Candida albicans Extracellular Vesicles Upregulate Nrg1 Transcription Repressor to Inhibit Self-Hyphal Development and Candidemia
Abstract
1. Introduction
2. Results
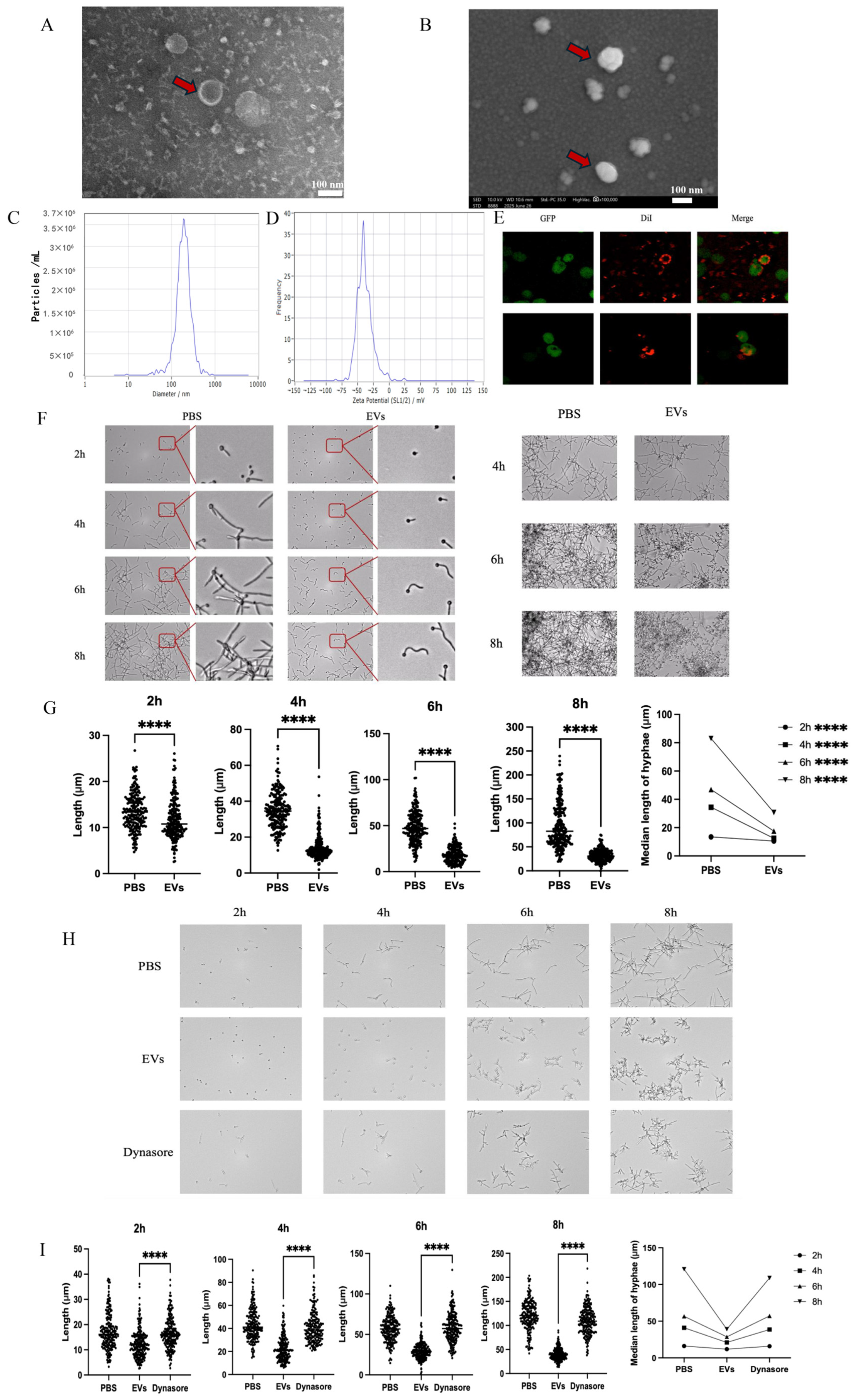
2.1. Extracellular Vesicles Inhibited C. albicans Hyphal Development
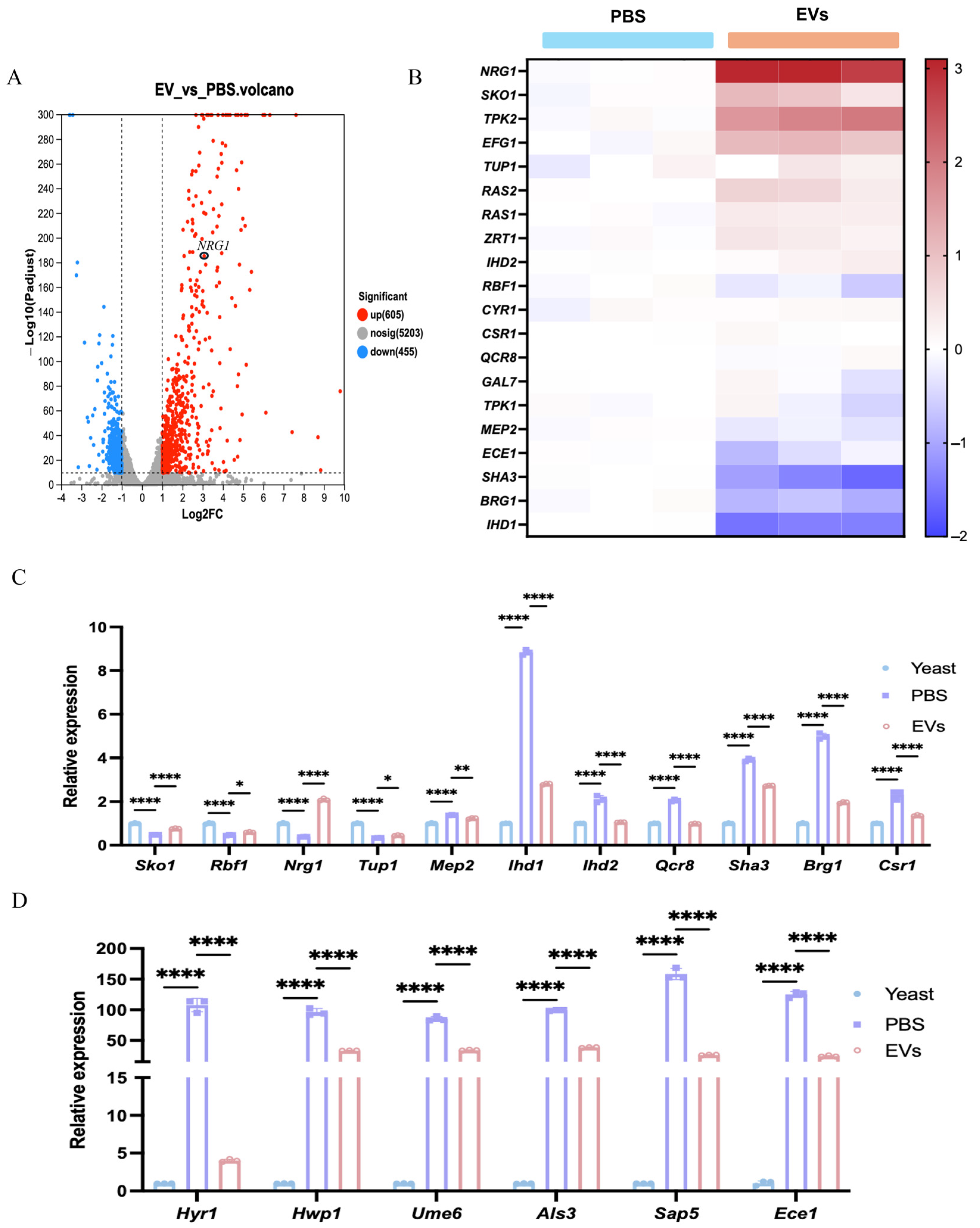
2.2. Extracellular Vesicles Shifted the Expression of Hyphal-Related Genes from C. albicans Transcriptome
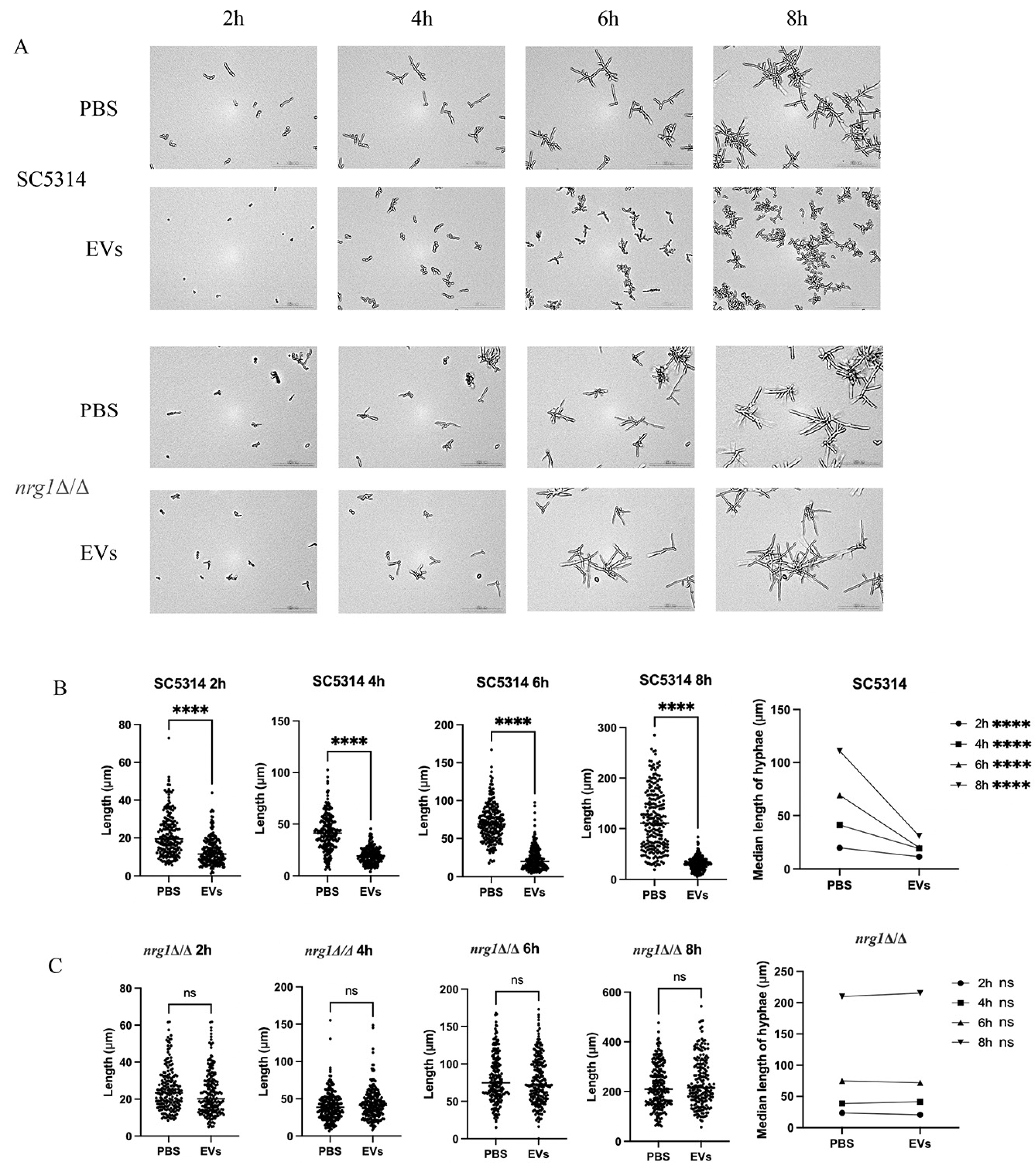
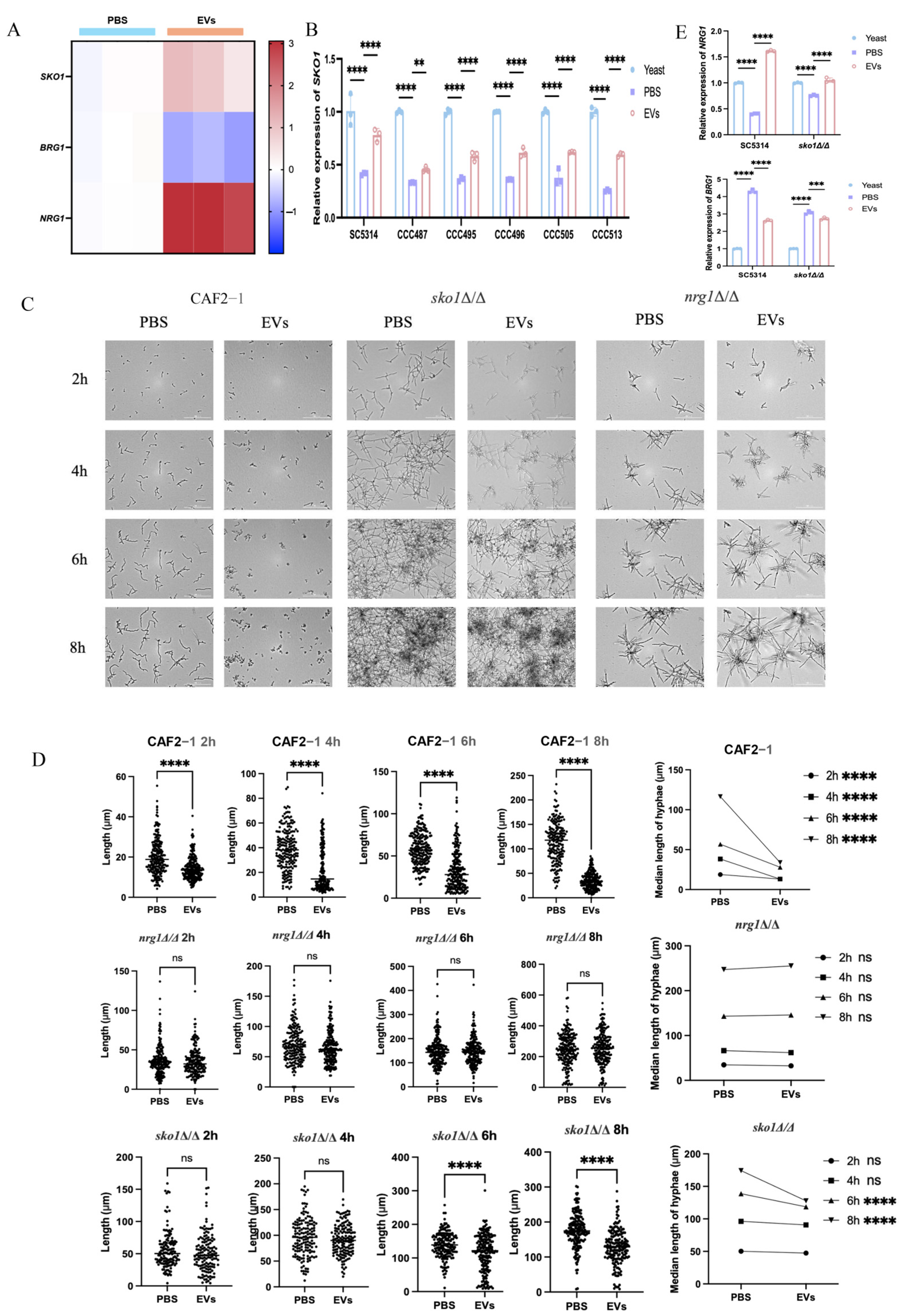
2.3. Extracellular Vesicles Inhibited C. albicans Hyphal Development Through the Upregulation of NRG1
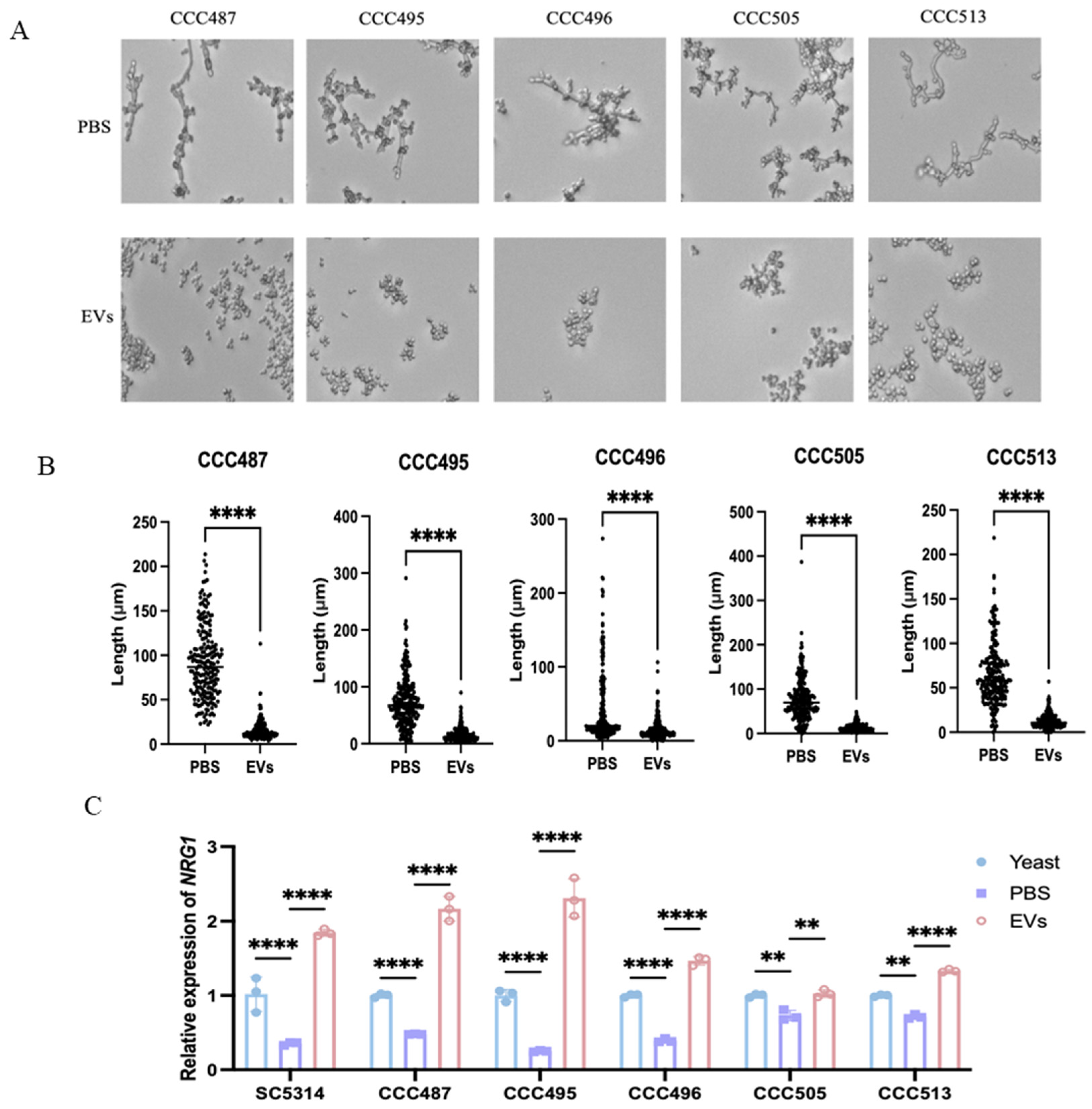
2.4. Extracellular Vesicles Upregulated NRG1 to Inhibit the Hyphal Development of Clinical C. albicans Isolates
2.5. Extracellular Vesicles Mediated Upregulation of SKO1 Correlates with Enhanced NRG1 Expression
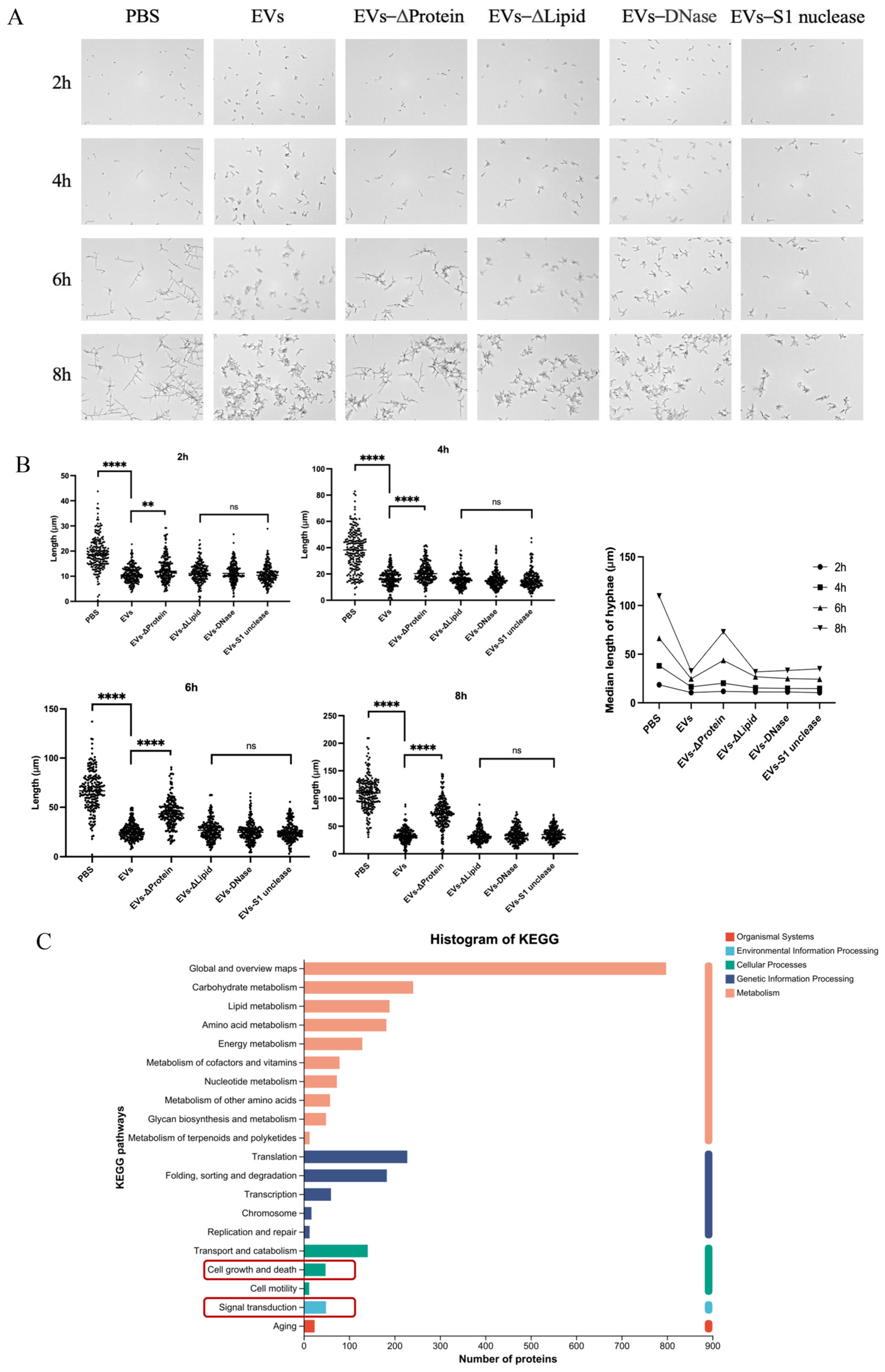
2.6. Protein Components Within EVs Contributed to Hyphal-Inhibitory Effects
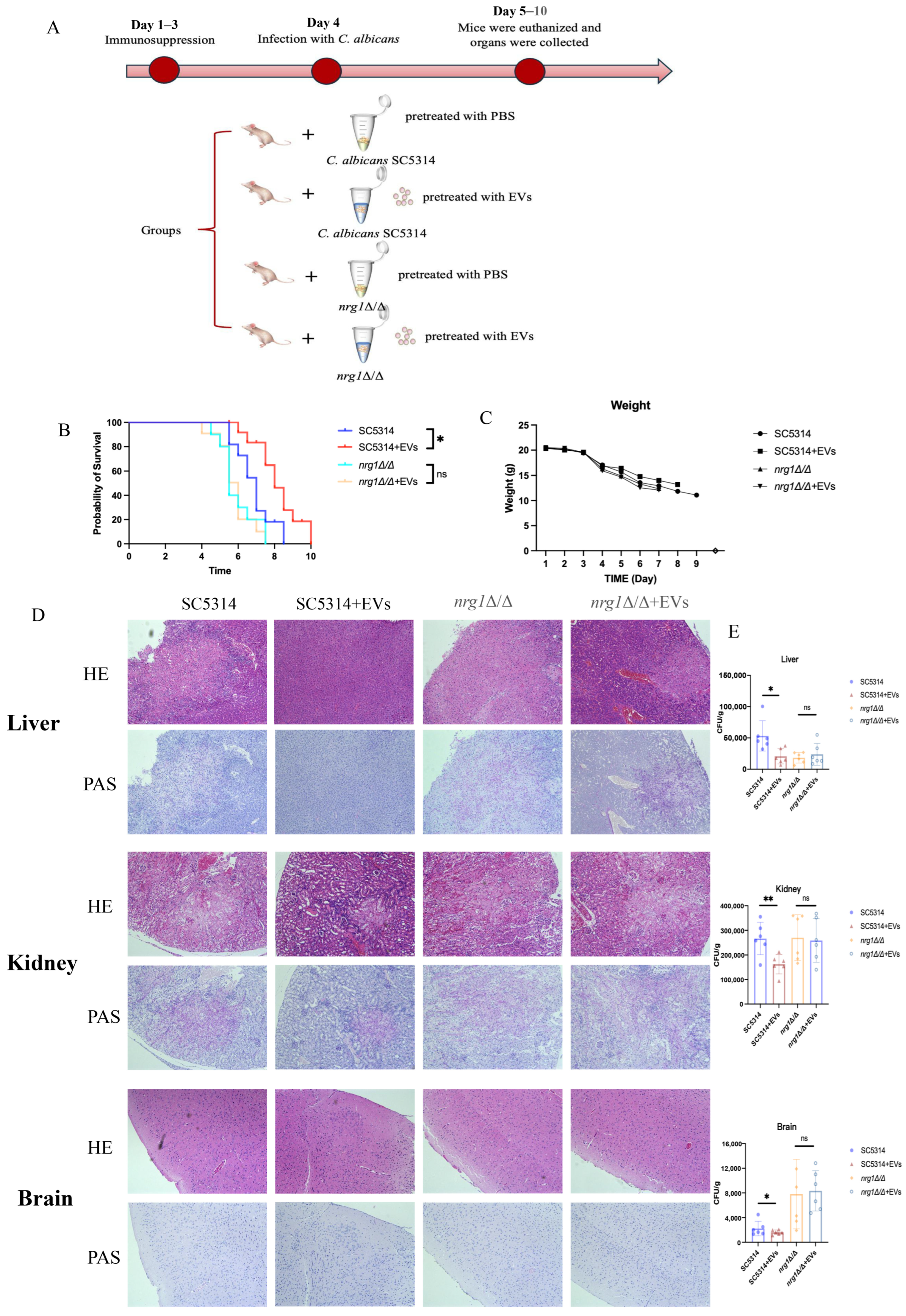
2.7. Extracellular Vesicles Reduced the Pathogenesis of C. albicans Through NRG1 in Candidemia Mice
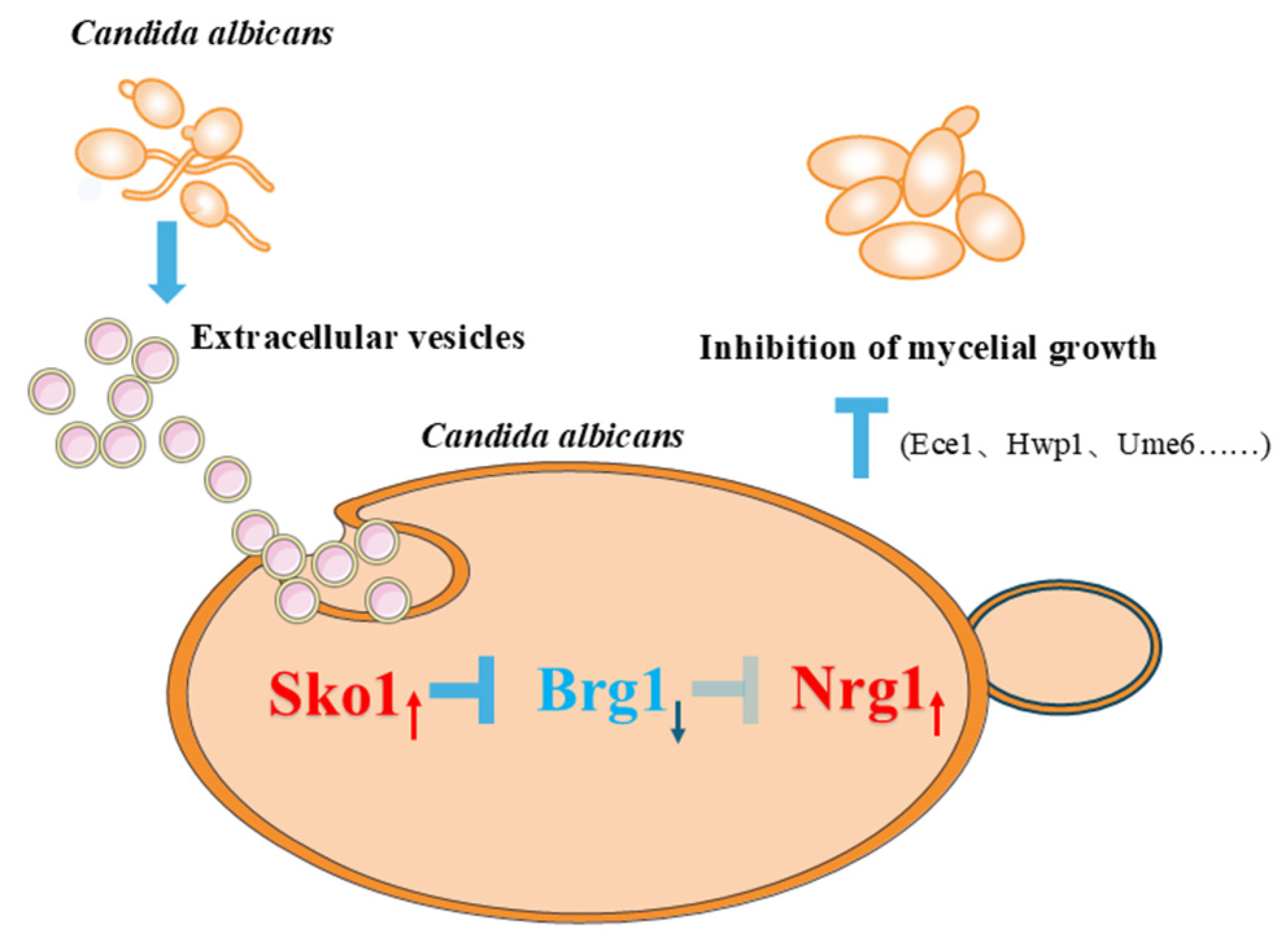
3. Discussion
4. Materials and Methods
4.1. Strains and Cultural Conditions
4.2. Isolation of Extracellular Vesicles
4.3. Characterization of EVs
4.4. Confocal Imaging
4.5. Real-Time Imaging and Distribution of C. albicans Hyphal Length
4.6. Dynasore-Treated C. albicans
4.7. Transcriptome Analysis
4.8. Real-Time RT-PCR
4.9. Selective Depletion of EV Components
4.10. EV Cargo Protein Profiling
4.11. Murine Model of Systemic Infection
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schroeder, J.A.; Wilson, C.M.; Pappas, P.G. Invasive Candidiasis. Infect. Dis. Clin. N. Am. 2025, 39, 93–119. [Google Scholar] [CrossRef]
- Ho, J.; Camilli, G.; Griffiths, J.S.; Richardson, J.P.; Kichik, N.; Naglik, J.R. Candida albicans and candidalysin in inflammatory disorders and cancer. Immunology 2021, 162, 11–16. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Gow, N.A.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.; Parmanu, P.K.; Sharma, M. Mechanisms of antifungal resistance and developments in alternative strategies to combat Candida albicans infection. Arch. Microbiol. 2024, 206, 95. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO Fungal Priority Pathogens List to Guide Research, Development and Public Health Action; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Chow, E.W.L.; Pang, L.M.; Wang, Y. From Jekyll to Hyde: The Yeast-Hyphal Transition of Candida albicans. Pathogens 2021, 10, 859. [Google Scholar] [CrossRef]
- Gow, N.A.; van de Veerdonk, F.L.; Brown, A.J.; Netea, M.G. Candida albicans morphogenesis and host defence: Discriminating invasion from colonization. Nat. Rev. Microbiol. 2011, 10, 112–122. [Google Scholar] [CrossRef]
- Chen, H.; Zhou, X.; Ren, B.; Cheng, L. The regulation of hyphae growth in Candida albicans. Virulence 2020, 11, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Lu, Y.; Liu, H. Reduced TOR signaling sustains hyphal development in Candida albicans by lowering Hog1 basal activity. Mol. Biol. Cell 2013, 24, 385–397. [Google Scholar] [CrossRef]
- Su, C.; Yu, J.; Sun, Q.; Liu, Q.; Lu, Y. Hyphal induction under the condition without inoculation in Candida albicans is triggered by Brg1-mediated removal of NRG1 inhibition. Mol. Microbiol. 2018, 108, 410–423. [Google Scholar] [CrossRef]
- Uppuluri, P.; Pierce, C.G.; Thomas, D.P.; Bubeck, S.S.; Saville, S.P.; Lopez-Ribot, J.L. The transcriptional regulator Nrg1p controls Candida albicans biofilm formation and dispersion. Eukaryot. Cell 2010, 9, 1531–1537. [Google Scholar] [CrossRef]
- Kim, S.; Kim, S.H.; Kweon, E.; Kim, J. Apoptotic Factors, CaNma111 and CaYbh3, Function in Candida albicans Filamentation by Regulating the Hyphal Suppressors, Nrg1 and Tup1. J. Microbiol. 2023, 61, 403–409. [Google Scholar] [CrossRef]
- Lu, Y.; Su, C.; Liu, H. Candida albicans hyphal initiation and elongation. Trends Microbiol. 2014, 22, 707–714. [Google Scholar] [CrossRef]
- Gavandi, T.; Patil, S.; Basrani, S.; Yankanchi, S.; Chougule, S.; Karuppayil, S.M.; Jadhav, A. MIG1, TUP1 and NRG1 mediated yeast to hyphal morphogenesis inhibition in Candida albicans by ganciclovir. Braz. J. Microbiol. 2024, 55, 2047–2056. [Google Scholar] [CrossRef]
- Lu, Y.; Su, C.; Wang, A.; Liu, H. Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance. PLoS Biol. 2011, 9, e1001105, Correction in PLoS Biol. 2011, 9, 10.1371/annotation/7b97b9ec-881a-4940-83ab-01f5318fd819. [Google Scholar] [CrossRef]
- Pascual-Ahuir, A.; Posas, F.; Serrano, R.; Proft, M. Multiple levels of control regulate the yeast cAMP-response element-binding protein repressor Sko1p in response to stress. J. Biol. Chem. 2001, 276, 37373–37378. [Google Scholar] [CrossRef]
- Tomás-Cobos, L.; Casadomé, L.; Mas, G.; Sanz, P.; Posas, F. Expression of the HXT1 low affinity glucose transporter requires the coordinated activities of the HOG and glucose signalling pathways. J. Biol. Chem. 2004, 279, 22010–22019. [Google Scholar] [CrossRef]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrügger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404, Correction in J. Extracell. Vesicles 2024, 13, e12451. [Google Scholar] [CrossRef]
- Oliveira, D.L.; Nakayasu, E.S.; Joffe, L.S.; Guimarães, A.J.; Sobreira, T.J.; Nosanchuk, J.D.; Cordero, R.J.; Frases, S.; Casadevall, A.; Almeida, I.C.; et al. Characterization of yeast extracellular vesicles: Evidence for the participation of different pathways of cellular traffic in vesicle biogenesis. PLoS ONE 2010, 5, e11113. [Google Scholar] [CrossRef]
- Kulig, K.; Rudolphi-Szydlo, E.; Barbasz, A.; Surowiec, M.; Wronowska, E.; Kowalik, K.; Satala, D.; Bras, G.; Barczyk-Woznicka, O.; Karnas, E.; et al. Functional properties of Candida albicans extracellular vesicles released in the presence of the antifungal drugs amphotericin B, fluconazole and caspofungin. Microbiology 2025, 171, 001565. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Nakayasu, E.S.; Oliveira, D.L.; Nimrichter, L.; Nosanchuk, J.D.; Almeida, I.C.; Casadevall, A. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot. Cell 2008, 7, 58–67. [Google Scholar] [CrossRef]
- Bielska, E.; May, R.C. Extracellular vesicles of human pathogenic fungi. Curr Opin. Microbiol. 2019, 52, 90–99. [Google Scholar] [CrossRef]
- Kulig, K.; Wronowska, E.; Juszczak, M.; Zawrotniak, M.; Karkowska-Kuleta, J.; Rapala-Kozik, M. Host cell responses to Candida albicans biofilm-derived extracellular vesicles. Front. Cell Infect. Microbiol. 2024, 14, 1499461. [Google Scholar] [CrossRef]
- Kabani, M.; Melki, R. Sup35p in Its Soluble and Prion States Is Packaged inside Extracellular Vesicles. mBio 2015, 6, e01017-15. [Google Scholar] [CrossRef]
- Zhang, L.; Chi, J.; Wu, H.; Xia, X.; Xu, C.; Hao, H.; Liu, Z. Extracellular vesicles and endothelial dysfunction in infectious diseases. J. Extracell. Biol. 2024, 3, e148, Correction in J. Extracell. Biol. 2025, 4, e70070. [Google Scholar] [CrossRef]
- Oliveira, D.L.; Freire-de-Lima, C.G.; Nosanchuk, J.D.; Casadevall, A.; Rodrigues, M.L.; Nimrichter, L. Extracellular vesicles from Cryptococcus neoformans modulate macrophage functions. Infect. Immun. 2010, 78, 1601–1609. [Google Scholar] [CrossRef]
- Kuhn, D.A.; Vanhecke, D.; Michen, B.; Blank, F.; Gehr, P.; Petri-Fink, A.; Rothen-Rutishauser, B. Different endocytotic uptake mechanisms for nanoparticles in epithelial cells and macrophages. Beilstein J. Nanotechnol. 2014, 5, 1625–1636. [Google Scholar] [CrossRef]
- Vargas, G.; Rocha, J.D.; Oliveira, D.L.; Albuquerque, P.C.; Frases, S.; Santos, S.S.; Nosanchuk, J.D.; Gomes, A.M.; Medeiros, L.C.; Miranda, K.; et al. Compositional and immunobiological analyses of extracellular vesicles released by Candida albicans. Cell. Microbiol. 2015, 17, 389–407. [Google Scholar] [CrossRef]
- Bitencourt, T.A.; Rezende, C.P.; Quaresemin, N.R.; Moreno, P.; Hatanaka, O.; Rossi, A.; Martinez-Rossi, N.M.; Almeida, F. Extracellular Vesicles from the Dermatophyte Trichophyton interdigitale Modulate Macrophage and Keratinocyte Functions. Front. Immunol. 2018, 9, 2343. [Google Scholar] [CrossRef]
- Bielska, E.; Sisquella, M.A.; Aldeieg, M.; Birch, C.; O’Donoghue, E.J.; May, R.C. Pathogen-derived extracellular vesicles mediate virulence in the fatal human pathogen Cryptococcus gattii. Nat. Commun. 2018, 9, 1556. [Google Scholar] [CrossRef]
- Baltazar, L.M.; Zamith-Miranda, D.; Burnet, M.C.; Choi, H.; Nimrichter, L.; Nakayasu, E.S.; Nosanchuk, J.D. Concentration-dependent protein loading of extracellular vesicles released by Histoplasma capsulatum after antibody treatment and its modulatory action upon macrophages. Sci. Rep. 2018, 8, 8065. [Google Scholar] [CrossRef]
- Monari, C.; Bistoni, F.; Vecchiarelli, A. Glucuronoxylomannan exhibits potent immunosuppressive properties. FEMS Yeast Res. 2006, 6, 537–542. [Google Scholar] [CrossRef]
- Yu, B.; Wang, Q.; Zhang, L.; Lin, J.; Feng, Z.; Wang, Z.; Gu, L.; Tian, X.; Luan, S.; Li, C.; et al. Ebselen improves fungal keratitis through exerting anti-inflammation, anti-oxidative stress, and antifungal effects. Redox Biol. 2024, 73, 103206. [Google Scholar] [CrossRef]
- Martínez-López, R.; Hernáez, M.L.; Redondo, E.; Calvo, G.; Radau, S.; Pardo, M.; Gil, C.; Monteoliva, L. Candida albicans Hyphal Extracellular Vesicles Are Different from Yeast Ones, Carrying an Active Proteasome Complex and Showing a Different Role in Host Immune Response. Microbiol. Spectr. 2022, 10, e0069822. [Google Scholar] [CrossRef]
- Kulig, K.; Rapala-Kozik, M.; Karkowska-Kuleta, J. Extracellular vesicle production: A bidirectional effect in the interplay between host and Candida fungi. Curr. Res. Microb. Sci. 2024, 7, 100255. [Google Scholar] [CrossRef]
- Brown, L.; Wolf, J.M.; Prados-Rosales, R.; Casadevall, A. Through the wall: Extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 2015, 13, 620–630. [Google Scholar] [CrossRef]
- Huang, S.H.; Wu, C.H.; Chang, Y.C.; Kwon-Chung, K.J.; Brown, R.J.; Jong, A. Cryptococcus neoformans-derived microvesicles enhance the pathogenesis of fungal brain infection. PLoS ONE 2012, 7, e48570. [Google Scholar] [CrossRef]
- Ikeda, M.A.K.; de Almeida, J.R.F.; Jannuzzi, G.P.; Cronemberger-Andrade, A.; Torrecilhas, A.C.T.; Moretti, N.S.; da Cunha, J.P.C.; de Almeida, S.R.; Ferreira, K.S. Extracellular Vesicles From Sporothrix brasiliensis Are an Important Virulence Factor That Induce an Increase in Fungal Burden in Experimental Sporotrichosis. Front. Microbiol. 2018, 9, 2286. [Google Scholar] [CrossRef]
- Xu, Z.; Qiao, S.; Wang, Z.; Peng, C.; Hou, Y.; Liu, B.; Cao, G.; Wang, T. PMA1-containing extracellular vesicles of Candida albicans triggers immune responses and colitis progression. Gut Microbes 2025, 17, 2455508. [Google Scholar] [CrossRef]
- Trentin, G.; Bitencourt, T.A.; Guedes, A.; Pessoni, A.M.; Brauer, V.S.; Pereira, A.K.; Costa, J.H.; Fill, T.P.; Almeida, F. Mass Spectrometry Analysis Reveals Lipids Induced by Oxidative Stress in Candida albicans Extracellular Vesicles. Microorganisms 2023, 11, 1669. [Google Scholar] [CrossRef]
- Lee, S.; Tsavou, A.; Zarnowski, R.; Pforte, R.; Allert, S.; Krüger, T.; Kniemeyer, O.; Brakhage, A.A.; Bui, T.T.T.; Andes, D.R.; et al. Candida albicans biofilm extracellular vesicles deliver candidalysin to epithelial cell membranes and induce host cell responses. Infect. Immun. 2025, 93, e0040424. [Google Scholar] [CrossRef]
- Chen, T.; Feng, Y.; Sun, W.; Zhao, G.; Wu, H.; Cheng, X.; Zhao, F.; Zhang, L.; Zheng, Y.; Zhan, P.; et al. The nucleotide receptor STING translocates to the phagosomes to negatively regulate anti-fungal immunity. Immunity 2023, 56, 1727–1742.e6. [Google Scholar] [CrossRef] [PubMed]
- Kwaku, G.N.; Jensen, K.N.; Simaku, P.; Floyd, D.J.; Saelens, J.W.; Reardon, C.M.; Ward, R.A.; Basham, K.J.; Hepworth, O.W.; Vyas, T.D.; et al. Extracellular vesicles from diverse fungal pathogens induce species-specific and endocytosis-dependent immunomodulation. PLoS Pathog. 2025, 21, e1012879. [Google Scholar] [CrossRef]
- Brown Harding, H.; Kwaku, G.N.; Reardon, C.M.; Khan, N.S.; Zamith-Miranda, D.; Zarnowski, R.; Tam, J.M.; Bohaen, C.K.; Richey, L.; Mosallanejad, K.; et al. Candida albicans extracellular vesicles trigger type I IFN signalling via cGAS and STING. Nat. Microbiol. 2024, 9, 95–107. [Google Scholar] [CrossRef]
- Vargas, G.; Honorato, L.; Guimarães, A.J.; Rodrigues, M.L.; Reis, F.C.G.; Vale, A.M.; Ray, A.; Nosanchuk, J.D.; Nimrichter, L. Protective effect of fungal extracellular vesicles against murine candidiasis. Cell. Microbiol. 2020, 22, e13238. [Google Scholar] [CrossRef]
- Bitencourt, T.A.; Hatanaka, O.; Pessoni, A.M.; Freitas, M.S.; Trentin, G.; Santos, P.; Rossi, A.; Martinez-Rossi, N.M.; Alves, L.L.; Casadevall, A.; et al. Fungal Extracellular Vesicles Are Involved in Intraspecies Intracellular Communication. mBio 2022, 13, e0327221. [Google Scholar] [CrossRef]
- Yang, S.; Li, N.; Wu, H.; Zhang, M.; Wang, L.; Xiao, M.; Cheng, X.; Yu, Q. Extracellular vesicles of Candida albicans show dual effects on Enterococcus faecalis growth and virulence: A laboratory-based investigation. Int. Endod. J. 2025, 58, 613–626. [Google Scholar] [CrossRef]
- Zarnowski, R.; Sanchez, H.; Jaromin, A.; Zarnowska, U.J.; Nett, J.E.; Mitchell, A.P.; Andes, D. A common vesicle proteome drives fungal biofilm development. Proc. Natl. Acad. Sci. USA 2022, 119, e2211424119. [Google Scholar] [CrossRef]
- Zarnowski, R.; Sanchez, H.; Covelli, A.S.; Dominguez, E.; Jaromin, A.; Bernhardt, J.; Mitchell, K.F.; Heiss, C.; Azadi, P.; Mitchell, A.; et al. Candida albicans biofilm-induced vesicles confer drug resistance through matrix biogenesis. PLoS Biol. 2018, 16, e2006872. [Google Scholar] [CrossRef]
- Honorato, L.; de Araujo, J.F.D.; Ellis, C.C.; Piffer, A.C.; Pereira, Y.; Frases, S.; de Sousa Araújo, G.R.; Pontes, B.; Mendes, M.T.; Pereira, M.D.; et al. Extracellular Vesicles Regulate Biofilm Formation and Yeast-to-Hypha Differentiation in Candida albicans. mBio 2022, 13, e0030122. [Google Scholar] [CrossRef]
- Wei, Y.; Wang, Z.; Liu, Y.; Liao, B.; Zong, Y.; Shi, Y.; Liao, M.; Wang, J.; Zhou, X.; Cheng, L.; et al. Extracellular vesicles of Candida albicans regulate its own growth through the L-arginine/nitric oxide pathway. Appl. Microbiol. Biotechnol. 2023, 107, 355–367. [Google Scholar] [CrossRef]
- Cleary, I.A.; Lazzell, A.L.; Monteagudo, C.; Thomas, D.P.; Saville, S.P. BRG1 and NRG1 form a novel feedback circuit regulating Candida albicans hypha formation and virulence. Mol. Microbiol. 2012, 85, 557–573. [Google Scholar] [CrossRef]
- Talapko, J.; Juzbašić, M.; Matijević, T.; Pustijanac, E.; Bekić, S.; Kotris, I.; Škrlec, I. Candida albicans-The Virulence Factors and Clinical Manifestations of Infection. J. Fungi 2021, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Witchley, J.N.; Penumetcha, P.; Abon, N.V.; Woolford, C.A.; Mitchell, A.P.; Noble, S.M. Candida albicans Morphogenesis Programs Control the Balance between Gut Commensalism and Invasive Infection. Cell Host Microbe 2019, 25, 432–443.e6. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Solis, N.V.; Filler, S.G.; Mitchell, A.P. Functional Dichotomy for a Hyphal Repressor in Candida albicans. mBio 2023, 14, e0013423. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, P.; Thompson, A.; Xie, Z.; Kashleva, H.; Ganguly, S.; Mitchell, A.P.; Dongari-Bagtzoglou, A. Role of Bcr1-activated genes Hwp1 and Hyr1 in Candida albicans oral mucosal biofilms and neutrophil evasion. PLoS ONE 2011, 6, e16218. [Google Scholar] [CrossRef]
- Childers, D.S.; Kadosh, D. Filament condition-specific response elements control the expression of NRG1 and UME6, key transcriptional regulators of morphology and virulence in Candida albicans. PLoS ONE 2015, 10, e0122775. [Google Scholar] [CrossRef]
- Takagi, J.; Aoki, K.; Turner, B.S.; Lamont, S.; Lehoux, S.; Kavanaugh, N.; Gulati, M.; Valle Arevalo, A.; Lawrence, T.J.; Kim, C.Y.; et al. Mucin O-glycans are natural inhibitors of Candida albicans pathogenicity. Nat. Chem. Biol. 2022, 18, 762–773. [Google Scholar] [CrossRef]
- Liu, J.; Willems, H.M.E.; Sansevere, E.A.; Allert, S.; Barker, K.S.; Lowes, D.J.; Dixson, A.C.; Xu, Z.; Miao, J.; DeJarnette, C.; et al. A variant ECE1 allele contributes to reduced pathogenicity of Candida albicans during vulvovaginal candidiasis. PLoS Pathog. 2021, 17, e1009884. [Google Scholar] [CrossRef]
- Pourtalebi Jahromi, L.; Rothammer, M.; Fuhrmann, G. Polysaccharide hydrogel platforms as suitable carriers of liposomes and extracellular vesicles for dermal applications. Adv. Drug Deliv. Rev. 2023, 200, 115028. [Google Scholar] [CrossRef]
- Hanumantha Rao, K.; Paul, S.; Ghosh, S. N-acetylglucosamine Signaling: Transcriptional Dynamics of a Novel Sugar Sensing Cascade in a Model Pathogenic Yeast, Candida albicans. J. Fungi 2021, 7, 65. [Google Scholar] [CrossRef]
- Rauceo, J.M.; Blankenship, J.R.; Fanning, S.; Hamaker, J.J.; Deneault, J.S.; Smith, F.J.; Nantel, A.; Mitchell, A.P. Regulation of the Candida albicans cell wall damage response by transcription factor Sko1 and PAS kinase Psk1. Mol. Biol. Cell 2008, 19, 2741–2751. [Google Scholar] [CrossRef]
- Heredia, M.Y.; Ikeh, M.A.C.; Gunasekaran, D.; Conrad, K.A.; Filimonava, S.; Marotta, D.H.; Nobile, C.J.; Rauceo, J.M. An expanded cell wall damage signaling network is comprised of the transcription factors Rlm1 and Sko1 in Candida albicans. PLoS Genet. 2020, 16, e1008908. [Google Scholar] [CrossRef]
- Wakade, R.S.; Kramara, J.; Wellington, M.; Krysan, D.J. Candida albicans Filamentation Does Not Require the cAMP-PKA Pathway In Vivo. mBio 2022, 13, e0085122. [Google Scholar] [CrossRef] [PubMed]
- García-Sánchez, S.; Mavor, A.L.; Russell, C.L.; Argimon, S.; Dennison, P.; Enjalbert, B.; Brown, A.J. Global roles of Ssn6 in Tup1- and Nrg1-dependent gene regulation in the fungal pathogen, Candida albicans. Mol. Biol. Cell 2005, 16, 2913–2925. [Google Scholar] [CrossRef]
- Ruben, S.; Garbe, E.; Mogavero, S.; Albrecht-Eckardt, D.; Hellwig, D.; Häder, A.; Krüger, T.; Gerth, K.; Jacobsen, I.D.; Elshafee, O.; et al. Ahr1 and Tup1 Contribute to the Transcriptional Control of Virulence-Associated Genes in Candida albicans. mBio 2020, 11, e00206-20. [Google Scholar] [CrossRef] [PubMed]
- Proft, M.; Struhl, K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell 2002, 9, 1307–1317. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.H.; Zafar, H.; Ponde, N.O.; Hepworth, O.W.; Sihra, D.; Aggor, F.E.Y.; Ainscough, J.S.; Ho, J.; Richardson, J.P.; Coleman, B.M.; et al. IL-36 and IL-1/IL-17 Drive Immunity to Oral Candidiasis via Parallel Mechanisms. J. Immunol. 2018, 201, 627–634. [Google Scholar] [CrossRef]
- Ye, X.; Liu, Y.; Chen, D.; Liao, B.; Wang, J.; Shen, J.; Gou, L.; Zhou, Y.; Zhou, X.; Liao, G.; et al. Moxidectin elevates Candida albicans ergosterol levels to synergize with polyenes against oral candidiasis. Appl. Microbiol. Biotechnol. 2024, 108, 509. [Google Scholar] [CrossRef]
- Alonso-Monge, R.; Román, E.; Arana, D.M.; Prieto, D.; Urrialde, V.; Nombela, C.; Pla, J. The Sko1 protein represses the yeast-to-hypha transition and regulates the oxidative stress response in Candida albicans. Fungal Genet. Biol. 2010, 47, 587–601. [Google Scholar] [CrossRef]
- Gerami-Nejad, M.; Berman, J.; Gale, C.A. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast 2001, 18, 859–864. [Google Scholar] [CrossRef]
- Vyas, V.K.; Bushkin, G.G.; Bernstein, D.A.; Getz, M.A.; Sewastianik, M.; Barrasa, M.I.; Bartel, D.P.; Fink, G.R. New CRISPR Mutagenesis Strategies Reveal Variation in Repair Mechanisms among Fungi. mSphere 2018, 3, e00154-18. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D. Yeast transformation by the LiAc/SS carrier DNA/PEG method. Methods Mol. Biol. 2014, 1163, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Halder, V.; Porter, C.B.M.; Chavez, A.; Shapiro, R.S. Design, execution, and analysis of CRISPR-Cas9-based deletions and genetic interaction networks in the fungal pathogen Candida albicans. Nat. Protoc. 2019, 14, 955–975, Correction in Nat. Protoc. 2019, 14, 2595. [Google Scholar] [CrossRef]
- Karkowska-Kuleta, J.; Kulig, K.; Karnas, E.; Zuba-Surma, E.; Woznicka, O.; Pyza, E.; Kuleta, P.; Osyczka, A.; Rapala-Kozik, M.; Kozik, A. Characteristics of Extracellular Vesicles Released by the Pathogenic Yeast-Like Fungi Candida glabrata, Candida parapsilosis and Candida tropicalis. Cells 2020, 9, 1722. [Google Scholar] [CrossRef]
- Zong, Y.W.; Cheng, X.Y.; Liao, B.Y.; Ye, X.C.; Liu, T.P.; Zhou, X.D.; Li, J.Y.; Cheng, L.; Xu, W.Y.; Ren, B. The dynamic landscape of parasitemia dependent intestinal microbiota shifting and the correlated gut transcriptome during Plasmodium yoelii infection. Microbiol. Res. 2022, 258, 126994. [Google Scholar] [CrossRef]
- Hu, Y.; Niu, Y.; Ye, X.; Zhu, C.; Tong, T.; Zhou, Y.; Zhou, X.; Cheng, L.; Ren, B. Staphylococcus aureus Synergized with Candida albicans to Increase the Pathogenesis and Drug Resistance in Cutaneous Abscess and Peritonitis Murine Models. Pathogens 2021, 10, 1036. [Google Scholar] [CrossRef]
- Kong, L.X.; Wang, Z.; Shou, Y.K.; Zhou, X.D.; Zong, Y.W.; Tong, T.; Liao, M.; Han, Q.; Li, Y.; Cheng, L.; et al. The FnBPA from methicillin-resistant Staphylococcus aureus promoted development of oral squamous cell carcinoma. J. Oral Microbiol. 2022, 14, 2098644. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, H.; Zhou, X.; Luo, H.; Tang, F.; Yang, J.; Alterovitz, G.; Cheng, L.; Ren, B. Lovastatin synergizes with itraconazole against planktonic cells and biofilms of Candida albicans through the regulation on ergosterol biosynthesis pathway. Appl. Microbiol. Biotechnol. 2018, 102, 5255–5264. [Google Scholar] [CrossRef]
- Ma, J.; Chen, T.; Wu, S.; Yang, C.; Bai, M.; Shu, K.; Li, K.; Zhang, G.; Jin, Z.; He, F.; et al. iProX: An integrated proteome resource. Nucleic Acids Res. 2019, 47, D1211–D1217. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Ma, J.; Liu, Y.; Chen, Z.; Xiao, N.; Lu, Y.; Fu, Y.; Yang, C.; Li, M.; Wu, S.; et al. iProX in 2021: Connecting proteomics data sharing with big data. Nucleic Acids Res. 2022, 50, D1522–D1527. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhou, Z.; Mo, L.; Guo, Q.; Peng, X.; Hu, T.; Zhou, X.; Ren, B.; Xu, X. Fluphenazine antagonizes with fluconazole but synergizes with amphotericin B in the treatment of candidiasis. Appl. Microbiol. Biotechnol. 2019, 103, 6701–6709. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Wei, Y.; Zhou, Y.; Li, B.; Wang, Z.; Liao, B.; Wang, J.; Zhou, J.; Zong, Y.; Chen, D.; Shen, J.; et al. Candida albicans Extracellular Vesicles Upregulate Nrg1 Transcription Repressor to Inhibit Self-Hyphal Development and Candidemia. Int. J. Mol. Sci. 2026, 27, 495. https://doi.org/10.3390/ijms27010495
Wei Y, Zhou Y, Li B, Wang Z, Liao B, Wang J, Zhou J, Zong Y, Chen D, Shen J, et al. Candida albicans Extracellular Vesicles Upregulate Nrg1 Transcription Repressor to Inhibit Self-Hyphal Development and Candidemia. International Journal of Molecular Sciences. 2026; 27(1):495. https://doi.org/10.3390/ijms27010495
Chicago/Turabian StyleWei, Yu, Yujie Zhou, Bolei Li, Zheng Wang, Binyou Liao, Jiannan Wang, Jingzhi Zhou, Yawen Zong, Ding Chen, Jiawei Shen, and et al. 2026. "Candida albicans Extracellular Vesicles Upregulate Nrg1 Transcription Repressor to Inhibit Self-Hyphal Development and Candidemia" International Journal of Molecular Sciences 27, no. 1: 495. https://doi.org/10.3390/ijms27010495
APA StyleWei, Y., Zhou, Y., Li, B., Wang, Z., Liao, B., Wang, J., Zhou, J., Zong, Y., Chen, D., Shen, J., Shi, Y., Zhou, X., Liao, G., Gou, L., Zhu, Z., Cheng, L., & Ren, B. (2026). Candida albicans Extracellular Vesicles Upregulate Nrg1 Transcription Repressor to Inhibit Self-Hyphal Development and Candidemia. International Journal of Molecular Sciences, 27(1), 495. https://doi.org/10.3390/ijms27010495






