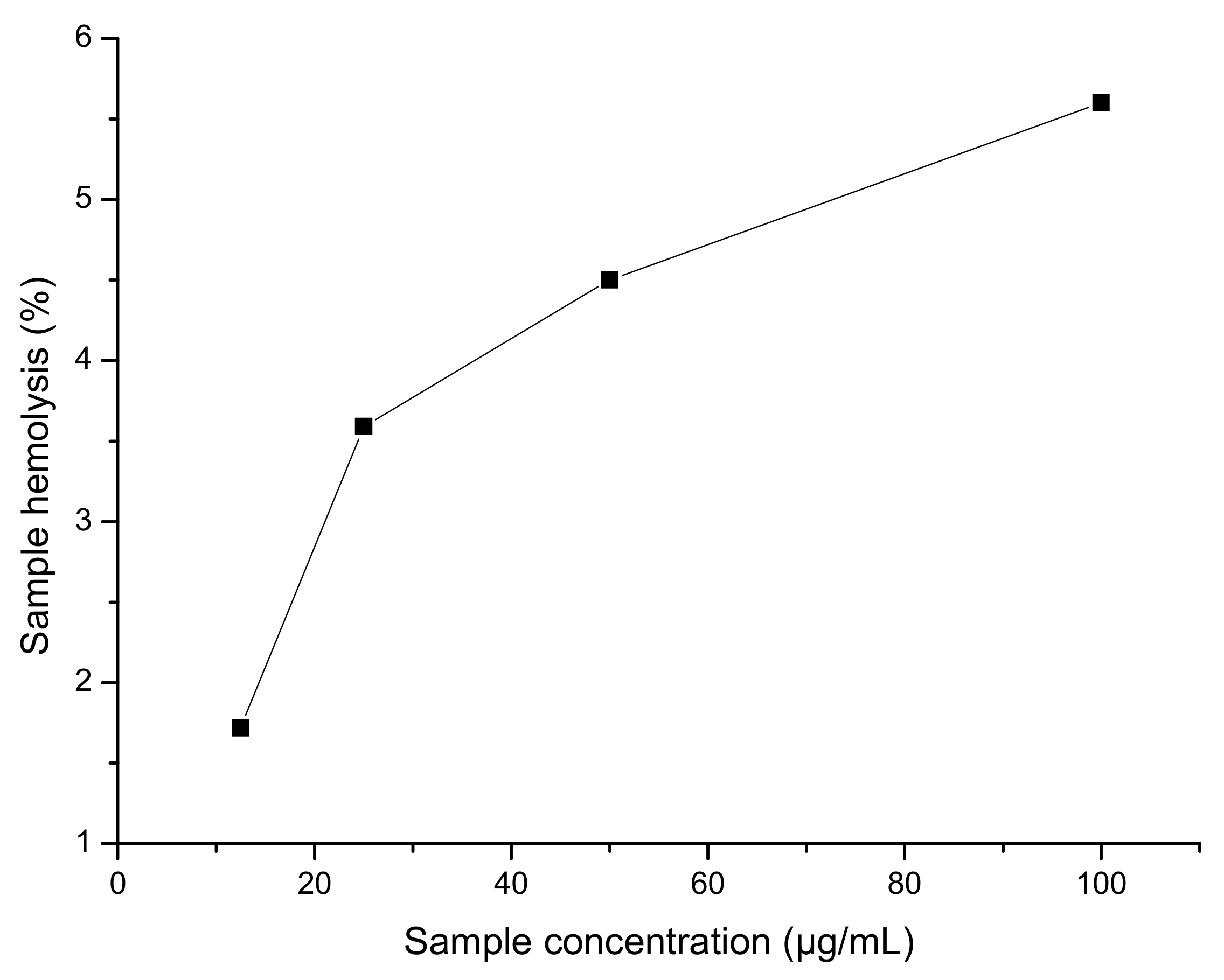
2.2.4. Hemolysis Assay
3. Discussion
3.1. Chemistry
General Information of 3-{[(2,3-Dichlorophenyl)amino]methyl}-5-(furan-2-ylmethylidene)-1,3-thiazolidine-2,4-dione
3.2. Biology
3.2.1. Anticancer Activity
Cytotoxicity Assessment Using Methyl Thiazolyl Tetrazolium (MTT) Assay
3.2.2. Anti-Inflammatory Activity
Evaluation of Anti-Inflammatory Activity by Inhibition of Albumin Denaturation
3.2.3. Hemolysis Assay
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, X.K.; Sun, T.; Li, Y.J.; Wang, Y.H.; Li, Y.J.; Yang, L.D.; Feng, D.; Zhao, M.G.; Wu, Y.M. A novel thiazolidinediones ATZD2 rescues memorydeficits in a rat model of type 2 diabetes through antioxidant and anti-inflammation. Oncotarget 2017, 8, 107409–107422. [Google Scholar] [PubMed]
- Ozen, C.; Unlusoy, M.C.; Aliary, N.; Ozturk, M.; Bozdag-Dundar, O. Thiazolidinedione or Rhodanine: A study on synthesis and anticancer activity comparison of novel thiazole derivatives. J. Pharm. Sci. 2017, 20, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Valadbeigi, E.; Ghodsi, S. Synthesis and characterization of some new thiazolidinedione derivatives containing a coumarin moiety for their antibacterial and antifungal activities. Med. Chem. 2017, 7, 178–185. [Google Scholar] [CrossRef]
- Naim, M.J.; Alam, M.J.; Nawaz, F.; Naidu, V.G.; Aaghaz, S.; Sahu, M.; Siddiqui, N.; Alam, O. Synthesis, molecular docking and anti-diabetic evaluation of 2,4- thiazolidinedione based amide derivatives. Bioorg. Chem. 2017, 73, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Galli, A.; Mello, T.; Ceni, E.; Surrenti, E.; Surrenti, C. The potential of antidiabetic thiazolidinediones for anticancer therapy. Expert Opin. Investig. Drugs 2006, 9, 15. [Google Scholar] [CrossRef] [PubMed]
- Marc, G.; Stana, A.; Oniga, S.D.; Pirnau, A.; Vlase, L.; Oniga, O. New phenolic derivatives of thiazolidine-2,4-dione with antioxidant and antiradical properties: Synthesis, characterization, in vitro evaluation and quantum studies. Molecules 2019, 24, 2060. [Google Scholar] [CrossRef] [PubMed]
- Omeragic, A.; Kara-Yacoubian, N.; Kelschenbach, J.; Sahin, C.; Cummins, C.L.; Volsky, D.J.; Bendayan, R. Peroxisome proliferator-activated receptor-gamma agonists exhibit anti-inflammatory and antiviral effects in an EcoHIV mouse model. Sci. Rep. 2019, 9, 9428. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Dachineni, R.; Ai, G.; Kumar, R.D.; Sadhu, S.S.; Tummala, H.; Bhat, J. Cyclin A2 and CDK2 as novel targets of aspirin and salicylic acid: A potential role in cancer prevention. Mol. Cancer Res. 2016, 14, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Raissa, K.C.; Jamerson, F.; Moreira, H.A.; Pinto, O.G.; Camargo, L.T.; Naves, P.L.; Camargo, A.J.; Ribeiro, L.; Ramos, L.M. Synthesis, antimicrobial activity and structure-activity relationship of some5-arylidene-thiazolidine-2,4-dione derivatives. J. Braz. Chem. Soc. 2019, 30, 164–172. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid calorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Mizushima, Y.; Kobayashi, M. Interaction of anti-inflammatory drugs with serum proteins, especially with some biologically active proteins. J. Pharma. Pharmac. 1968, 20, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Sakat, S.; Preeti, T.; Juvekar, A. In vitro anti-inflammatory activity of aqueous and methanol extracts of erythrina indica Lam leave. Pharmacologyonline 2009, 3, 221–229. [Google Scholar]
- Sashidhara, K.V.; Rao, K.B.; Kushwara, P.; Modukuru, R.K.; Singh, P.; Shukla, P.K.; Chopra, S.; Pasupeleti, M. Novel chalcone-thiazole-hybrids as potent inhibitors of drug resistance Staphylococcus aureus. ACS Med. Chem. Lett. 2015, 6, 809–813. [Google Scholar] [CrossRef] [PubMed]
| Sample Code | IC50 a (µM) |
|---|---|
| 5 | 42.30 |
| Cisplatin | 21.42 |
| Sample | Concentration (µM) | Cytotoxicity (Mean ± SD) |
|---|---|---|
| 5 | 5 10 25 50 100 | 0.19 ± 0.13 5.44 ± 0.13 15.53 ± 0.37 32.63 ± 1.86 70.20 ± 0.71 |
| Compound/Concentration | 20 µg/mL | 40 µg/mL | 60 µg/mL | 80 µg/mL | 100 µg/mL | IC50 (µg/mL) |
|---|---|---|---|---|---|---|
| 5 ** | 45.63 ± 1.02 | 60.30 ± 1.05 | 79.06 ± 1.08 | 89.02 ± 1.04 | 97.08 ± 1.40 | 48.30 |
| Diclofenac sodium ** | 42.98 ± 1.28 | 57.08 ± 1.6 | 75.54 ± 1.6 | 83.56 ± 0.96 | 94.60 ± 0.90 | 35.03 |
| Sample | Sample Concentration (µg/mL) | Sample Hemolysis (%) |
|---|---|---|
| 5 | 100 50 25 12.5 | 5.60 4.50 3.59 1.72 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uwabagira, N.; Sarojini, B.K. 3-{[(2,3-Dichlorophenyl)amino]methyl}-5-(furan-2-ylmethylidene)-1,3-thiazolidine-2,4-dione. Molbank 2019, 2019, M1083. https://doi.org/10.3390/M1083
Uwabagira N, Sarojini BK. 3-{[(2,3-Dichlorophenyl)amino]methyl}-5-(furan-2-ylmethylidene)-1,3-thiazolidine-2,4-dione. Molbank. 2019; 2019(4):M1083. https://doi.org/10.3390/M1083
Chicago/Turabian StyleUwabagira, Nadine, and Balladka Kunhana Sarojini. 2019. "3-{[(2,3-Dichlorophenyl)amino]methyl}-5-(furan-2-ylmethylidene)-1,3-thiazolidine-2,4-dione" Molbank 2019, no. 4: M1083. https://doi.org/10.3390/M1083
APA StyleUwabagira, N., & Sarojini, B. K. (2019). 3-{[(2,3-Dichlorophenyl)amino]methyl}-5-(furan-2-ylmethylidene)-1,3-thiazolidine-2,4-dione. Molbank, 2019(4), M1083. https://doi.org/10.3390/M1083




