N-[2-(1H-Indol-3-yl)-1-(5-thioxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)ethyl]-4-methylbenzenesulfonamide
Abstract
:1. Introduction
2. Results
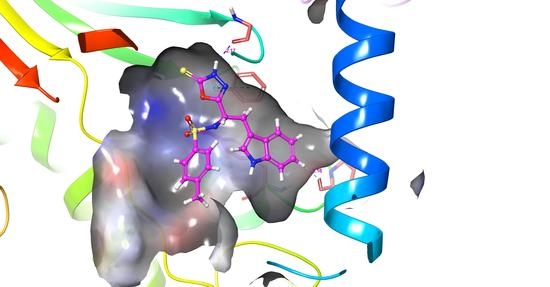
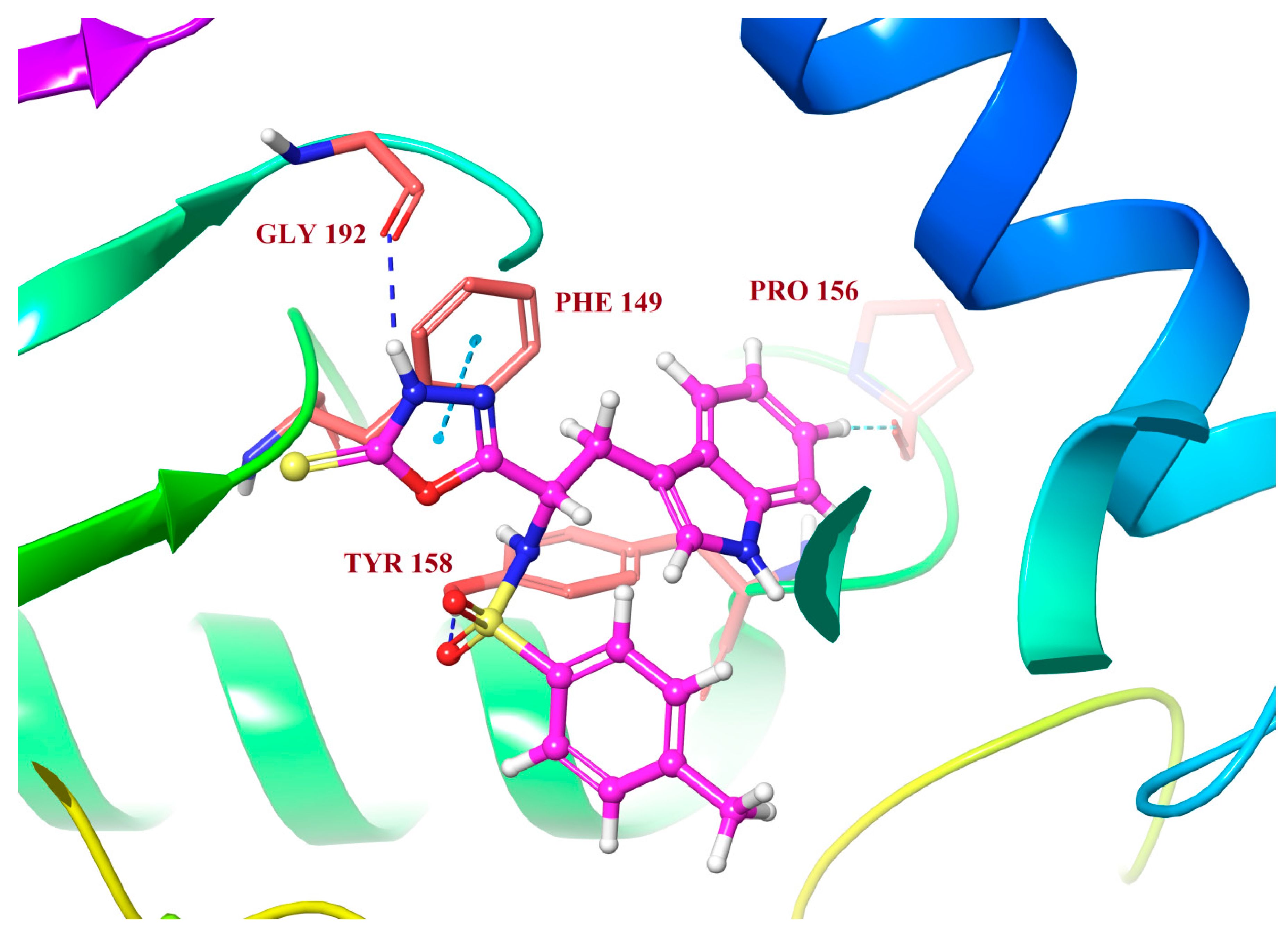
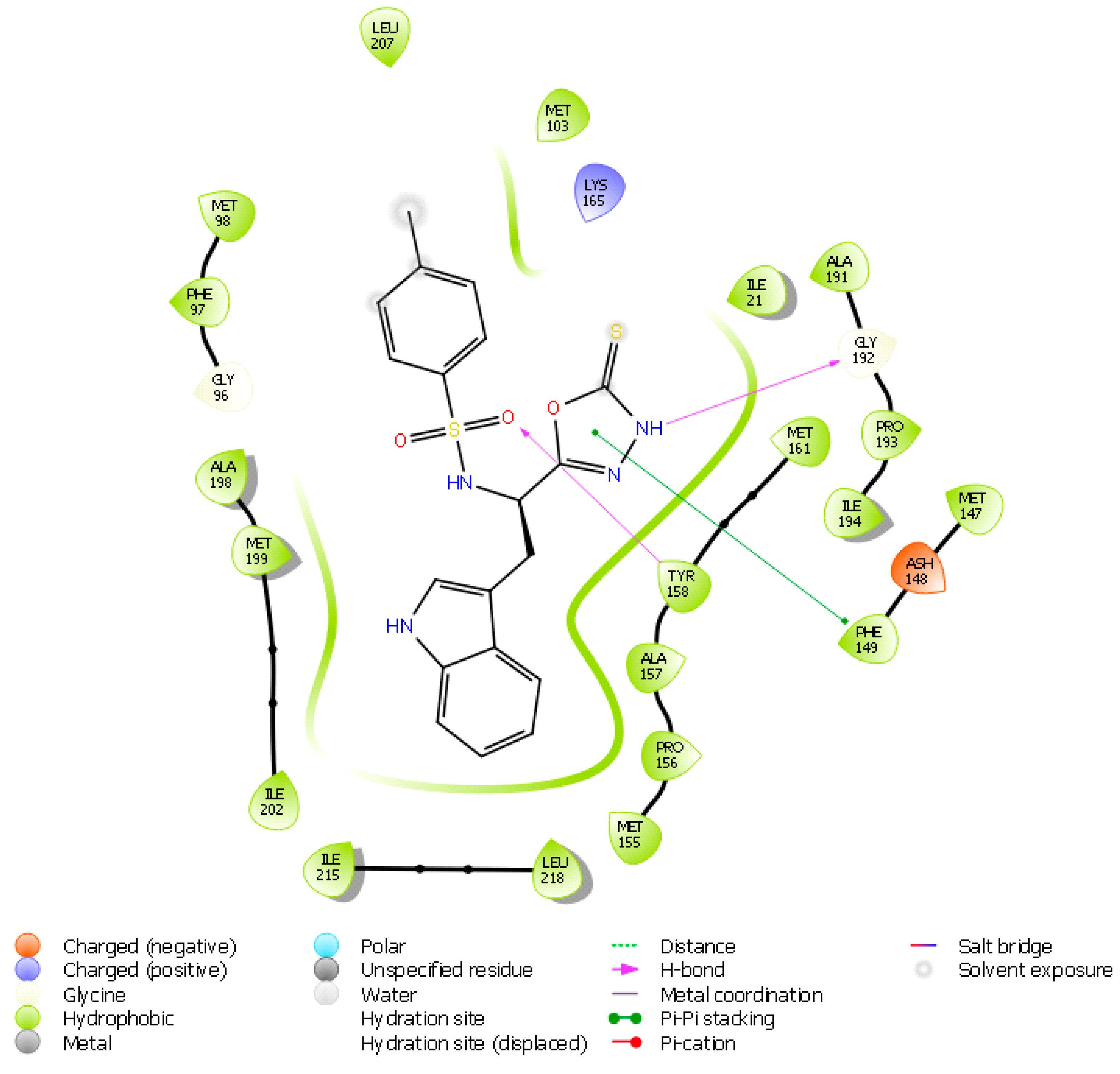
Molecular Docking
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Adil, A.O.; Mebrouk, K.; Sarah, A. 1,3,4-Oxadiazole, 1,3,4-thiadiazole and 1,2,4-triazole derivatives as potential antibacterial agents. Arab. J. Chem. 2014. [Google Scholar] [CrossRef]
- Ates, O.; Kocabalkanli, A.; Cesur, N.; Otuk, G. Synthesis andantimicrobial activity of some 5-aryl-2-[(N,N-disubstitutedthiocarbamoylthio)acylamino]-1,3,4-oxadiazoles. Il Farmaco 1998, 53, 541–544. [Google Scholar] [CrossRef]
- Li, Z.; Zhan, P.; Liu, X. 1,3,4-Oxadiazole: A Privileged Structure in Antiviral Agents. Mini-Rev. Med. Chem. 2011, 11, 1130–1142. [Google Scholar] [CrossRef] [PubMed]
- Maria, A.M.; Isabel, C.G.V.; Julie, V.E.; Richard, K.G.; Candice, S.M.; Eva, M.M.; Aaron, K.; Yulia, O.; Lindsay, F.; Anisa, G.; et al. Synthesis and biological evaluation of aryl-oxadiazoles as inhibitors of Mycobacterium tuberculosis. Bioorg. Med. Chem. Lett. 2018, 28, 1758–1764. [Google Scholar]
- Ahmed, S.A.; Hamdy, M.A.; Nadia, M.M.; Mahmoud, A.E. Novel 5-(2-hydroxyphenyl)-3-substituted-2,3-dihydro-1,3,4-oxadiazole-2-thione derivatives: Promising anticancer agents. Bioorg. Med. Chem. 2006, 14, 1236–1246. [Google Scholar]
- Musser, J.H.; Brown, R.E.; Love, B.; Bailey, K.; Jones, H.; Kahen, R.; Huang, F.; Khandwala, A.; Leibowitz, M.; Sonnino-Goldman, P.; et al. Synthesis of 2-(2,3-dihydro-2-oxo-1,3,4-oxadiazol-5-yl) benzo heterocycles. A novel series of orally activeantiallergic agents. J. Med. Chem. 1984, 27, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Sawhney, S.N.; Sharma, P.K. Synthesis and anti-inflammatory activity of some 3-heterocycle-1,2-benzisothiazoles. Bioorg. Med. Chem. Lett. 1993, 3, 1551–1554. [Google Scholar] [CrossRef]
- Zarghi, A.; Tabatabai, S.A.; Faizi, M.; Ahadian, A.; Navabi, P.; Zanganesh, V.; Shafiee, A. Synthesis and anticonvulsant activity of new 2-substituted-5-(2-benzyloxyphenyl)-1,3,4-oxadiazoles. Bioorg. Med. Chem. Lett. 2005, 15, 1863–1865. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Joshi, G.K.; Shakya, A.K.; Agarwal, R.K.; Patnaik, G.K. Pharmacological screening of few new 2-(substitutedacetyl)amino-5-alkyl-1,3,4-oxadiazoles. Ind. J. Physiol. Pharmacol. 1992, 36, 247–250. [Google Scholar]
- Desai, N.C.; Hardik, S.; Amit, T.; Kandarp, B.; Laxman, N.; Vijay, M.K.; Prakash, C.J.; Dhiman, S. Synthesis, biological evaluation and molecular docking study ofsome novel indole and pyridine based 1,3,4-oxadiazole derivatives aspotential antitubercular agents. Bioorg. Med. Chem. Lett. 2016, 26, 1776–1783. [Google Scholar] [CrossRef] [PubMed]
- Espinal, M.A. The global situation of MDR-TB. Tuberculosis 2003, 83, 44–51. [Google Scholar] [CrossRef]
- Goldberg, D.E.; Siliciano, R.F.; Jacobs, W.R. Outwitting evolution: fightingdrug-resistant TB, malaria, and HIV. Cell 2012, 148, 1271–1283. [Google Scholar] [CrossRef] [PubMed]
- Tashfeen, A.; Shahid, H.; Khalid, M.K.; Muhammad, I.C. Syntheses, urease inhibition, and antimicrobial studies of some chiral3-substituted-4-amino-5-thioxo-1H,4H-1,2,4-triazoles. Med. Chem. 2008, 4, 539–543. [Google Scholar]
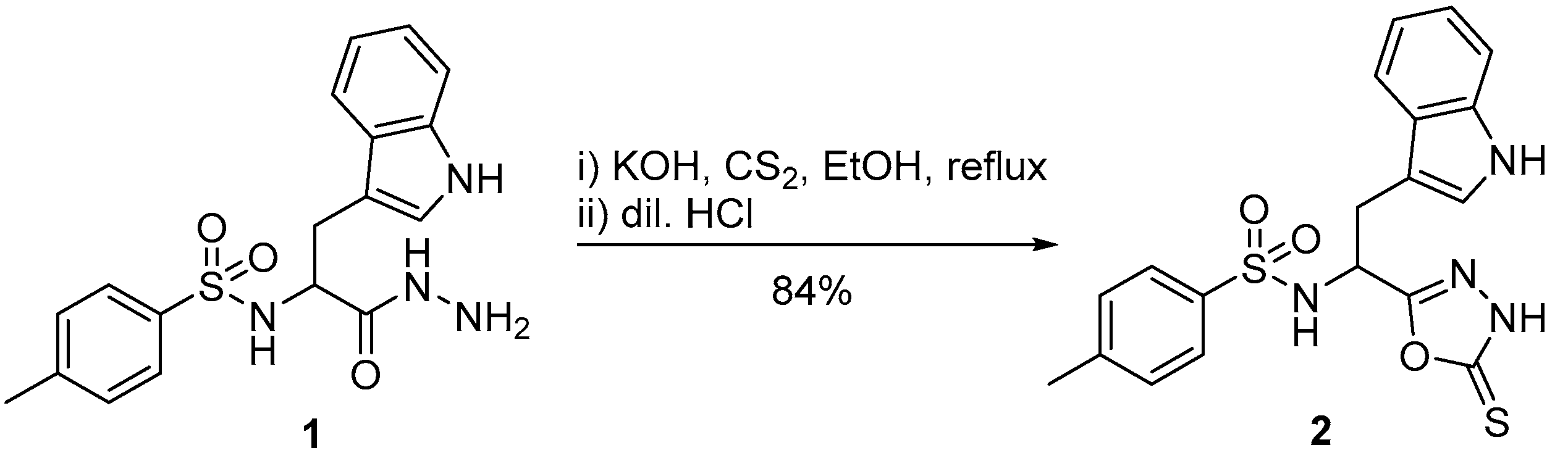
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Purushotham, N.; Poojary, B. N-[2-(1H-Indol-3-yl)-1-(5-thioxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)ethyl]-4-methylbenzenesulfonamide. Molbank 2018, 2018, M1008. https://doi.org/10.3390/M1008
Purushotham N, Poojary B. N-[2-(1H-Indol-3-yl)-1-(5-thioxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)ethyl]-4-methylbenzenesulfonamide. Molbank. 2018; 2018(3):M1008. https://doi.org/10.3390/M1008
Chicago/Turabian StylePurushotham, Nikil, and Boja Poojary. 2018. "N-[2-(1H-Indol-3-yl)-1-(5-thioxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)ethyl]-4-methylbenzenesulfonamide" Molbank 2018, no. 3: M1008. https://doi.org/10.3390/M1008
APA StylePurushotham, N., & Poojary, B. (2018). N-[2-(1H-Indol-3-yl)-1-(5-thioxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)ethyl]-4-methylbenzenesulfonamide. Molbank, 2018(3), M1008. https://doi.org/10.3390/M1008