1. Introduction
Migraine is a highly prevalent and disabling primary headache disorder that imposes a substantial burden on individuals and health systems worldwide. Analyses from the Global Burden of Disease (GBD) 2019 study confirmed migraine as the second leading cause of years lived with disability globally and the leading cause among young and middle-aged women [
1]. Using GBD 2021 data, more recent assessments estimate that approximately 1.16–1.20 billion people—around one-seventh of the world’s population—were living with migraine in 2021, with the highest age-standardized rates in high-sociodemographic-index regions and a disproportionate burden among women and working-age adults [
2,
3]. Over the past three decades, global migraine prevalence and migraine-attributable disability-adjusted life years have risen by more than 50%, indicating that demographic expansion and changing exposures have outpaced therapeutic gains [
3,
4]. Cost-of-illness studies further show that migraine is associated with high direct health-care expenditures and substantial productivity losses, leading to an annual economic burden in the United States that is conservatively estimated in the tens of billions of US dollars [
5,
6]. Together, these observations underscore the need for deeper mechanistic insights to support more effective prevention and treatment strategies [
7].
Current models view migraine as a complex disorder arising from the interaction of neuronal hyperexcitability, activation of the trigeminovascular system, neurogenic inflammation, and context-dependent vascular changes, with cortical spreading depression (CSD) acting as a key initiator or amplifier in migraine with aura [
8,
9]. Across the premonitory and headache phases, hypothalamic activation preceding pain onset, release of calcitonin gene-related peptide (CGRP) and other neuropeptides from trigeminal afferents, meningeal neurovascular–neuroimmune crosstalk, and subsequent central sensitization together shape the characteristic unilateral throbbing pain, sensory hypersensitivity, and autonomic symptoms [
9,
10,
11]. Marked clinical heterogeneity—between migraine with and without aura and among patients with prominent autonomic, immune-linked, or affective features—supports conceptualizing migraine as a multisystem brain disorder that variably recruits central nociceptive circuits and peripheral sensory–autonomic inputs [
12,
13].
In recent years, with the advancement of the Human Genome Project and the construction of large-scale biological databases, our understanding of the genetic architecture of migraine has made significant progress [
14,
15]. However, research on how migraine operates through expression quantitative trait loci (eQTLs) and splicing quantitative trait loci (sQTLs) in human tissues, as well as through the regulation of specific cell types, gene expression networks, and signaling pathways, remains a weak link in current studies. Traditional single-level research methods are inadequate for systematically revealing its multi-scale mechanisms; thus, there is an urgent need to integrate genetic data with multi-omics functional data.
To systematically dissect the complex mechanisms of migraine, this study is based on large-scale population genetic summary data, integrating multi-level data such as single-cell RNA sequencing (scRNA-seq), spatial transcriptomics, and single-cell-level chromatin accessibility information, employing cutting-edge computational methods like cross-omics integration analysis to comprehensively analyze the biological basis of migraine in systemic, cellular, and molecular dimensions. Specifically, we aim not only to identify risk gene loci, validate their tissue association information, and reveal their regulatory effects in different cell types but also to focus on the specific roles of candidate pathways such as regulation in the overlapping mechanisms, as well as the spatial expression distribution of risk genes at the embryonic level. This study seeks to uncover the key molecules mediating migraine and their functional connections, thereby providing solid theoretical foundations and data support for future targeted therapies, biomarker discovery, and early identification of high-risk populations. Ultimately, we hope these findings can offer new perspectives and strategies for unraveling migraine.
3. Discussion
Our study provides a comprehensive multi-scale perspective on how common genetic risk factors map onto the biology of migraine. Consistent with the view of migraine as a complex neurovascular and neuroinflammatory disorder involving neurons, glia, vasculature, and immune elements [
16,
17,
18,
19], we found that migraine-associated variants converge on pathways and tissues far beyond the brain alone. This aligns with growing evidence that migraine pathophysiology extends to systemic dysregulation, including immune activation and metabolic disturbances. A recent scoping review summarized consistent abnormalities in pro- and anti-inflammatory cytokines and other immune mediators in people with migraine, both during and outside attacks [
18], while a multi-omics Mendelian randomization analysis proposed a “lactylation–immune regulatory axis” in which lactate-derived histone and protein lactylation modulates immune traits and migraine risk [
20]. Epidemiologically, several immune-mediated diseases, such as rheumatoid arthritis, show an elevated subsequent risk of migraine [
21]. At the same time, neuromodulatory peptides like CGRP and pituitary adenylate cyclase-activating polypeptide (PACAP) bridge vascular, neural, and immune signaling and are now key drug targets [
22]. Notably, in a real-world cohort of patients with migraine and concomitant immune disorders, combined use of anti-CGRP monoclonal antibodies and disease-modifying immunomodulatory therapy was associated with better migraine outcomes in this real-world cohort compared with monotherapy [
23]. Together, these observations support a neuroimmune–metabolic framework for migraine and underscore the need to integrate central and systemic perspectives in mechanistic studies and therapeutic strategies.
Genome-wide association studies have firmly established that migraine is highly polygenic. The largest migraine meta-analysis to date identified 123 risk loci and showed that migraine-associated variants are enriched in both vascular tissues and central nervous system cell types, reinforcing a neurovascular model of the disorder [
14]. However, translating these loci into specific effector genes, tissues, and pathways requires functional context. Our work addresses this gap by integrating GWASs with tissue QTLs, single-cell transcriptomic and multi-omics data, chromatin accessibility, and spatial transcriptomics to construct a system-wide atlas of migraine genetic risk from organs to cell types and molecular mechanisms. This complements more focused efforts that emphasize particular biological layers. For example, a recent single-cell multi-omics framework highlighted immune cell subsets and transcriptional programs that may drive migraine and suggested repurposable immune-modulating therapeutics [
24]. While that study concentrated on peripheral immune cells, our integrative strategy systematically surveyed diverse tissues and annotated cell types to build a broader map.
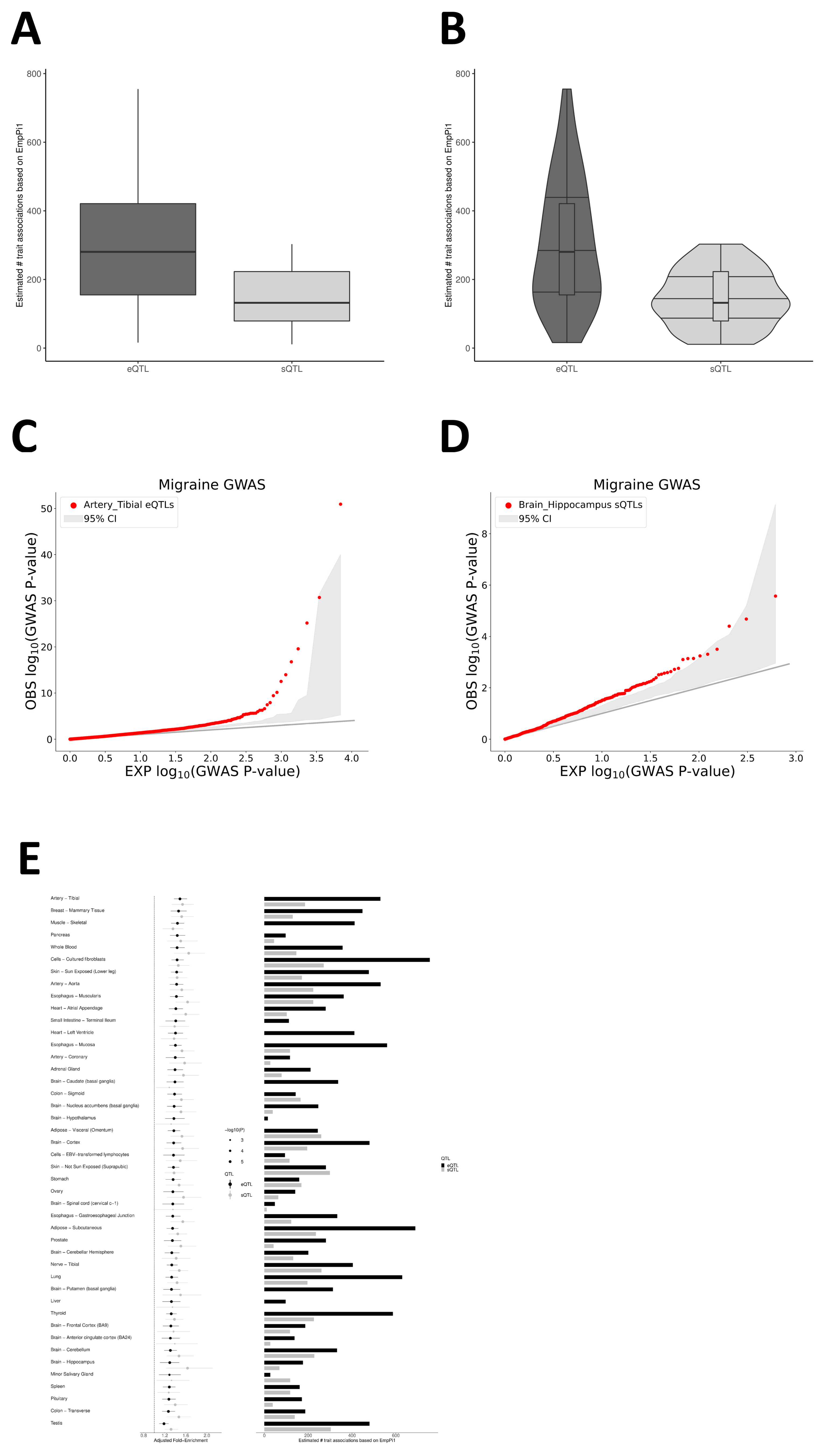
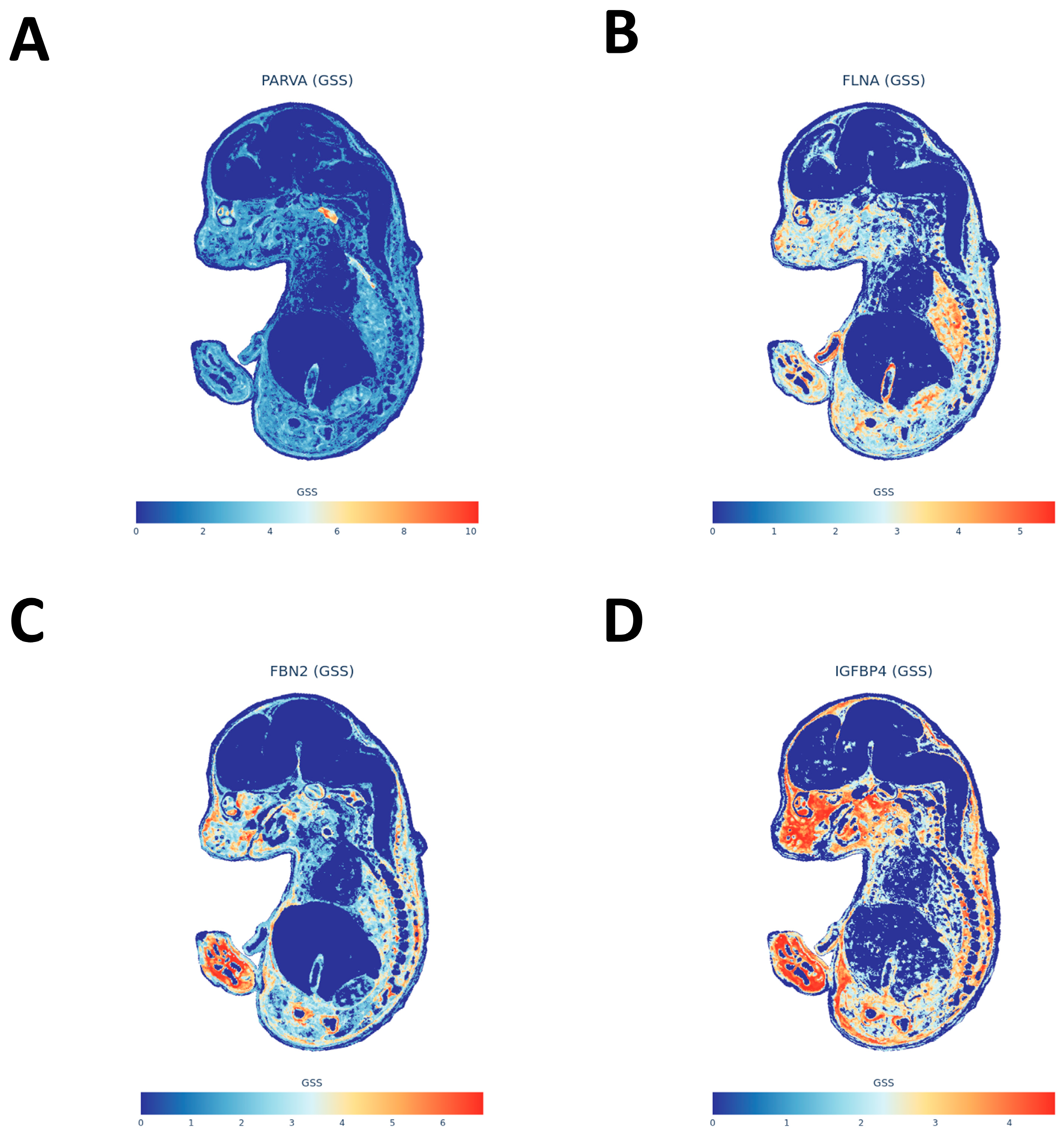
At the tissue level, we observed that migraine risk variants are non-randomly concentrated in eQTLs and sQTLs from peripheral arteries (e.g., tibial artery and aorta), heart, tibial nerve, selected brain regions, and whole blood. This pattern suggests that common migraine variants exert substantial effects on vascular and smooth muscle biology, as well as on peripheral sensory- and central pain-modulatory circuits. The finding that arterial and myocardial tissues show the most robust eQTL co-localization and pathway enrichment aligns with prior GWAS-based evidence of vascular involvement [
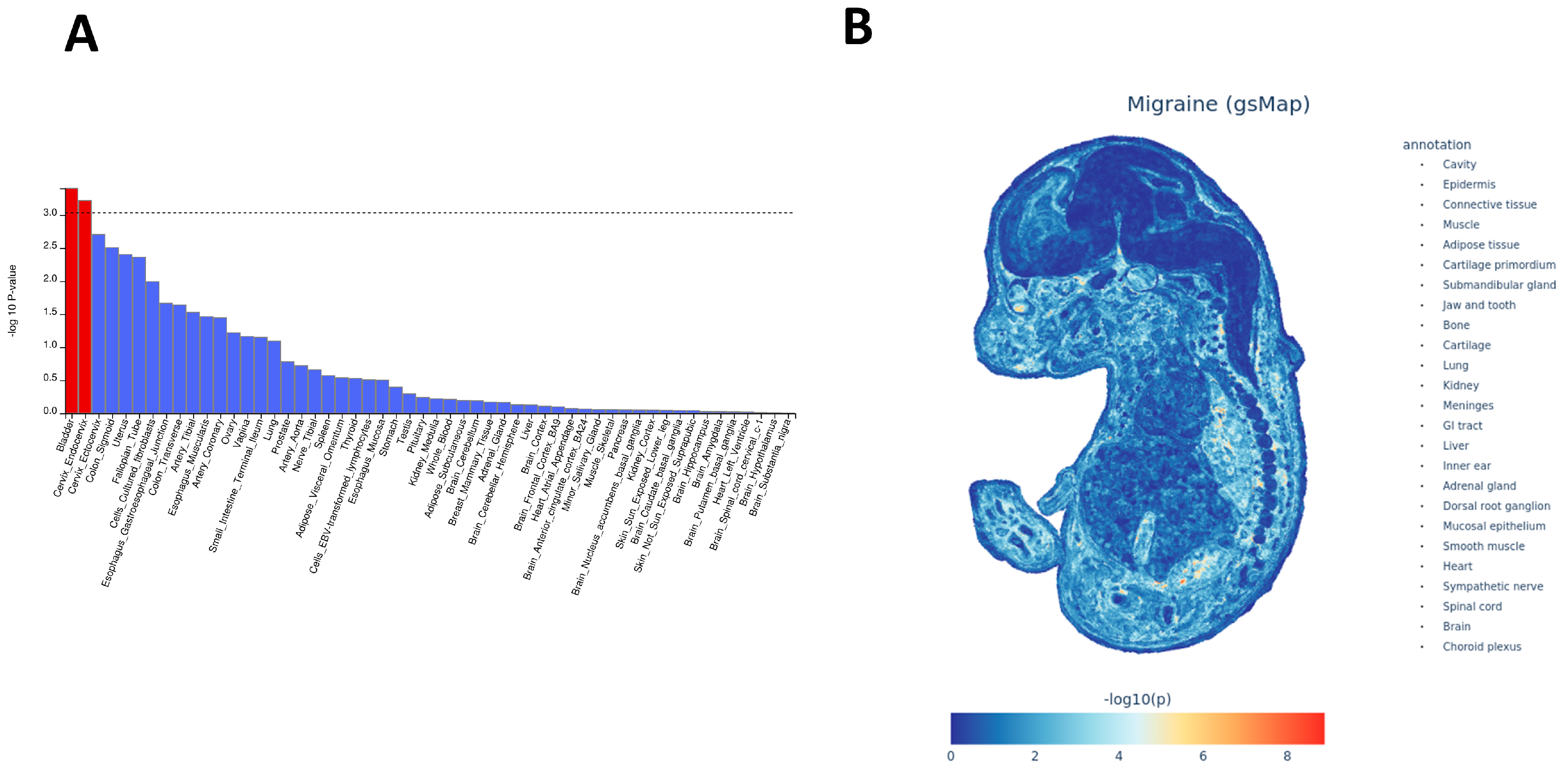
14] and underscores the importance of the vascular wall as a site where migraine risk alleles manifest their effects. Our MAGMA further highlighted tissues that combine mucosal, smooth muscle, and autonomic-innervation features—such as the bladder and the cervix endocervix. We interpret these findings with caution, as the biological link between these organs and migraine is not fully established. Rather than implying direct organ pathology, these signals likely reflect shared underlying biological properties, such as smooth muscle contractility and autonomic regulation. Consistent with this view, sub-threshold but top-ranked tissues included esophageal muscularis and other gastrointestinal mucosal structures. Complementary spatial transcriptomic mapping using the MOSTA atlas likewise pointed to visceral smooth muscle, peripheral autonomic and sensory ganglia, and craniofacial/meningeal interfaces. We acknowledge that migraine typically manifests in adolescence or adulthood; thus, these embryonic spatial signals should be interpreted through the lens of “developmental priming.” We hypothesize that genetic risk variants expressed during the organogenesis of the trigeminovascular system and autonomic ganglia may compromise their structural integrity or wiring. This developmental “imprinting” could establish a latent neurovascular vulnerability that predisposes individuals to migraine later in life, consistent with the developmental origins of health and disease framework. These signals are notable because gastrointestinal and genitourinary symptoms are common in migraine, and irritable bowel syndrome (IBS) in particular shows bidirectional comorbidity with migraine, with odds ratios around two to two-and-a-half in both directions in the meta-analysis [
25]. Such patterns are compatible with a shared gut–brain/autonomic dysregulation, in which mucosa–smooth muscle–vascular tone axes are co-affected in migraine and IBS [
26,
27,
28]. In addition, we observed spatial enrichment in regions corresponding to cranial meninges, trigeminal nerve territories, and related craniofacial mesenchyme, consonant with the established trigeminovascular model in which meningeal vessels and their innervation are central generators of migraine pain [
29]. Taken together, the tissue and spatial results support a view of migraine as a multisystem disorder engaging peripheral vascular and mucosal organs, autonomic and sensory nodes, and specific central structures, rather than a cortex-restricted brain disease.
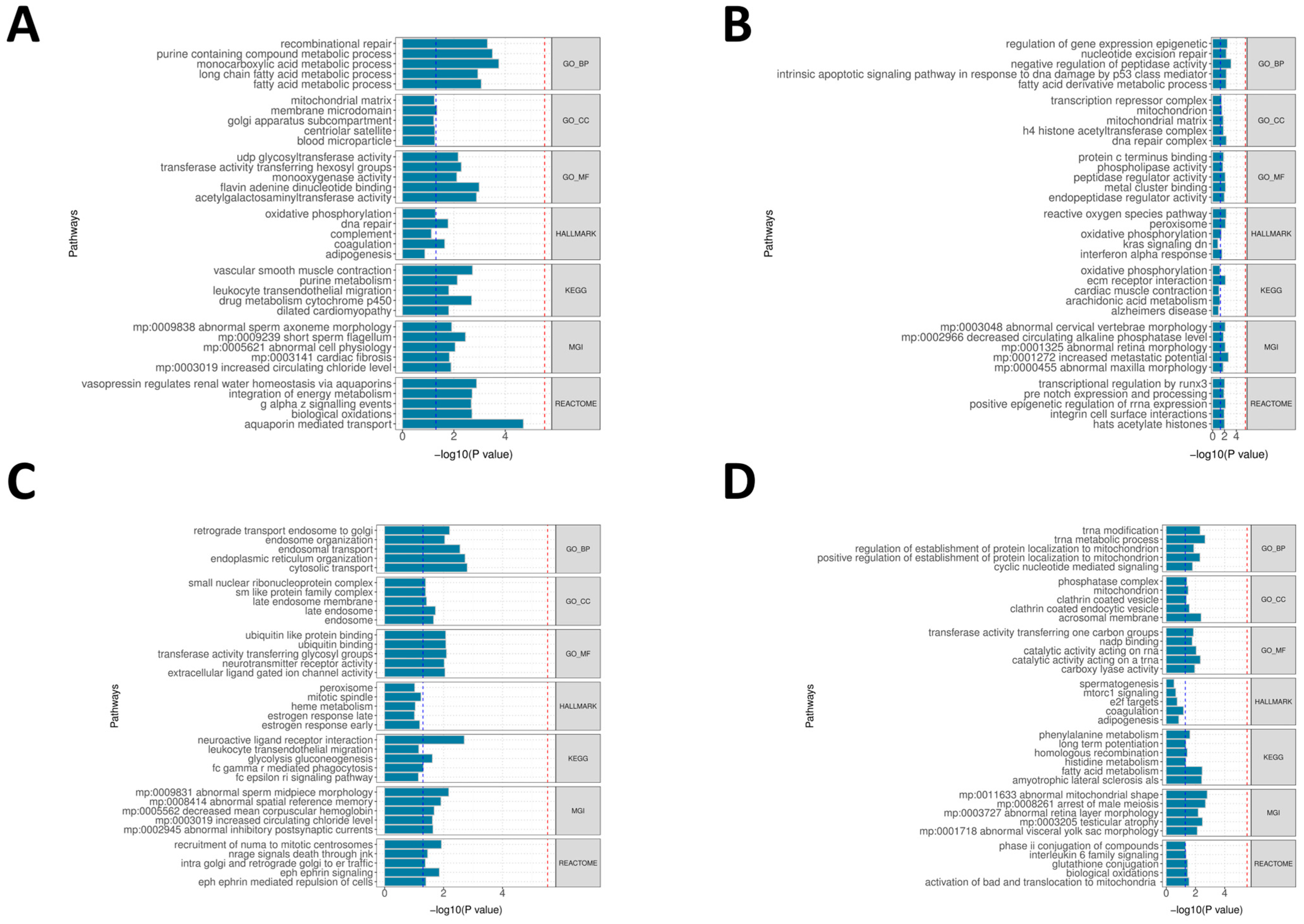
The pleiotropic nature of migraine genetics was further evident at the pathway and cell-type levels. Gene set enrichment analyses in migraine-relevant tissues (arteries, heart, tibial nerve, selected brain regions, and whole blood) highlighted several recurring biological themes: (i) vascular smooth muscle contraction and cytoskeletal or extracellular matrix processes, implicating regulation of vessel tone and structural integrity; (ii) metabolic and mitochondrial pathways, including oxidative phosphorylation and monocarboxylate/lipid metabolism, consistent with a role for bioenergetic stress and metabolic by-products in modulating neural excitability and neuroinflammation; (iii) neuroactive ligand–receptor interaction and ion-channel activity, highlighting neurotransmitter, neuropeptide, and neuromodulator systems; and (iv) immune and inflammatory pathways, including leukocyte activation and interferon signaling, indicating that both innate and adaptive immune processes contribute to migraine susceptibility. These pathway-level findings resonate with current pathophysiological models in which vascular reactivity, CGRP/PACAP signaling, mitochondrial function, and inflammatory mediators all shape migraine attack thresholds and phenotypes [
16,
30,
31]. In particular, the enrichment of immune pathways is in line with comprehensive evidence from clinical and experimental studies that migraine is associated with altered cytokine profiles, complement components, and immune cell function [
18,
32].
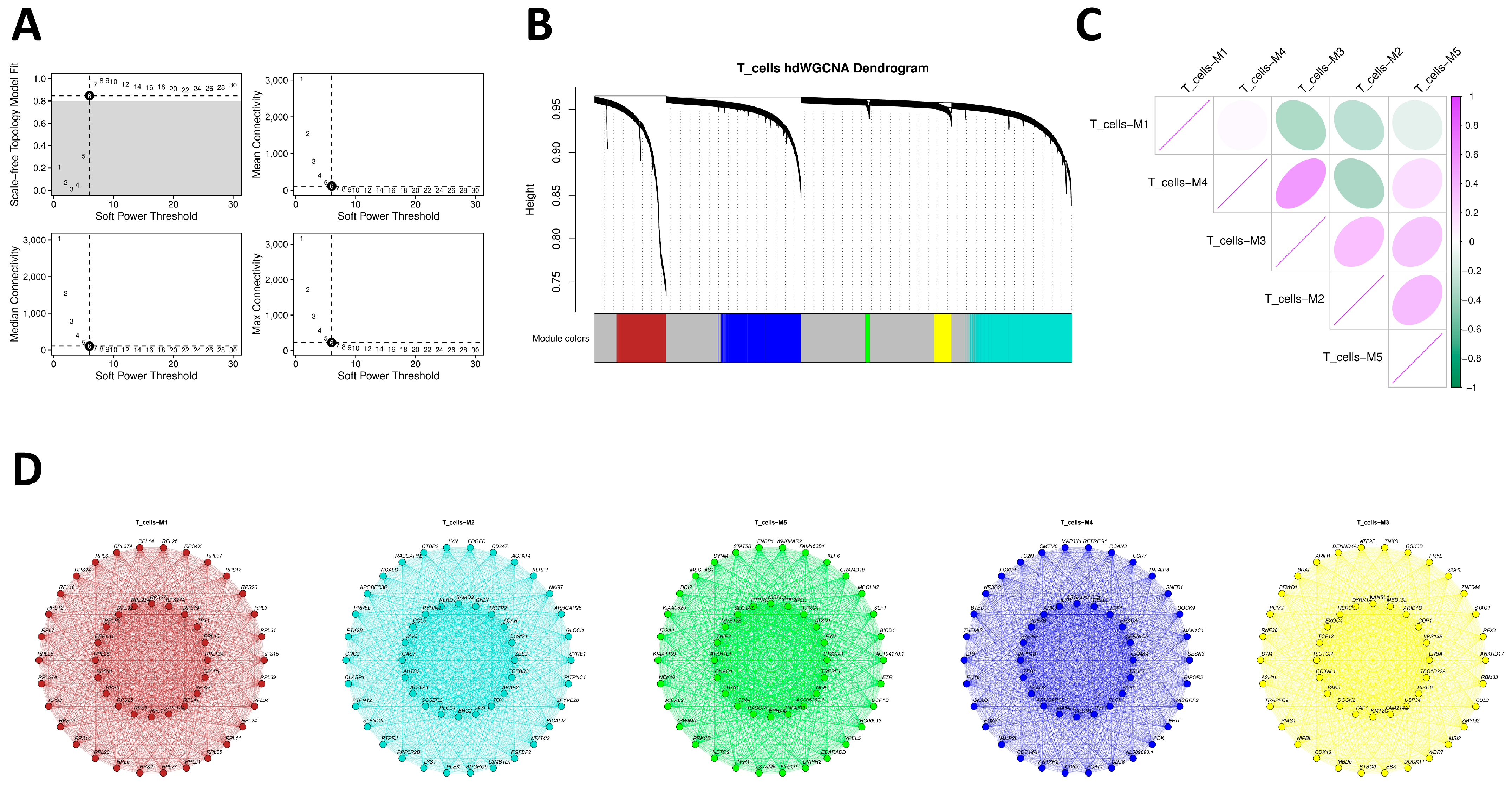
In terms of specific cell types, our heritability partitioning and cell-type enrichment analyses consistently prioritized vascular wall cell lineages. Using CELLECT, migraine-associated variants showed significant enrichment in multiple endothelial subtypes, vascular smooth muscle cells, pericytes, and related fibroblast/myofibroblast populations across independent reference panels, whereas brain cell-type signals were more modest and selective, with nominal enrichment in an inhibitory interneuron subset. This pattern supports the concept of a “neurovascular unit” in which vessel wall cells and local neural/glial elements jointly integrate genetic risk [
33,
34]. The inhibitory interneuron signal also aligns with evidence for cortical hyperexcitability and impaired inhibition, particularly in migraine with aura, and suggests that genetic variants influencing inhibitory circuits may modulate thresholds for spreading depolarization and sensory hypersensitivity [
35,
36].
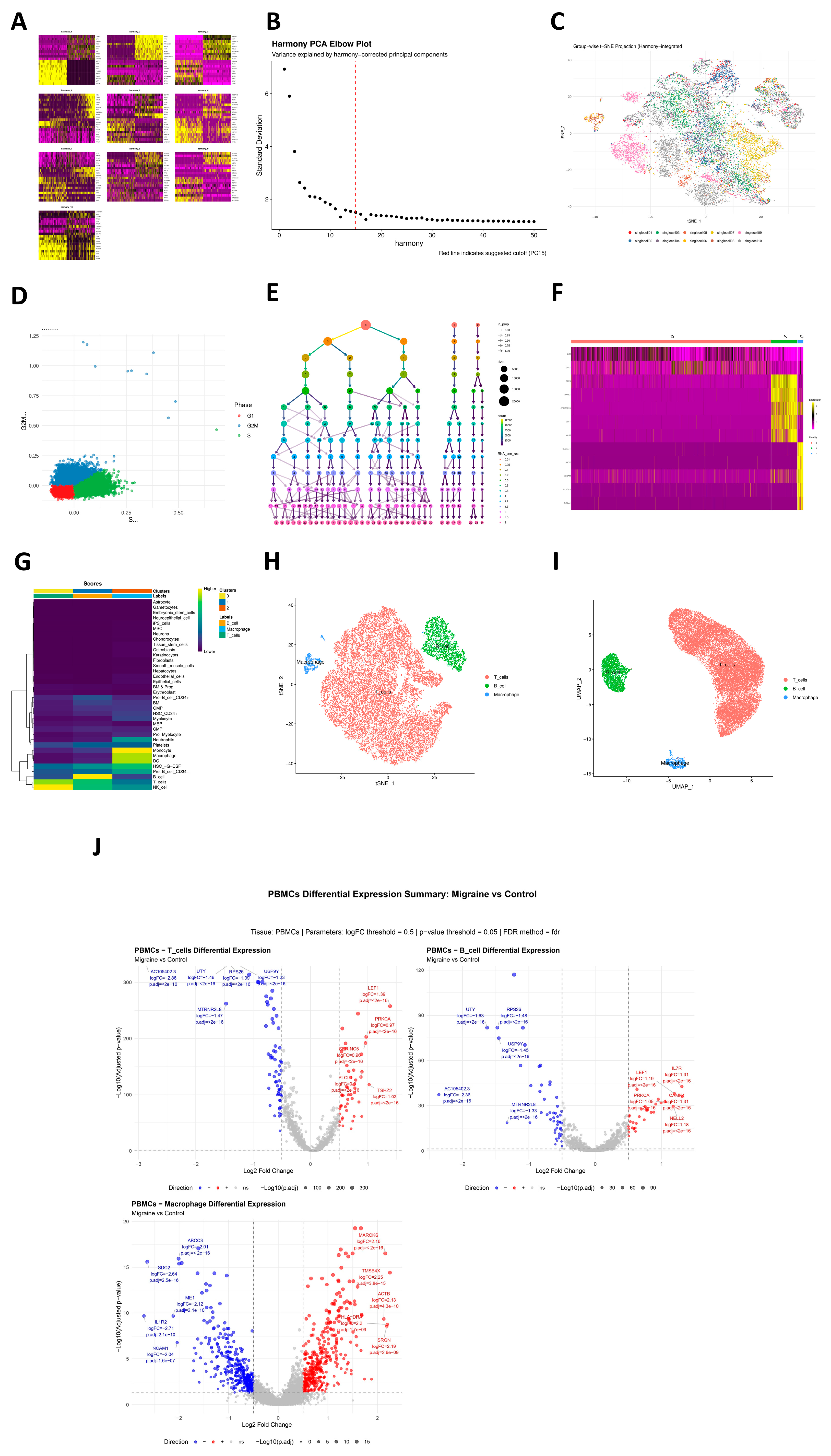
By contrast, our analyses did not identify significant heritability enrichment in any specific peripheral blood immune cell type, and ECLIPSER did not detect BH-significant enrichment of GWAS signals in PBMC-derived regulatory annotations. This absence of strong enrichment in circulating leukocytes is consistent with a recent Mendelian randomization study that found no evidence for a causal effect of major autoimmune diseases on migraine risk or for substantial genetic correlation between them [
37], despite the epidemiological co-occurrence of migraine with conditions such as rheumatoid arthritis [
21]. Reflecting this, our integrated prioritization system assigned a lower priority index (0.5) to PBMC subsets compared with vascular lineages (1.0). However, the lack of global heritability enrichment does not preclude specific risk genes from functioning within immune cells. Nonetheless, hdWGCNA in T cells revealed co-expression modules enriched for cytokine signaling, cytotoxicity, and T-cell receptor signaling, within which some co-localized genes (such as PTK2B) reside. These modules may reflect genetically primed immune programs that are activated under specific stimuli in migraine, a possibility that warrants further functional work and dovetails with clinical data on the benefit of immune-targeted co-therapies in selected patients [
22].
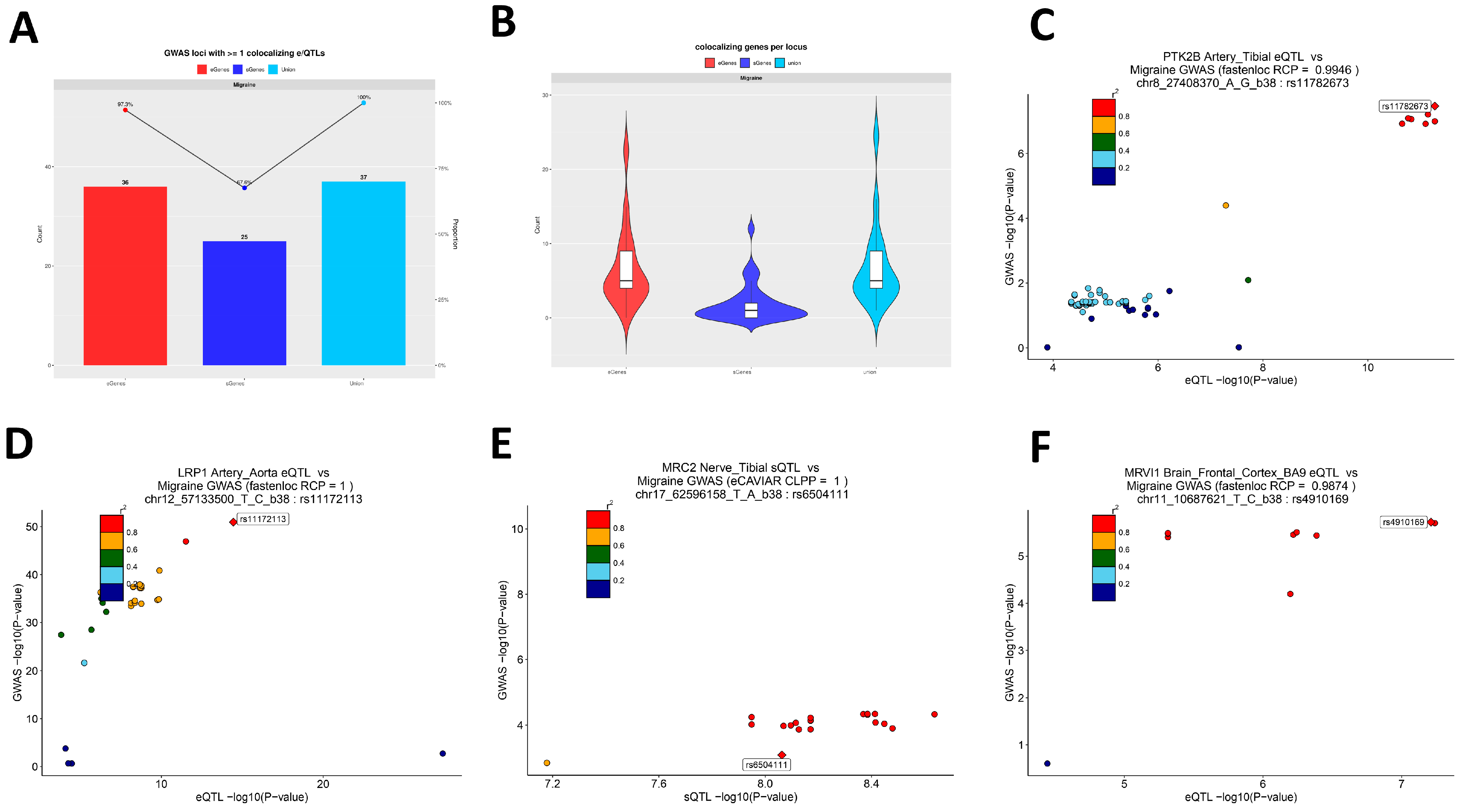
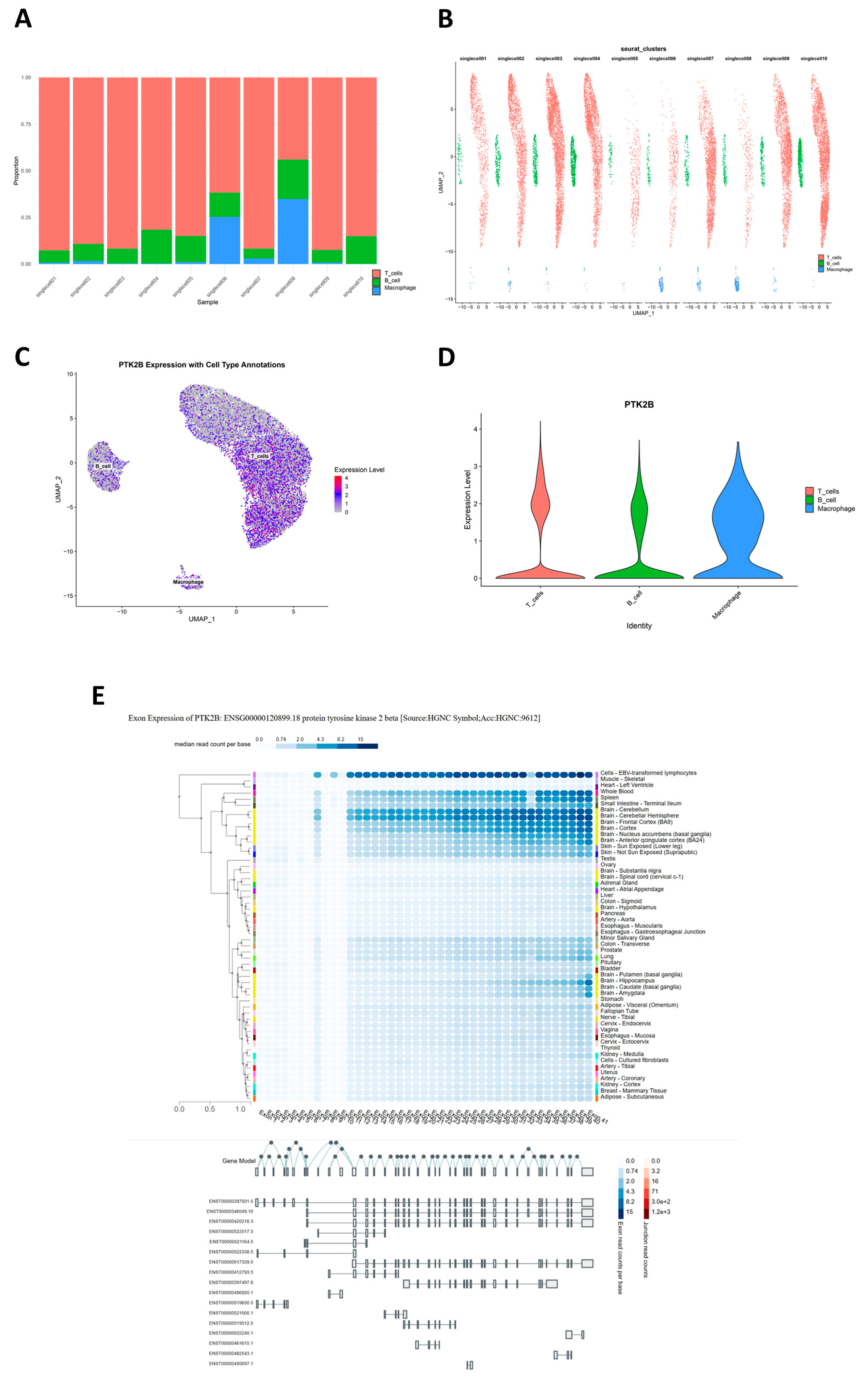
By integrating FUMA locus discovery with Bayesian co-localization (eCAVIAR, fastENLOC) across 49 GTEx tissues, we were able to nominate candidate effector genes for nearly all migraine GWAS loci. At 36 of 37 loci, we identified at least one co-localizing eQTL gene and at 25 loci at least one sQTL gene, with most loci harboring multiple plausible effectors. This supports a model in which common variants act largely through perturbations of gene expression or splicing in specific tissues. Importantly, many of the prioritized genes have functions that are highly compatible with migraine biology. As a representative example of this multi-omics convergence, PTK2B (encoding the non-receptor tyrosine kinase PYK2) showed strong co-localization with eQTLs in arterial and myocardial tissues as well as sQTLs in multiple cortical regions and was embedded within an activation/cytotoxicity module in T cells. PYK2 is a calcium-sensitive kinase involved in integrin, neurotransmitter, and immune signaling, offering a mechanistic link among vascular, neural, and immune compartments that is coherent with our multi-scale findings [
38,
39]. LRP1, which we identified through arterial eQTL co-localization at a genome-wide significant locus, encodes a multifunctional endocytic receptor that influences lipid trafficking, endothelial barrier function, and cell signaling [
40]. Recent work has shown that the LRP1–SHP2 pathway modulates TRPV1 sensitivity in peripheral nociceptors and that amyloid β1–42 can alter this axis to change pain thresholds [
41]. These data provide convergent support for LRP1 as a modulator of nociceptive processing at the neurovascular–neural interface, consistent with its prioritization in our vascular-focused analyses. Another example is MRVI1, which we found to co-localize with cortical eQTLs at a migraine risk locus. MRVI1 is involved in NO–cGMP–IP3 receptor signaling and regulates intracellular calcium release in smooth muscle cells and neurons, placing it in a pathway central to vasodilation and excitability [
42,
43,
44]. Taken together, such genes illustrate how integrating GWAS with tissue QTLs and cell-level data can refine effector candidates from broad loci to specific molecules with testable roles in migraine pathophysiology.
Our findings also align with and extend recent integrative genomics work that has begun to propose druggable targets in migraine. A multi-omics Mendelian randomization study integrating GWAS, eQTL, and protein quantitative trait loci (pQTL) data highlighted GSTM4, a glutathione S-transferase gene involved in oxidative stress responses, as a promising therapeutic target [
45]. Separately, machine learning-driven analysis of gene programs in migraine identified key transcription factors and regulatory modules enriched for synaptic and calcium-signaling pathways, with additional involvement of inflammatory processes [
46]. Moreover, analysis of a familial migraine–epilepsy phenotype combined with GWAS information has implicated NCOR2, a nuclear corepressor that coordinates inflammatory and neuronal gene expression programs, as a candidate gene linking neuronal hyperexcitability and migraine susceptibility [
47]. These independent studies converge with our observation that migraine genes cluster in networks governing ion channels, second-messenger cascades, synaptic function, and immune–metabolic regulation. Importantly, several clinically validated targets, such as CALCA/CALCB (CGRP ligands) and HTR1F (5-HT1F receptor), reside in migraine loci in the large GWAS [
14]. The fact that unbiased genetic and functional datasets repeatedly point to targets already proven by pharmacology lends weight to newly prioritized genes like PTK2B, LRP1, MRVI1, and GSTM4, which may represent the next generation of mechanistic and therapeutic candidates.
Several limitations of our approach warrant consideration. First, most of the QTL and single-cell reference datasets we used were generated from non-migraine donors and represent baseline regulatory architecture rather than disease or attack states. While such resources are indispensable for mapping genetic effects, they cannot capture dynamic transcriptomic or epigenomic changes during migraine attacks, nor can they reflect the disease-driven remodeling of tissues. Future studies incorporating genotype-informed omics in migraine patients—ideally sampling key tissues such as trigeminal ganglia, meninges, and hypothalamus during relevant phases—will be crucial to adding disease-context specificity. Second, tissue coverage remains incomplete. Critical migraine-relevant structures, including meningeal arteries, trigeminal ganglia, specific brainstem nuclei, and hypothalamic subnuclei, are absent or sparsely represented in current eQTL and single-cell QTL catalogs. Our reliance on proxies (e.g., tibial nerve for peripheral nociceptors and major arteries for meningeal vessels) introduces uncertainty and may obscure highly localized effects. Expanding QTL and single-cell resources to these specialized tissues and cell types is therefore a priority for the field. Third, our single-cell analyses of immune cells were limited to PBMCs in a modest-sized case–control cohort. While we did not observe clear genetic enrichment in peripheral immune subsets, this does not exclude important roles for tissue-resident immune cells, nor does it capture non-genetic drivers of immune activation in migraine. Integrating larger, ethnically diverse cohorts with the multi-omics single-cell profiling of central and peripheral neuroimmune interfaces will help resolve these questions. Fourth, the gsMap-based projection of human migraine GWAS data onto a mouse embryonic spatial atlas is inherently indirect. While this approach effectively identifies the developmental origins of susceptible tissues, embryonic expression patterns may only partially recapitulate adult tissue architecture and disease-relevant states. Therefore, these spatial enrichments likely reflect the genetic establishment of structural susceptibility (e.g., vascular tone or neural connectivity) rather than the active pathophysiology of a migraine attack. We therefore interpret spatial hotspots as hypothesis-generating, pointing to candidate organ systems (e.g., craniofacial mesenchyme, sympathetic chain, and visceral smooth muscle) rather than as definitive localization. Finally, our analyses, like all polygenic studies, describe probabilistic shifts in pathway activity and tissue liability, not deterministic outcomes. Migraine risk alleles increase susceptibility but operate in concert with environmental exposures, hormonal factors, and epigenetic mechanisms. Experimental validation—through gene editing, pharmacologic perturbation, and in vivo models—is essential to confirming causal roles for the genes and pathways we have prioritized and to delineate their interactions.
4. Materials and Methods
4.1. Study Design and Ethics
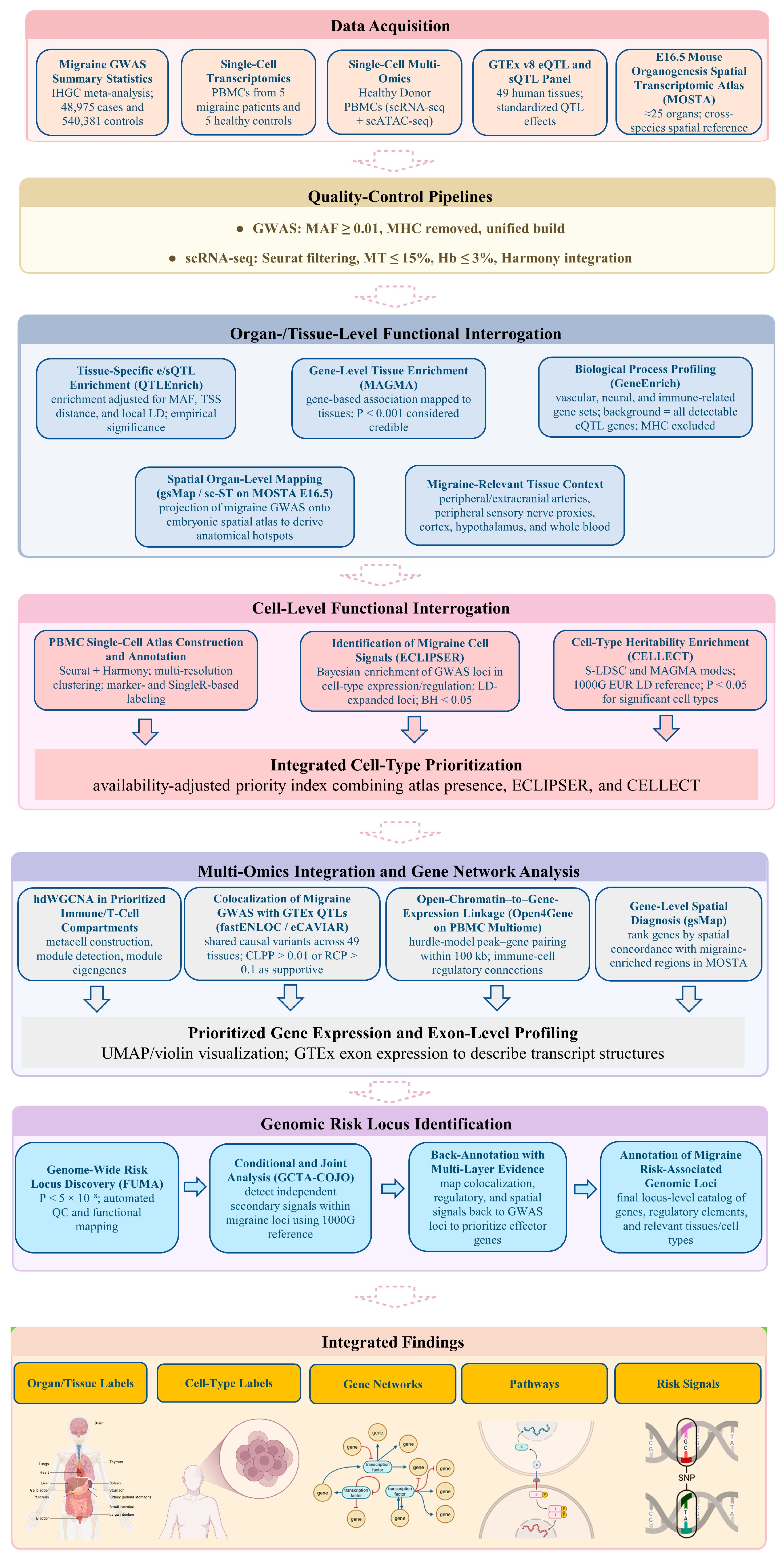
All data included in this study complied with relevant ethical standards. An overview of the study design and analysis pipeline is outlined in
Figure 9.
4.2. Data Sources
4.2.1. Migraine Genome-Wide Association Summary Statistics
We obtained summary-level genome-wide association study (GWAS) statistics for migraine from the large-scale meta-analysis conducted by the International Headache Genetics Consortium (IHGC), as reported by Hautakangas et al. in 2022 [
14]. This dataset combines four major cohorts of European ancestry (IHGC2016, UK Biobank, Manchester, UK, GeneRISK, Budapest, Hungary, and HUNT), comprising 48,975 individuals with migraine and 540,381 migraine-free controls. Migraine case status in the contributing cohorts was defined either by self-reported physician-diagnosed migraine or by meeting the International Classification of Headache Disorders criteria. Association analyses within each cohort were adjusted for age, sex, and population structure prior to meta-analysis. Due to data-sharing restrictions, individual-level and cohort-level summary statistics from the 23andMe cohort were excluded from the version of the dataset used in our study to protect participant privacy. All contributing studies obtained approval from local institutional review boards or ethics committees, and all participants provided informed consent. We analyzed only de-identified, aggregate-level summary statistics and did not access any individual-level genotype or clinical data.
4.2.2. Single-Cell Transcriptomic Data
The single-cell transcriptomic data analyzed in this study were generated from an ethics-approved case–control clinical cohort. Adult participants were recruited across multiple neurology and otolaryngology centers in Spain and included individuals diagnosed with migraine as well as neurologically healthy volunteers without a history of migraine. All participants provided written informed consent prior to blood collection, and the study protocol was approved by the institutional ethics committees of the participating hospitals. Peripheral blood was drawn, and peripheral blood mononuclear cells (PBMCs) were isolated and processed under standardized conditions. Isolated cells/nuclei were encapsulated using the 10× Genomics Chromium Next GEM platform to generate single-cell RNA sequencing (scRNA-seq) and single-cell assay for transposase-accessible chromatin using sequencing (scATAC-seq) libraries. Libraries were sequenced on Illumina high-throughput platforms, and raw data were processed with the Cell Ranger/Cell Ranger ARC pipeline to produce gene-by-cell count matrices for downstream single-cell expression analysis. For downstream analyses in the present work, we used the PBMC single-cell transcriptome matrices from migraine cases and healthy controls within this cohort (GSE269117), comprising five individuals with migraine and five healthy control individuals [
48].
4.2.3. Healthy Human Peripheral Blood Mononuclear Cell (PBMC) Single-Cell Multi-Omics (ATAC + Gene Expression) Dataset
Using 10× Genomics Chromium single-cell multi-omics technology, PBMCs from a 25-year-old healthy female donor were analyzed. After removing granulocytes via flow cytometry sorting, cell nuclei were isolated according to the 10× Genomics standard protocol (CG000365 Rev A), yielding approximately 11,909 high-quality cells. The experiment followed the Chromium Next GEM single-cell multi-omics protocol (CG000338 Rev A) to construct paired ATAC-seq and gene expression libraries. Sequencing was performed on the Illumina NovaSeq 6000 (Illumina, San Diego, CA, USA) platform using a paired-end dual-index strategy (gene expression library cycles: 28-10-10-90; ATAC library cycles: 50-8-16-49). The final high-quality dataset included gene expression data with a median of 1826 genes and 3776 unique molecular identifier (UMI) counts per cell, as well as chromatin accessibility data with a median of 13,486 high-quality fragments per cell. A total of 108,377 open chromatin peaks and 15,494 genes were detected, with 85,468 successful peak–gene linkages, providing a reliable multi-omics dataset for investigating immune cell heterogeneity and epigenetic regulatory mechanisms.
4.3. Quality Control
Prior to analyzing the GWAS data, we implemented a series of quality-control (QC) steps to ensure data accuracy and reliability. The detailed methods for GWAS data QC are as follows: To ensure representativeness and reduce false positives, we first calculated the minor allele frequency (MAF) for each single-nucleotide polymorphism (SNP). By setting an MAF threshold of ≥0.01, low-frequency variants that could introduce substantial uncertainty were filtered out. SNPs below this threshold were removed to minimize bias and enhance data reliability. Due to the high linkage disequilibrium (LD) in the major histocompatibility complex (MHC) region (located on chromosome 6), which may affect GWAS analysis accuracy, SNPs in this region were removed. To ensure consistency in genome build and data format, we converted the GWAS data into a unified version and standard format for further analysis. For single-cell transcriptomic data, we applied a systematic QC workflow. First, after loading and initializing the data using the Seurat package, we calculated the percentages of mitochondrial genes (marked by “^MT-”) and hemoglobin genes (including HBA1, HBA2, HBB, etc.) to assess cell quality. Strict filtering criteria were then applied to retain high-quality cells, requiring each cell to have at least 1000 UMIs, between 200 and 5000 detected genes, a mitochondrial gene percentage ≤ 15%, and a hemoglobin gene percentage ≤ 3%. Dimensionality reduction was performed using principal component analysis (PCA), t-distributed stochastic neighbor embedding (t-SNE), and uniform manifold approximation and projection (UMAP), with quality evaluated via heatmaps and elbow plots. To eliminate batch effects from sample sources, we used the Harmony algorithm for integration correction (parameters: theta = 2, lambda = 1).
4.4. Tissue-Specific e/sQTL Enrichment Analysis Using QTLEnrich
Using Genotype-Tissue Expression (GTEx) v8 data across 49 tissues (splicing quantitative trait loci [sQTLs]/expression quantitative trait loci [eQTLs]), we evaluated migraine-related tissue-level associations. QTLEnrich is a rank- and permutation-based method for assessing whether phenotype associations are enriched in eQTLs and sQTLs in specific tissues, quantifying the statistical significance of these enrichments. This method accounts for three potential factors: MAF, distance to the target gene transcription start site, and local LD. Adjusted fold enrichment and enrichment p-value were used to evaluate the significance of the QTLEnrich method. To assess the significance and reliability of the e/sQTL enrichment analysis results in QTLEnrich, we first obtained the p-value for each SNP’s association with the phenotype. These p-values were then -log10-transformed for more intuitive visualization. Next, we plotted a quantile–quantile (QQ) plot with the x-axis as the −log10 of theoretical p-values (assuming that all SNPs are unrelated to the phenotype and follow a uniform distribution) and the y-axis as the −log10 of observed p-values.
4.5. Tissue-Specific Enrichment Analysis Using MAGMA
To expand on the QTLEnrich enrichment, we employed Multi-marker Analysis of GenoMic Annotation (MAGMA) enrichment analysis to explore migraine-related genomic features. First, migraine data were formatted for MAGMA. We then used the MAGMA tool for gene-level enrichment analysis, considering p < 0.001 indicative of credible tissues.
4.6. Single-Cell Spatial Transcriptomics for Migraine Tissue Enrichment Specificity
To investigate migraine, this phase integrated single-cell spatial transcriptomics (sc-ST) data with GWAS statistics to map migraine-related cellular spatial distribution patterns at single-cell resolution. We used a genetically informed spatial mapping of cells for complex traits (gsMap) algorithm, which integrates cross-species analyses of mouse embryo/brain tissues, rhesus macaque cerebral cortex, and human GWAS data to identify spatial distribution patterns of disease-related cell populations. Its core principle involves mapping trait-related genes derived from GWASs to spatially resolved cells based on their expression patterns, thereby evaluating associations between specific anatomical regions and complex traits at the cellular level. Based on the spatial transcriptomic atlas of mouse embryos at embryonic day 16.5 (E16.5; covering 25 organs), we generated migraine enrichment specificity maps along with gene spatial expression maps, establishing a single-cell resolution spatial pathogenic mechanism atlas for migraine.
4.7. Analysis of Migraine-Related Tissue Biological Functional Processes Using GeneEnrich
We employed gene set enrichment analysis to analyze genetic variants and expression profiles associated with migraine. This study used the GeneEnrich tool, applying hypergeometric and permutation tests to evaluate the enrichment of candidate gene sets in neurological/vascular regulation/immune-related biological functions or phenotype gene sets. To reduce tissue-specific bias, we used all detectable genes with eQTLs in each tissue as the background set, excluding genes in the MHC region, and calculated empirical p-values via permutation tests. This study focused on tissues and tissue proxies highly relevant to migraine pathophysiology, including peripheral and extracranial vascular-related tissues (Artery—Tibial, Artery—Aorta, and Artery—Coronary), peripheral sensory nerve proxies (Nerve—Tibial, approximating trigeminal sensory neurons/nociceptive afferent pathways), cerebral cortical regions (Brain—Cortex and Brain—Anterior cingulate cortex [BA24], reflecting cortical excitability and pain integration), and hypothalamus (Brain—Hypothalamus, reflecting autonomic/neuroendocrine-driven prodromal migraine regulation), while including Whole Blood to assess systemic immune and inflammatory contributions. The functional gene sets were sourced from Gene Ontology, Reactome, Kyoto Encyclopedia of Genes and Genomes (KEGG), Molecular Signatures Database, and Mouse Genome Informatics-related biological function databases. Statistical significance was determined as empirical p < 0.05 within each database for nominal significance.
4.8. Single-Cell Transcriptomic Atlas Annotation for Identification of Migraine Cell Signals
In this study, we conducted detailed analysis of pre-processed single-cell data, first using Harmony-corrected dimensionality reduction results for multi-dimensional visualization, including principal component heatmaps, elbow plots, and t-SNE and Harmony dimensionality reduction projection visualizations. We also evaluated cell-cycle distribution by quantifying S-phase and G2M-phase scores. Based on the Harmony dimensionality reduction results, we constructed cell neighborhoods and applied a multi-resolution strategy (13 resolutions from 0.01 to 3.0) for clustering, determining the optimal resolution of 0.01 via clustering trees to obtain stable cell subpopulations. To identify molecular features of each cell cluster, we used the FindAllMarkers function (minimum expression proportion of 25%, log-fold change threshold of 0.25) to screen cluster-specific highly expressed genes and generated heatmaps based on marker genes. Cell-type annotation was achieved using the SingleR algorithm for automated annotation.
4.9. Identification of Migraine Cell Signals Using ECLIPSER
Enrichment of Causal Loci and Identification of Pathogenic cells in Single Cell Expression and Regulation data (ECLIPSER) was configured with background GWAS locus scores using Bayesian Fisher’s exact test. With background GWAS locus sets as controls, we estimated cell-type-specific enrichment fold and p-values for each trait (GWAS locus set), tissue, and cell-type combination, where the cell-type specificity threshold was set to the 95th percentile of background locus scores. The Bayesian method estimates the 95% confidence interval (CI) for enrichment fold, suitable for traits with few loci or no loci exceeding the enrichment threshold, based on multi-level annotation data from functional genomics platforms. The specific workflow first determines the target trait and its significantly associated genetic loci based on GWAS results and then expands original signals using LD proxy relationships (r2 > 0.8) to comprehensively cover potential functional variants. In the preparation phase, Wilcoxon tests were used for inter-group differential expression analysis within each cell type, with key parameters: minimum cells per group = 3, minimum gene expression proportion = 10%, and log2 fold-change significance threshold = 0.5. For genes meeting differential expression criteria in specific cell types (adjusted p < 0.05 and absolute log2 fold-change > 0.5), ECLIPSER significance was controlled using Benjamini–Hochberg (BH) correction, with BH ≤ 0.05 cell types deemed statistically significant enriched clusters.
4.10. Identification of Migraine Cell Signals Using CELLECT
To assess the contribution of cell-type-specific expression to disease heritability, we used the CELL-type Expression-specific integration for Complex Traits (CELLECT) framework with two complementary methods: stratified LD score regression (S-LDSC) based on heritability and MAGMA gene analysis based on gene sets. We applied S-LDSC to analyze GWAS summary statistics for migraine. The cell-type datasets defining the analysis baseline were sourced from the tabula_muris-test and mousebrain-test datasets. LD scores were calculated using European samples from 1000 Genomes Project Phase 3 as the reference panel. p < 0.05 was considered significant enrichment. The MAGMA gene set analysis module tested correlations between gene-level association statistics and average gene expression levels in specific cell types. MAGMA analyzed paired combinations for given cell types to identify characteristic association signals independent of other important cell types. Similar to S-LDSC, p < 0.05 was considered significant enrichment.
4.11. Integration of Multi-Dimensional Single-Cell Evidence for Cell Prioritization
To prioritize putative disease-relevant cell types in migraine, we integrated three complementary sources: (i) a PBMC single-cell atlas (presence of annotated clusters); (ii) ECLIPSER (cell-type enrichment of GWAS signals in PBMC regulatory annotations); and (iii) CELLECT (including S-LDSC and MAGMA-based cell-type-specific expression). Each evidence source contributed one equal weight; to avoid coverage bias, non-applicable methods were flagged NA and excluded from the denominator, yielding an availability-adjusted priority index (priority index = positive/applicable). The atlas scored by presence; ECLIPSER and CELLECT scored by nominal significance (p < 0.05).
4.12. Weighted Gene Co-Expression Network Analysis in Preferred Cells to Identify Core Module Genes
Given the limited sample size (n = 10) and the lack of global heritability enrichment in the ECLIPSER analysis, we consider the following PBMC-based findings as exploratory. To describe molecular co-expression modules in the immune compartment, we performed high-dimensional weighted gene co-expression network analysis (hdWGCNA) on annotated T cells derived from the combined dataset of migraine cases and healthy controls. This pooling strategy was employed to maximize cell numbers for the construction of robust, reference-level co-expression networks independent of transient disease states. First, the annotated single-cell Seurat object was loaded, and the target cell type was set for analysis. The WGCNA function initialized the WGCNA analysis environment, selecting genes expressed in at least 5% of cells as the candidate gene set. MetacellsByGroups constructed metacells based on cell type and sample source (k = 25) to reduce single-cell data sparsity while preserving biological variation, followed by normalization and standardization of metacell expression data. After PCA dimensionality reduction, Harmony correction was employed for batch effects from sample sources. To construct the gene co-expression network, SetDatExpr extracted the expression matrix for the target cell type, and TestSoftPowers tested soft-threshold powers to select an appropriate soft threshold for building a signed topology overlap matrix network. Dynamic tree cutting identified co-expression modules and calculated module eigengenes. Further analyses examined module associations with cell types, calculated module connectivity, and renamed modules for enhanced interpretability.
4.13. Migraine Genomic Risk Loci Analysis
In this study, we used the Functional Mapping and Annotation (FUMA) platform for identification and annotation of genomic risk loci. First, we uploaded summary statistics files from the migraine GWAS, including SNP identifiers and genomic LD format reference values. The FUMA platform performed initial QC on these GWAS results, removing missing values and low-quality SNPs. By setting an appropriate significance threshold (p < 5 × 10−8), we screened genomic risk loci associated with migraine.
4.14. Conditional Analysis of Migraine Genomic Risk Loci
We performed conditional analysis on high-precision risk loci obtained from migraine risk loci to investigate whether independent secondary signals co-localized with significant GWAS signals within loci. Specifically, using the lead variant of each locus as the conditioning variable, we applied the Genome-wide Complex Trait Analysis (GCTA)-COJO tool for conditional association analysis on summary statistics. Variant allele frequencies required for analysis were sourced from the 1000 Genomes Project.
4.15. eCAVIAR Analysis of Migraine Genomic Loci
To identify high-confidence genes and regulatory mechanisms (eQTL/sQTL) potentially associated with migraine common risk loci, we used the Bayesian co-localization method eCAVIAR. This method evaluates whether co-occurring GWAS and e/sQTL signals tag the same causal variant or haplotype, accounting for local LD and allelic heterogeneity. eCAVIAR includes fine-mapping functionality and can handle large GWAS summary statistics without genotype data. In the analysis, we assumed up to two independent causal variants per locus. Inputs were Z-scores (beta divided by standard error) for variants from GWAS and GTEx e/sQTL studies. The LD window around each GWAS lead variant was defined as the chromosomal region containing variants with r2 > 0.1 (calculated using 1000 Genomes Project Phase 3 corresponding population as reference panel), extended by 50,000 bp on each side. GWAS–e/sQTL–tissue combinations with eCAVIAR co-localization posterior probability (CLPP) > 0.01 were considered significant.
4.16. fastENLOC Analysis of Migraine Genomic Loci
To finely identify genes and regulatory mechanisms (eQTL/sQTL) mediating associations at common risk loci for migraine, we further used the Bayesian co-localization method fastENLOC. This method uses its embedded Deterministic Approximation of Posteriors for Genetics (DAP-G) algorithm to fine-map GWAS and e/sQTLs separately, estimating posterior probabilities for each variant being causal and then assessing co-localization probability for shared causal variants without pre-setting an upper limit on independent causal variants per locus. Inputs were Z-scores (effect size divided by standard error) for variants from GWAS and GTEx e/sQTL summary data. The LD window for each GWAS lead variant was defined as the chromosomal region containing variants with r2 > 0.1 (using 1000 Genomes Project Phase 3 corresponding population as reference), extended by 50,000 bp on each side. We tested migraine GWAS loci against 49 GTEx tissues, with co-localization results being expressed as regional co-localization probability (RCP). As per method recommendation, GWAS–e/sQTL–tissue combinations with RCP > 0.1 were considered to have significant co-localization evidence.
4.17. Open Chromatin-to-Gene Expression Analysis Using Open4Gene
The purpose of open chromatin-to-gene expression analysis is to explore genes expressed in immune cells from normal tissues to determine whether stable genes in immune cells have potential associations with cells annotated in the migraine single-cell transcriptomic atlas. Open4Gene is a hurdle model-based statistical method specifically designed to handle zero-inflation common in single-cell data [
49]. The analysis workflow includes the following key steps: First, scRNA-seq and scATAC-seq data were normalized, dimensionality-reduced, and clustered using Seurat and Signac packages. RNA expression matrices, ATAC peak matrices, and cell metadata (including cell-type annotations and technical covariates) were extracted from the Seurat object. ATAC peaks were then linked to gene promoter regions using a 100 kb window to define peak–gene pairs. A two-component hurdle model tested associations for each peak–gene pair: the zero component used binomial distribution (logit link) to model the relationship between ATAC openness and zero gene expression probability; the count component used truncated negative binomial distribution (log link) to model ATAC signal intensity and gene expression levels (when non-zero).
4.18. gsMap Gene-Level Spatial Diagnosis
After converting standardized migraine GWAS summary statistics (including variant positions, alleles, effect sizes, standard errors, p-values, and sample sizes) to the required trait input format for gsMap, we projected the trait onto the E16.5 mouse embryo Mouse Organogenesis Spatial Transcriptomic Atlas (MOSTA) as reference. gsMap first constructs disease-related spatial enrichment maps based on GWAS signals on the spatial atlas, then calculates consistency metrics for all detectable genes in the atlas with this spatial enrichment map, generating gene-level spatial diagnosis results. These include embryonic tissue/anatomical annotations for the gene, median gene-specificity score (Median_GSS), and Pearson correlation coefficient (PCC) between gene spatial expression and disease spatial enrichment. After analysis, genes were ranked by diagnosis scores for subsequent visualization.
4.19. Preferred-Gene Cell Expression Annotation and Exon Expression Analysis
Stacked bar charts and grouped UMAP were used to display cell composition proportions across samples, and specific genes (from fastENLOC analysis, eCAVIAR analysis, Open4Gene analysis, gsMap, and preferred cell weighted gene co-expression network analysis) were annotated for cell expression, including violin plots and UMAP expression projections. All analysis results were saved as high-quality image files and structured data tables. Using exon expression from the GTEx project, preferred genes were annotated for transcript types, structural features of each transcript, and expression distribution differences across human tissues.