Characterization of Xyloglucanase TpXEG12a from Talaromyces pinophilus
Abstract
1. Introduction
2. Results
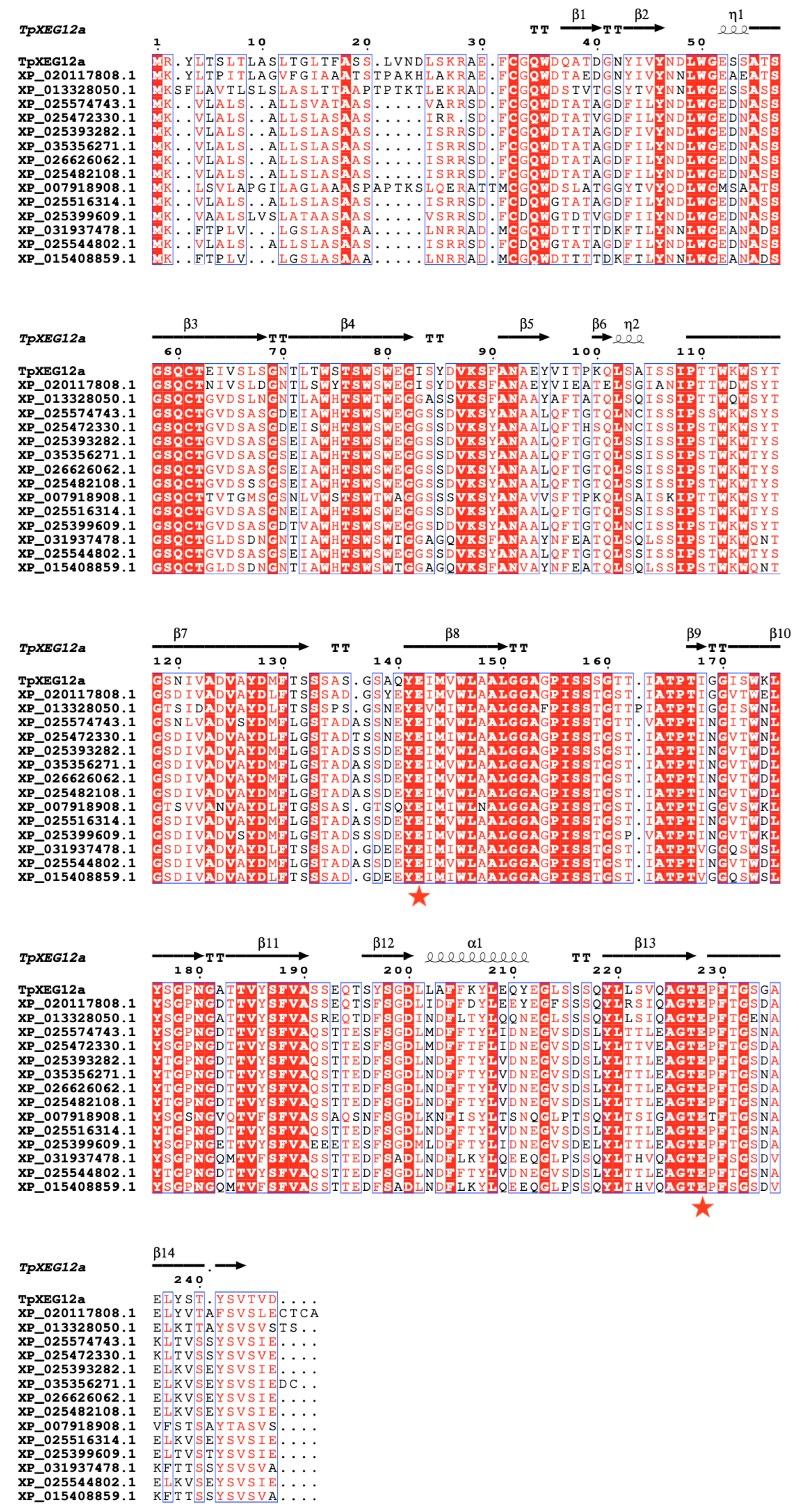
2.1. Phylogenetic Analysis and Sequence Alignment of TpXEG12a
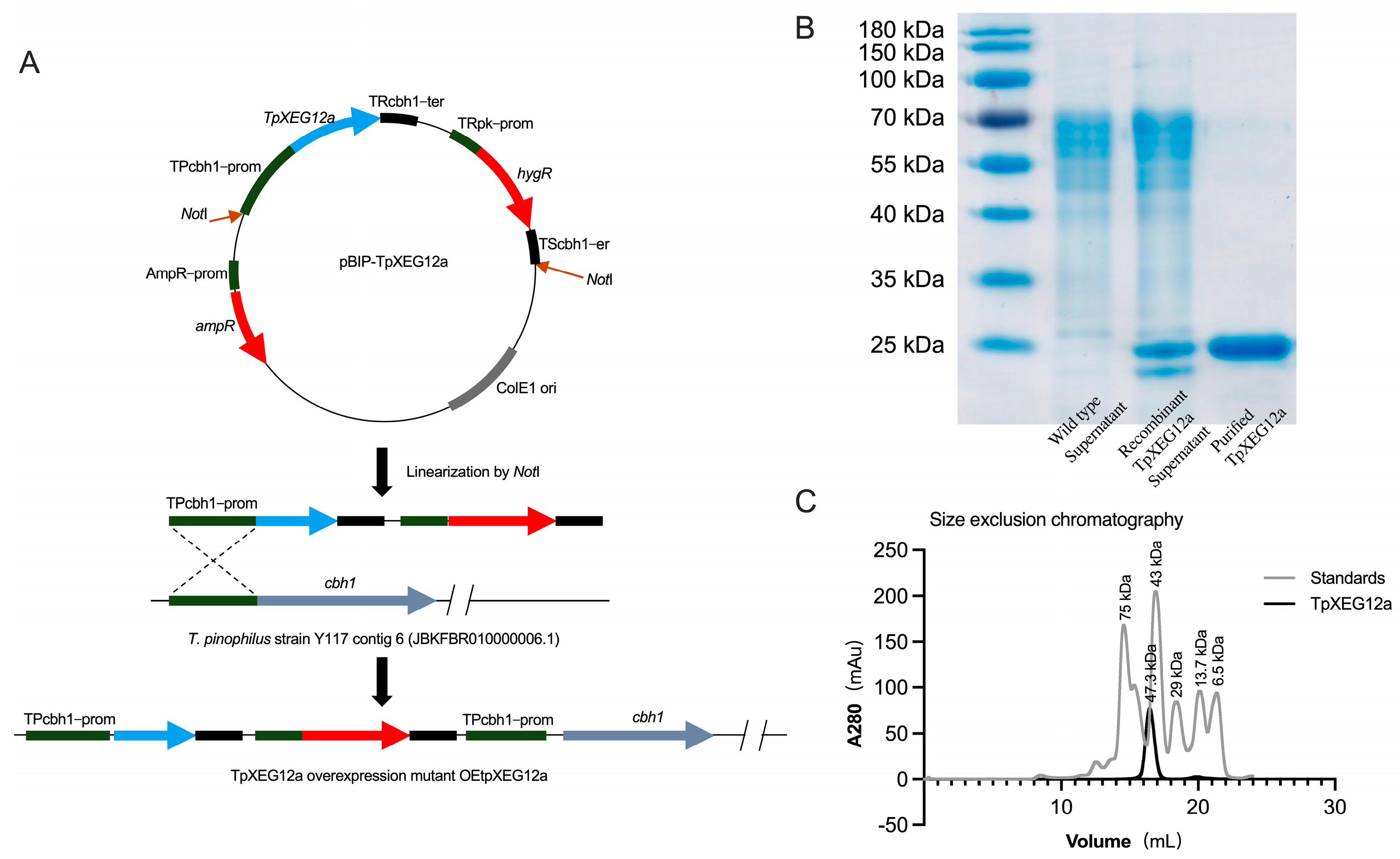
2.2. Expression and Purification of TpXEG12a
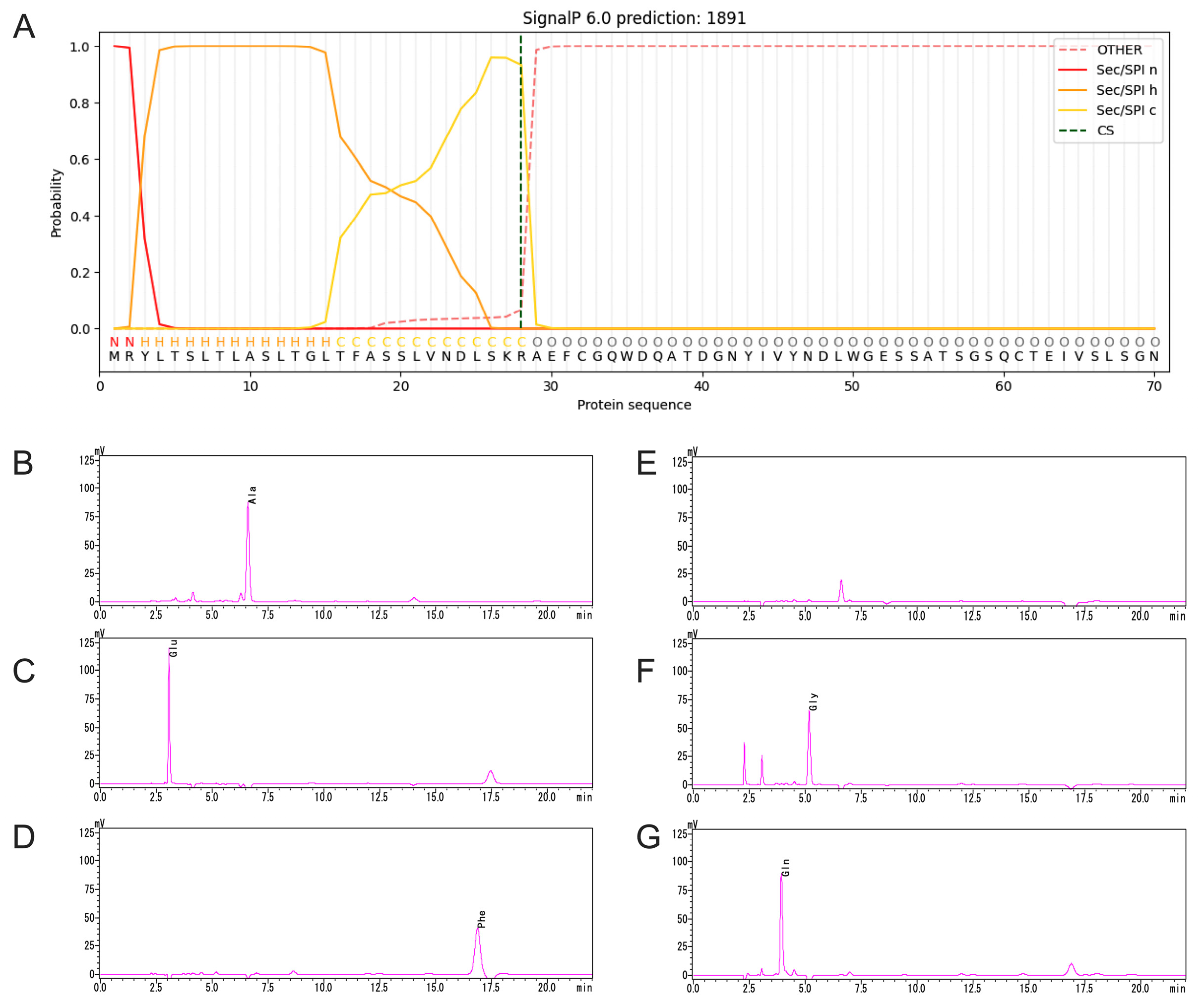
2.3. Signal Peptide of TpXEG12a
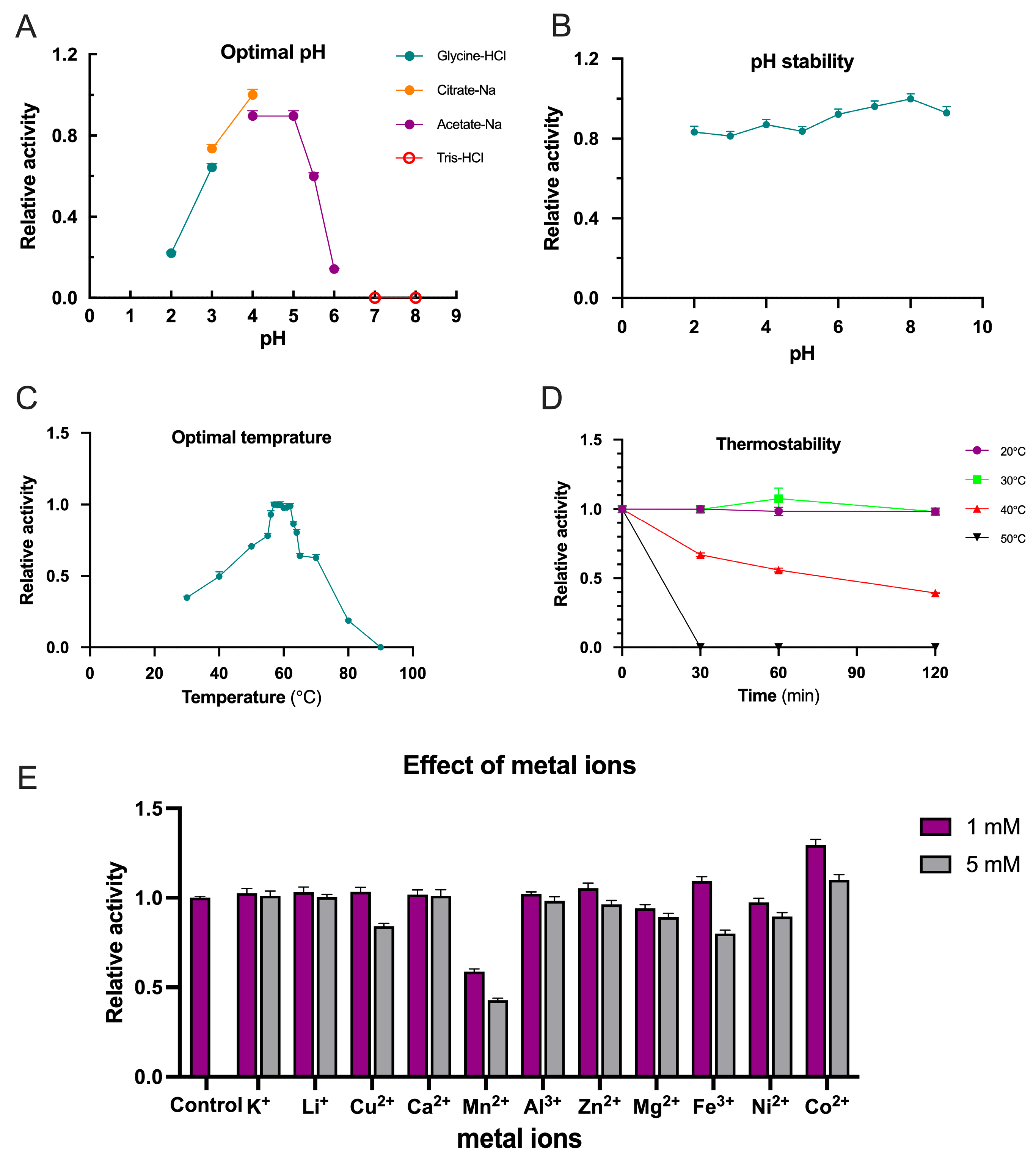
2.4. Biochemical Characterization of TpXEG12a
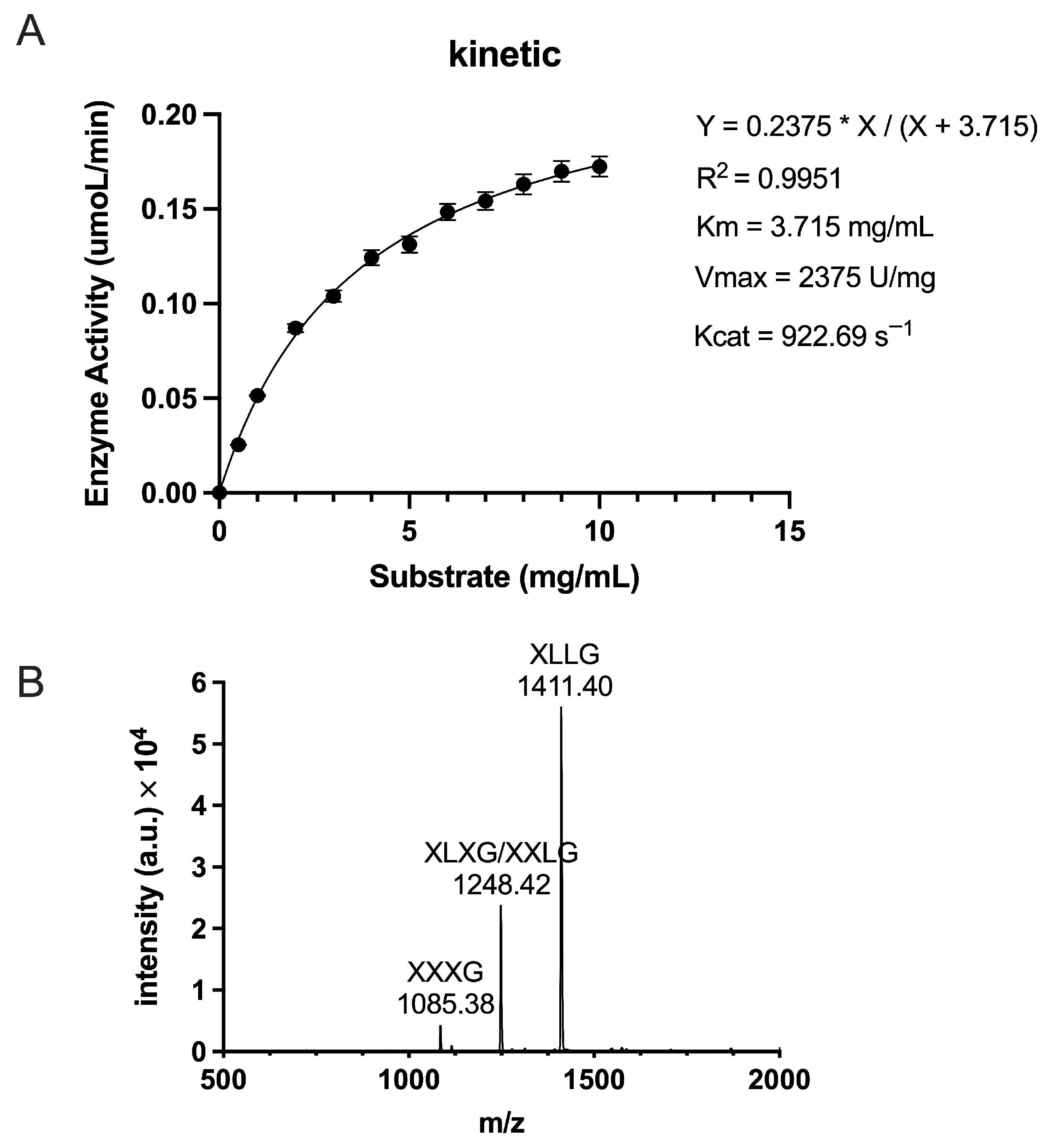
2.5. Enzymatic Kinetics and Hydrolysis Product Analysis of TpXEG12a
2.6. Substrate Specificity
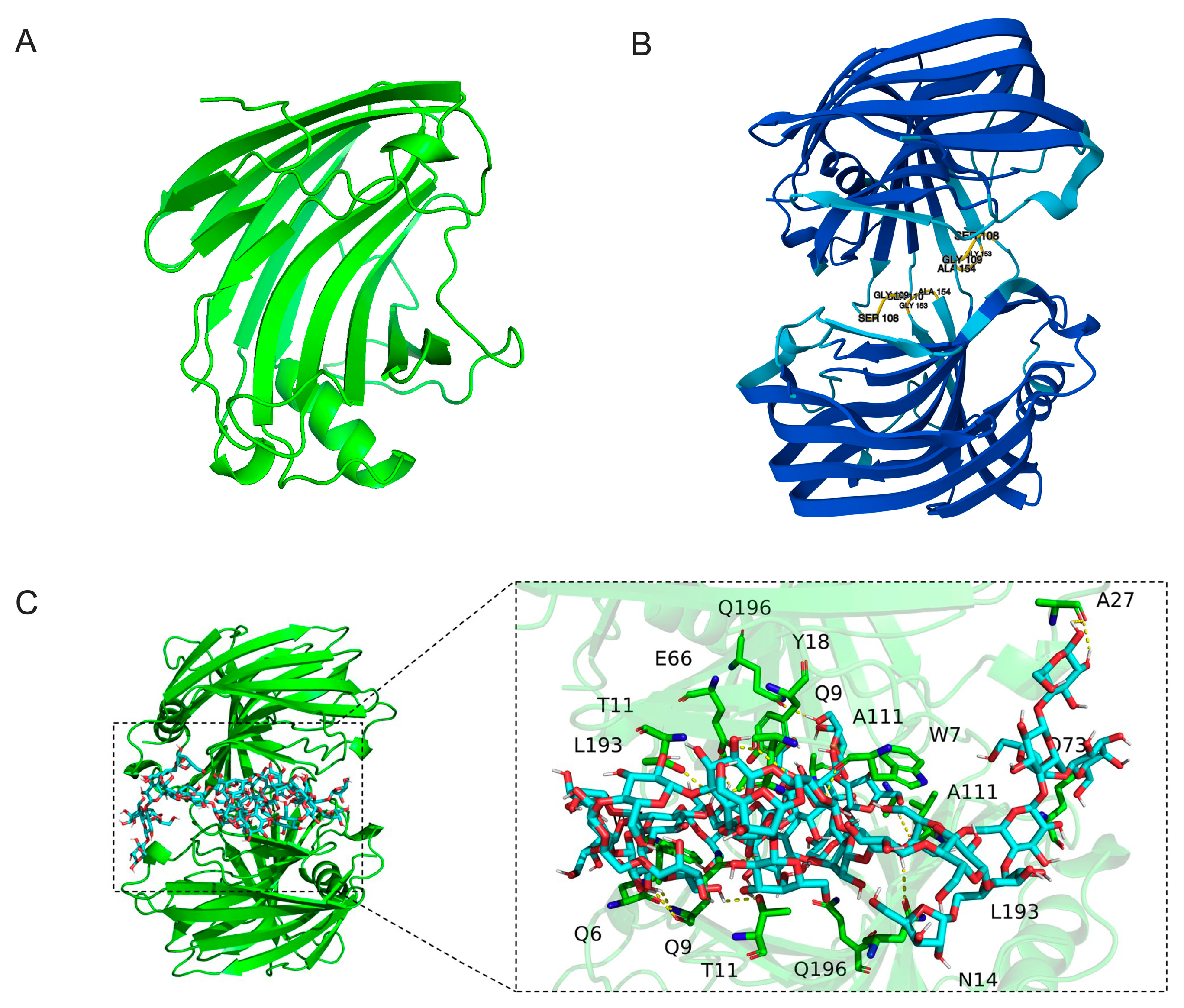
2.7. Structural Prediction, Molecular Docking, and Identification of Key Catalytic Residues of TpXEG12a
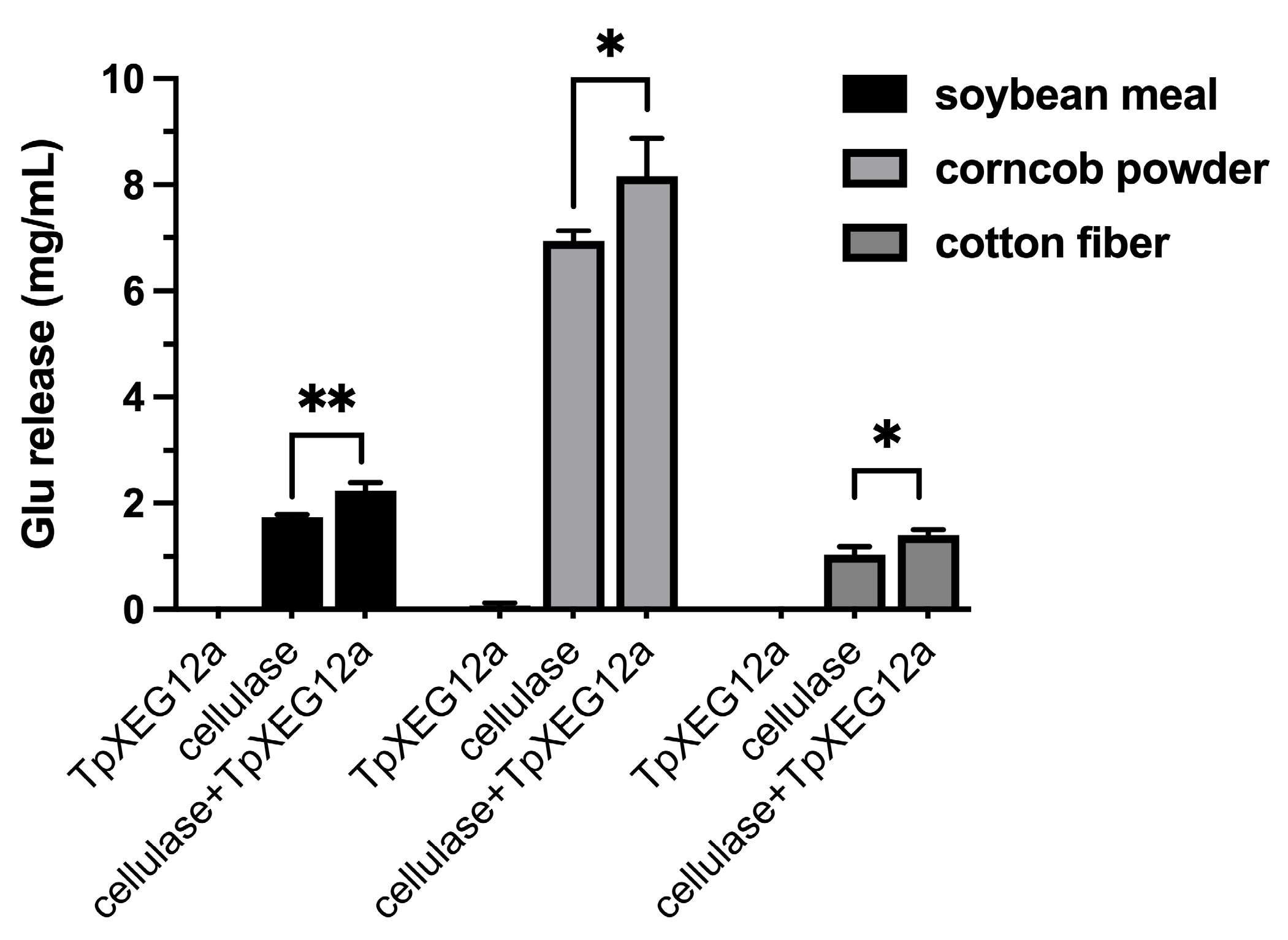
2.8. Synergistic Action Between TpXEG12a and Cellulase
3. Discussion
4. Materials and Methods
4.1. Strains, Plasmids, and Culture Conditions
4.2. Construction of TpXEG12a Overexpression Vector and Strain Transformation
4.3. Expression and Purification of Recombinant TpXEG12a
4.4. Signal Peptide Analysis and N-Terminal Sequencing
4.5. Enzyme Activity Assay
4.6. Substrate Specificity
4.7. Biochemical Characterization
4.8. Enzyme Kinetics
4.9. Analysis of Hydrolysis Products
4.10. Structural Prediction and Molecular Docking
4.11. Phylogenetic Analysis
4.12. Synergistic Degradation with Cellulase
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| XEG | xyloglucanase |
| GH12 | glycoside hydrolase family 12 |
| SEC | size-exclusion chromatography |
| CMC | carboxymethyl cellulose |
| XGO | xyloglucan oligosaccharides |
References
- Somerville, C.; Bauer, S.; Brininstool, G.; Facette, M.; Hamann, T.; Milne, J.; Osborne, E.; Paredez, A.; Persson, S.; Raab, T.; et al. Toward a systems approach to understanding plant cell walls. Science 2004, 306, 2206–2211. [Google Scholar] [CrossRef] [PubMed]
- Pauly, M.; Albersheim, P.; Darvill, A.; York, W.S. Molecular domains of the cellulose/xyloglucan network in the cell walls of higher plants. Plant J. 1999, 20, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.B.; Cosgrove, D.J. Xyloglucan and its interactions with other components of the growing cell wall. Plant Cell Physiol. 2015, 56, 180–194. [Google Scholar] [CrossRef]
- Baumann, M.J. Xyloglucan-Active Enzymes: Properties, Structures and Applications; KTH: Stockholm, Sweden, 2007. [Google Scholar]
- Master, E.R.; Zheng, Y.; Storms, R.; Tsang, A.; Powlowski, J. A xyloglucan-specific family 12 glycosyl hydrolase from Aspergillus niger: Recombinant expression, purification and characterization. Biochem. J. 2008, 411, 161–170. [Google Scholar] [CrossRef]
- Sandgren, M.; Stahlberg, J.; Mitchinson, C. Structural and biochemical studies of GH family 12 cellulases: Improved thermal stability, and ligand complexes. Prog. Biophys. Mol. Biol. 2005, 89, 246–291. [Google Scholar] [CrossRef]
- Li, K.; Barrett, K.; Agger, J.W.; Zeuner, B.; Meyer, A.S. Bioinformatics-based identification of GH12 endoxyloglucanases in citrus-pathogenic Penicillium spp. Enzyme. Microb. Technol. 2024, 178, 110441. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Chen, K.; Zhang, P.; Long, L.; Ding, S. Comparison of the Biochemical Properties and Roles in the Xyloglucan-Rich Biomass Degradation of a GH74 Xyloglucanase and Its CBM-Deleted Variant from Thielavia terrestris. Int. J. Mol. Sci. 2022, 23, 5276. [Google Scholar] [CrossRef] [PubMed]
- Matsuzawa, T.; Shimada, N.; Fujiwa, S. Production of Isoprimeverose from Xyloglucan Using Aspergillus oryzae. J. Appl. Glycosci. 2025, 72, 7201202. [Google Scholar] [CrossRef]
- Lang, W.; Watanabe, T.; Lee, C.; Tagami, T.; Li, F.; Yamamoto, T.; Tajima, K.; Borsali, R.; Takahashi, K.; Satoh, T.; et al. Fully biosourced amphiphilic block copolymer from tamarind seed xyloglucan and solanesol: Synthesis, aqueous self-assembly, and drug encapsulation. Carbohydr. Polym. 2025, 352, 123181. [Google Scholar] [CrossRef]
- Trudeau, E.D.; Brumer, H.; Berbee, M.L. Origins of xyloglucan-degrading enzymes in fungi. New Phytol. 2025, 245, 458–464. [Google Scholar] [CrossRef]
- Attia, M.A.; Nelson, C.E.; Offen, W.A.; Jain, N.; Davies, G.J.; Gardner, J.G.; Brumer, H. In vitro and in vivo characterization of three Cellvibrio japonicus glycoside hydrolase family 5 members reveals potent xyloglucan backbone-cleaving functions. Biotechnol. Biofuels 2018, 11, 45. [Google Scholar] [CrossRef]
- Baumann, M.J.; Eklof, J.M.; Michel, G.; Kallas, A.M.; Teeri, T.T.; Czjzek, M.; Brumer, H., III. Structural evidence for the evolution of xyloglucanase activity from xyloglucan endo-transglycosylases: Biological implications for cell wall metabolism. Plant Cell 2007, 19, 1947–1963. [Google Scholar] [CrossRef]
- Attia, M.A.; Brumer, H. Recent structural insights into the enzymology of the ubiquitous plant cell wall glycan xyloglucan. Curr. Opin. Struct. Biol. 2016, 40, 43–53. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Fry, S.C.; York, W.S.; Albersheim, P.; Darvill, A.; Hayashi, T.; Joseleau, J.P.; Kato, Y.; Lorences, E.P.; Maclachlan, G.A.; McNeil, M.; et al. An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol. Plant. 1993, 89, 1–3. [Google Scholar] [CrossRef]
- Gloster, T.M.; Ibatullin, F.M.; Macauley, K.; Eklof, J.M.; Roberts, S.; Turkenburg, J.P.; Bjornvad, M.E.; Jorgensen, P.L.; Danielsen, S.; Johansen, K.S.; et al. Characterization and three-dimensional structures of two distinct bacterial xyloglucanases from families GH5 and GH12. J. Biol. Chem. 2007, 282, 19177–19189. [Google Scholar] [CrossRef] [PubMed]
- Sandgren, M.; Gualfetti, P.J.; Shaw, A.; Gross, L.S.; Saldajeno, M.; Day, A.G.; Jones, T.A.; Mitchinson, C. Comparison of family 12 glycoside hydrolases and recruited substitutions important for thermal stability. Protein Sci. 2003, 12, 848–860. [Google Scholar] [CrossRef]
- Damasio, A.R.; Ribeiro, L.F.; Ribeiro, L.F.; Furtado, G.P.; Segato, F.; Almeida, F.B.; Crivellari, A.C.; Buckeridge, M.S.; Souza, T.A.; Murakami, M.T.; et al. Functional characterization and oligomerization of a recombinant xyloglucan-specific endo-beta-1,4-glucanase (GH12) from Aspergillus niveus. Biochim. Biophys. Acta 2012, 1824, 461–467. [Google Scholar] [CrossRef]
- Sun, P.; Li, X.; Dilokpimol, A.; Henrissat, B.; de Vries, R.P.; Kabel, M.A.; Makela, M.R. Fungal glycoside hydrolase family 44 xyloglucanases are restricted to the phylum Basidiomycota and show a distinct xyloglucan cleavage pattern. iScience 2022, 25, 103666. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, S.; Wu, X.; Liu, S.; Li, D.; Xu, H.; Gao, P.; Chen, G.; Wang, L. Subsite-specific contributions of different aromatic residues in the active site architecture of glycoside hydrolase family 12. Sci. Rep. 2015, 5, 18357. [Google Scholar] [CrossRef]
- Hernandez-Benitez, L.J.; Ramirez-Rodriguez, M.A.; Hernandez-Santoyo, A.; Rodriguez-Romero, A. A trimeric glycosylated GH45 cellulase from the red abalone (Haliotis rufescens) exhibits endo and exoactivity. PLoS ONE 2024, 19, e0301604. [Google Scholar] [CrossRef]
- Shallom, D.; Golan, G.; Shoham, G.; Shoham, Y. Effect of dimer dissociation on activity and thermostability of the alpha-glucuronidase from Geobacillus stearothermophilus: Dissecting the different oligomeric forms of family 67 glycoside hydrolases. J. Bacteriol. 2004, 186, 6928–6937. [Google Scholar] [CrossRef]
- Ruiz-Pernia, J.J.; Tunon, I.; Moliner, V.; Allemann, R.K. Why are some Enzymes Dimers? Flexibility and Catalysis in Thermotoga Maritima Dihydrofolate Reductase. ACS Catal. 2019, 9, 5902–5911. [Google Scholar] [CrossRef]
- Sinitsyna, O.A.; Fedorova, E.A.; Pravilnikov, A.G.; Rozhkova, A.M.; Skomarovsky, A.A.; Matys, V.Y.; Bubnova, T.M.; Okunev, O.N.; Vinetsky, Y.P.; Sinitsyn, A.P. Isolation and properties of xyloglucanases of Penicillium sp. Biochemistry 2010, 75, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Schiano-di-Cola, C.; Kolaczkowski, B.; Sorensen, T.H.; Christensen, S.J.; Cavaleiro, A.M.; Windahl, M.S.; Borch, K.; Morth, J.P.; Westh, P. Structural and biochemical characterization of a family 7 highly thermostable endoglucanase from the fungus Rasamsonia emersonii. FEBS J. 2020, 287, 2577–2596. [Google Scholar] [CrossRef] [PubMed]
- Contato, A.G.; Vici, A.C.; Pinheiro, V.E.; Oliveira TBd Freitas ENd Aranha, G.M.; Valvassora Junior, A.L.A.; Rechia, C.G.V.; Buckeridge, M.S.; Polizeli, M.d.L.T.d.M. Comparison of Trichoderma longibrachiatum xyloglucanase production using tamarind (Tamarindus indica) and jatoba (Hymenaea courbaril) seeds: Factorial design and immobilization on ionic supports. Fermentation 2022, 8, 510. [Google Scholar] [CrossRef]
- Matsuzawa, T.; Kameyama, A.; Nakamichi, Y.; Yaoi, K. Identification and characterization of two xyloglucan-specific endo-1,4-glucanases in Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2020, 104, 8761–8773. [Google Scholar] [CrossRef] [PubMed]
- Vitcosque, G.L.; Ribeiro, L.F.; de Lucas, R.C.; da Silva, T.M.; Ribeiro, L.F.; de Lima Damasio, A.R.; Farinas, C.S.; Goncalves, A.Z.; Segato, F.; Buckeridge, M.S.; et al. The functional properties of a xyloglucanase (GH12) of Aspergillus terreus expressed in Aspergillus nidulans may increase performance of biomass degradation. Appl. Microbiol. Biotechnol. 2016, 100, 9133–9144. [Google Scholar] [CrossRef]
- Furtado, G.P.; Santos, C.R.; Cordeiro, R.L.; Ribeiro, L.F.; de Moraes, L.A.; Damasio, A.R.; Polizeli Mde, L.; Lourenzoni, M.R.; Murakami, M.T.; Ward, R.J. Enhanced xyloglucan-specific endo-beta-1,4-glucanase efficiency in an engineered CBM44-XegA chimera. Appl. Microbiol. Biotechnol. 2015, 99, 5095–5107. [Google Scholar] [CrossRef]
- Ostby, H.; Varnai, A. Hemicellulolytic enzymes in lignocellulose processing. Essays Biochem. 2023, 67, 533–550. [Google Scholar] [CrossRef]
- Rodd, A.M.; Mawhinney, W.M.; Brumer, H. A scalable, chromatography-free, biocatalytic method to produce the xyloglucan heptasaccharide XXXG. Biotechnol. Biofuels Bioprod. 2024, 17, 116. [Google Scholar] [CrossRef] [PubMed]
- Grishutin, S.G.; Gusakov, A.V.; Markov, A.V.; Ustinov, B.B.; Semenova, M.V.; Sinitsyn, A.P. Specific xyloglucanases as a new class of polysaccharide-degrading enzymes. Biochim. Biophys. Acta 2004, 1674, 268–281. [Google Scholar] [CrossRef]
- Stanley, J.S.; Mansbridge, S.C.; Bedford, M.R.; Connerton, I.F.; Mellits, K.H. Prebiotic Xylo-Oligosaccharides Modulate the Gut Microbiome to Improve Innate Immunity and Gut Barrier Function and Enhance Performance in Piglets Experiencing Post-Weaning Diarrhoea. Microorganisms 2025, 13, 1760. [Google Scholar] [CrossRef]
- Zhou, Y.; Tang, S.; Lv, Y.; Zhang, D.; Huang, X.; Chen, Y.; Lai, C.; Yong, Q. The prebiotic impacts of galactose side-chain of tamarind xyloglucan oligosaccharides on gut microbiota. Heliyon 2024, 10, e37864. [Google Scholar] [CrossRef]
- Kaida, R.; Kaku, T.; Baba, K.; Oyadomari, M.; Watanabe, T.; Nishida, K.; Kanaya, T.; Shani, Z.; Shoseyov, O.; Hayashi, T. Loosening xyloglucan accelerates the enzymatic degradation of cellulose in wood. Mol. Plant 2009, 2, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Jacomini, D.; Bussler, L.; da-Conceicao Silva, J.L.; Maller, A.; Kadowaki, M.K.; Simao, R.C.G. Biopolishing of denim by the recombinant xylanase II of Caulobacter crescentus. Braz. J. Microbiol. 2023, 54, 1559–1564. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.R.; Lant, N.J.; Berlinguer-Palmini, R.; Burgess, J.G. Chemical mapping of xyloglucan distribution and cellulose crystallinity in cotton textiles reveals novel enzymatic targets to improve clothing longevity. Carbohydr. Polym. 2024, 339, 122243. [Google Scholar] [CrossRef]
- Marzoug, I.B.; Cheriaa, R. Effects optimization of bio-polishing industrial process parameters. J. Text. Sci. Technol. 2023, 9, 30–51. [Google Scholar] [CrossRef]
- Ravn, J.L.; Martens, H.J.; Pettersson, D.; Pedersen, N.R. Enzymatic solubilisation and degradation of soybean fibre demonstrated by viscosity, fibre analysis and microscopy. Int. J. Biol. 2015, 7, 1. [Google Scholar] [CrossRef]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP); Bampidis, V.; Azimonti, G.; Bastos, M.d.L.; Christensen, H.; Durjava, M.; Dusemund, B.; Kouba, M.; López-Alonso, M.; Puente, S.L.; et al. Safety and efficacy of a feed additive consisting of endo-1, 4-β xylanase, endo-1, 4-β-glucanase and xyloglucan-specific-endo-β-1, 4-glucanase produced by Trichoderma citrinoviride DSM 33578 (Huvezym® neXo) for all Suidae (Huvepharma EOOD). EFSA J. 2024, 22, e8643. [Google Scholar]
- Plouhinec, L.; Neugnot, V.; Lafond, M.; Berrin, J.G. Carbohydrate-active enzymes in animal feed. Biotechnol. Adv. 2023, 65, 108145. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.A.; Takasuka, T.E.; Deng, K.; Bianchetti, C.M.; Udell, H.S.; Prom, B.M.; Kim, H.; Adams, P.D.; Northen, T.R.; Fox, B.G. Multifunctional cellulase catalysis targeted by fusion to different carbohydrate-binding modules. Biotechnol. Biofuels 2015, 8, 220. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, A.C.; Scholfield, M.R.; Rowe, R.K.; Ford, M.C.; Alexander, A.T.; Mehl, R.A.; Ho, P.S. Increasing Enzyme Stability and Activity through Hydrogen Bond-Enhanced Halogen Bonds. Biochemistry 2018, 57, 4135–4147. [Google Scholar] [CrossRef]
- Wijma, H.J.; Floor, R.J.; Jekel, P.A.; Baker, D.; Marrink, S.J.; Janssen, D.B. Computationally designed libraries for rapid enzyme stabilization. Protein Eng. Des. Sel. 2014, 27, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, W.; Ji, P.; Zhao, Y.; Hua, C.; Han, C. Engineering the conserved and noncatalytic residues of a thermostable beta-1,4-endoglucanase to improve specific activity and thermostability. Sci. Rep. 2018, 8, 2954. [Google Scholar] [CrossRef]
- Li, Y.; Yue, S.; Li, P.; Zeng, J.; Guo, J.; Xiong, D.; Zhang, S.; Deng, T.; Yuan, L. Genome Sequencing and Comparative Genomics of the Hyper-Cellulolytic Fungus Talaromyces pinophilus Y117. J. Fungi 2025, 11, 614. [Google Scholar] [CrossRef]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gislason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Powlowski, J.; Mahajan, S.; Schapira, M.; Master, E.R. Substrate recognition and hydrolysis by a fungal xyloglucan-specific family 12 hydrolase. Carbohydr Res. 2009, 344, 1175–1179. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Nie, J.; Li, P.; Zhang, C.; Zeng, J.; Yue, S.; Guo, J.; Xiong, D.; Zhang, S.; Huang, G.; Yuan, L. Characterization of Xyloglucanase TpXEG12a from Talaromyces pinophilus. Int. J. Mol. Sci. 2026, 27, 294. https://doi.org/10.3390/ijms27010294
Nie J, Li P, Zhang C, Zeng J, Yue S, Guo J, Xiong D, Zhang S, Huang G, Yuan L. Characterization of Xyloglucanase TpXEG12a from Talaromyces pinophilus. International Journal of Molecular Sciences. 2026; 27(1):294. https://doi.org/10.3390/ijms27010294
Chicago/Turabian StyleNie, Junhui, Peng Li, Cheng Zhang, Jing Zeng, Siyuan Yue, Jianjun Guo, Dawei Xiong, Shuaiwen Zhang, Guochang Huang, and Lin Yuan. 2026. "Characterization of Xyloglucanase TpXEG12a from Talaromyces pinophilus" International Journal of Molecular Sciences 27, no. 1: 294. https://doi.org/10.3390/ijms27010294
APA StyleNie, J., Li, P., Zhang, C., Zeng, J., Yue, S., Guo, J., Xiong, D., Zhang, S., Huang, G., & Yuan, L. (2026). Characterization of Xyloglucanase TpXEG12a from Talaromyces pinophilus. International Journal of Molecular Sciences, 27(1), 294. https://doi.org/10.3390/ijms27010294





