RNA-Binding Proteins: Modulators of Canonical Wnt Signaling Pathway
Abstract
1. Introduction
2. RBPs in Regulation of Canonical Wnt Signaling
2.1. IGF2BP1
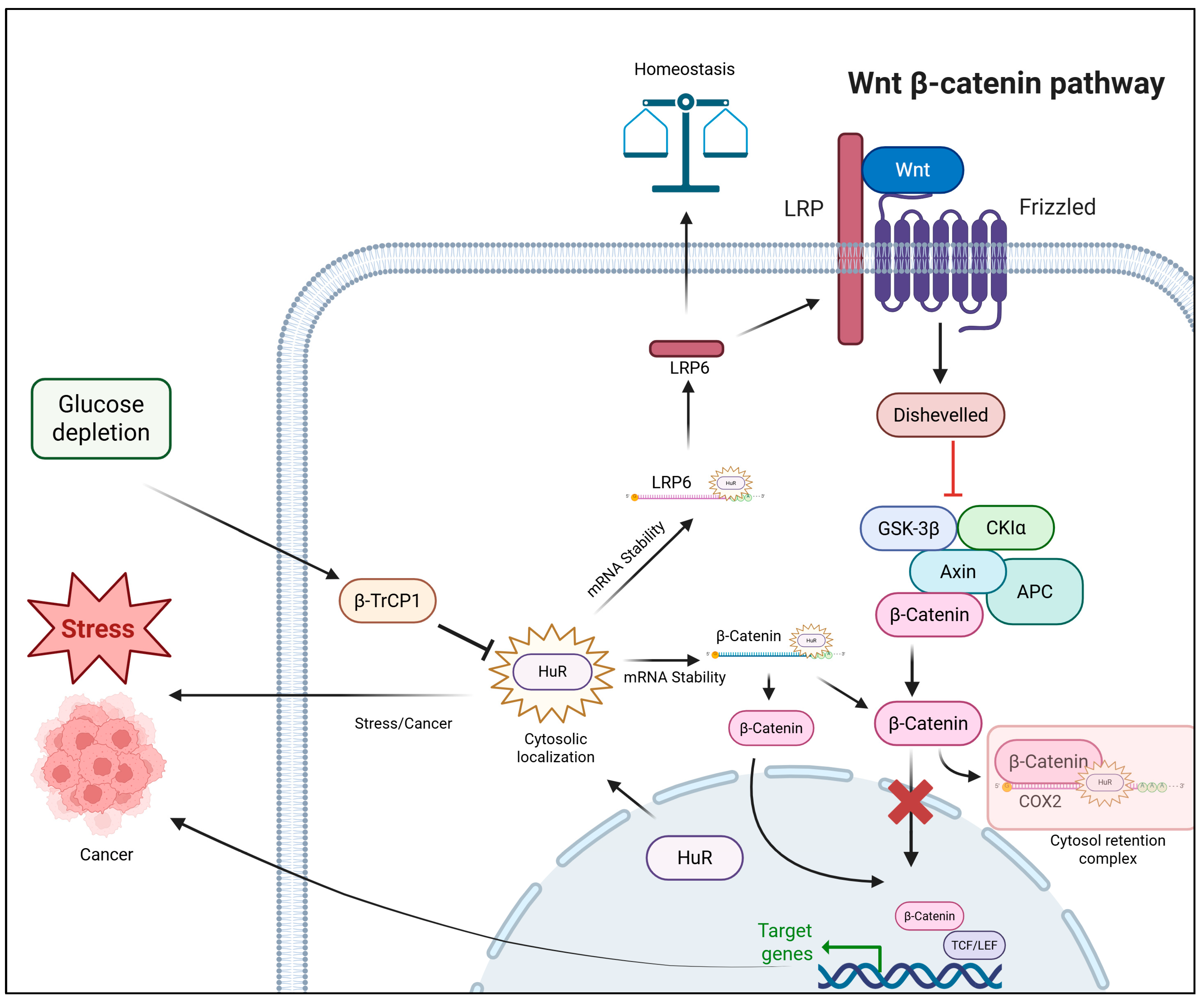
2.2. ELAVL1 (HuR)
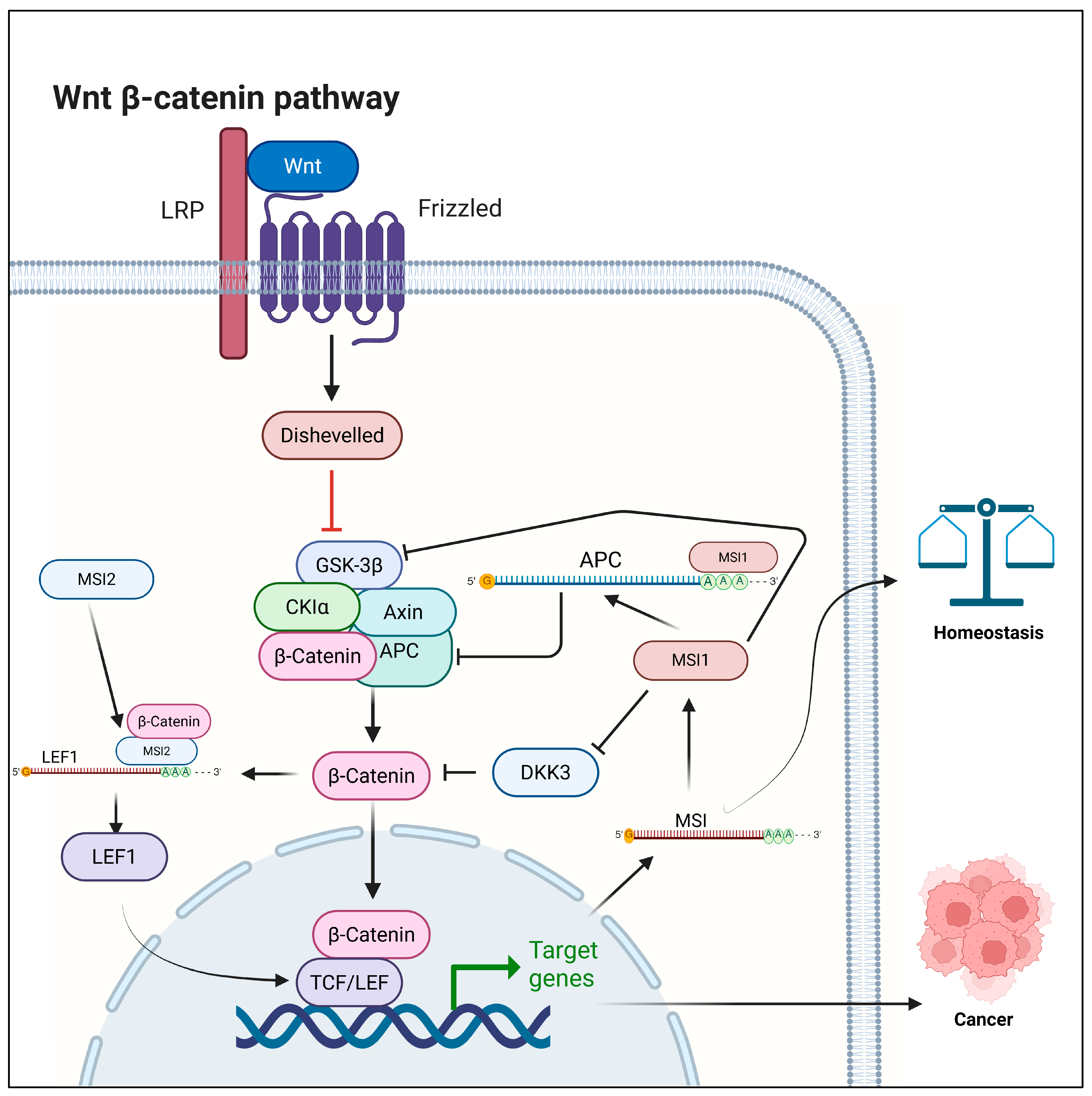
2.3. Musashi
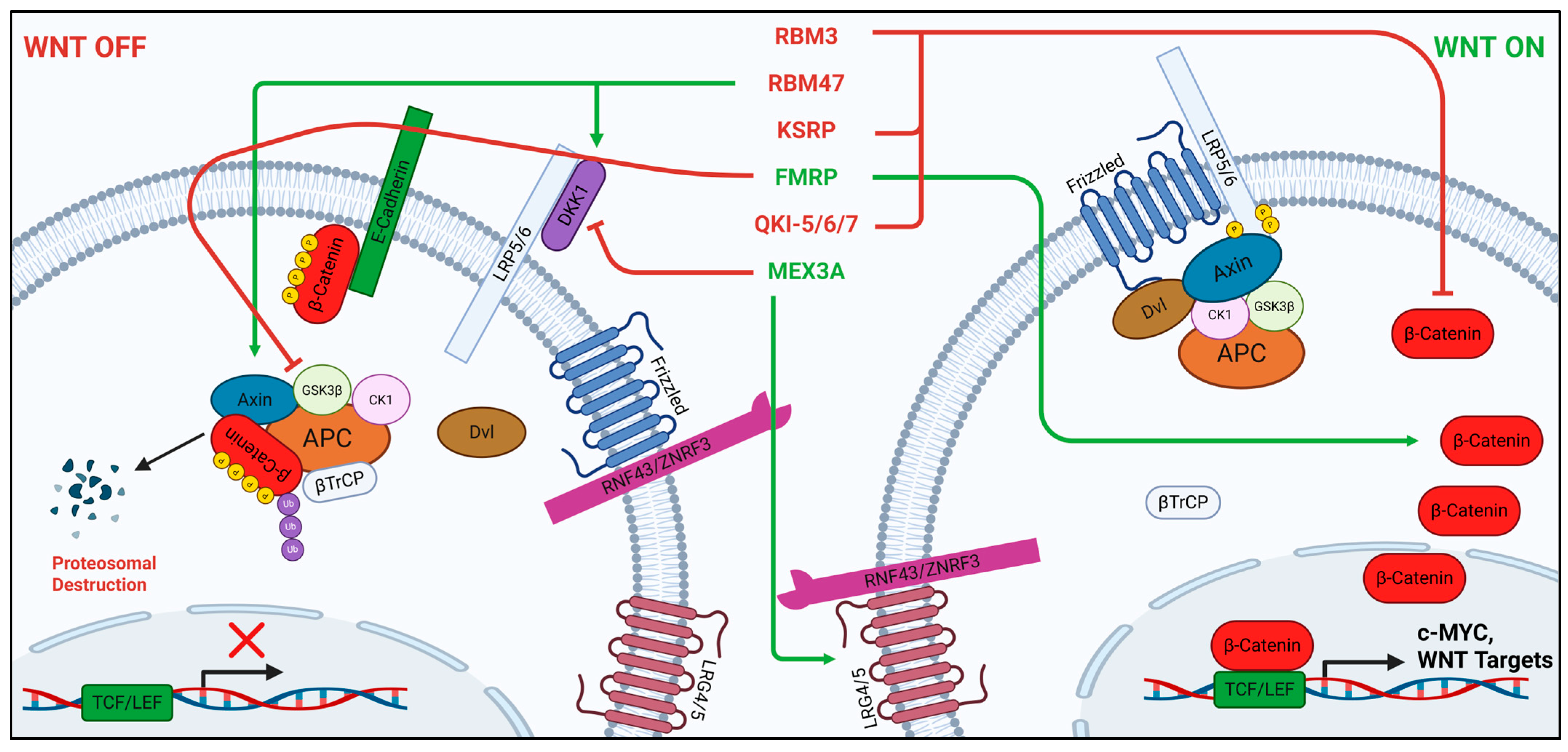
2.4. RBM Family
2.5. KSRP
2.6. FMRP
2.7. Quaking
2.8. MEX3A
3. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nusse, R.; Brown, A.; Papkoff, J.; Scambler, P.; Shackleford, G.; McMahon, A.; Moon, R.; Varmus, H. A new nomenclature for int-1 and related genes: The Wnt gene family. Cell 1991, 64, 231. [Google Scholar] [CrossRef] [PubMed]
- Niehrs, C.; Acebron, S.P. Mitotic and mitogenic Wnt signalling. EMBO J. 2012, 31, 2705–2713. [Google Scholar] [CrossRef] [PubMed]
- Nusse, R.; Clevers, H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef] [PubMed]
- Chae, W.J.; Bothwell, A.L.M. Canonical and Non-Canonical Wnt Signaling in Immune Cells. Trends Immunol. 2018, 39, 830–847. [Google Scholar] [CrossRef]
- Mao, J.; Wang, J.; Liu, B.; Pan, W.; Farr, G.H., 3rd; Flynn, C.; Yuan, H.; Takada, S.; Kimelman, D.; Li, L.; et al. Low-density lipoprotein receptor-related protein-5 binds to Axin and regulates the canonical Wnt signaling pathway. Mol. Cell 2001, 7, 801–809. [Google Scholar] [CrossRef]
- González-Sancho, J.M.; Aguilera, O.; García, J.M.; Pendás-Franco, N.; Peña, C.; Cal, S.; García de Herreros, A.; Bonilla, F.; Muñoz, A. The Wnt antagonist DICKKOPF-1 gene is a downstream target of β-catenin/TCF and is downregulated in human colon cancer. Oncogene 2005, 24, 1098–1103. [Google Scholar] [CrossRef]
- Tian, X.; Liu, Z.; Niu, B.; Zhang, J.; Tan, T.K.; Lee, S.R.; Zhao, Y.; Harris, D.C.; Zheng, G. E-cadherin/β-catenin complex and the epithelial barrier. J. Biomed. Biotechnol. 2011, 2011, 567305. [Google Scholar] [CrossRef]
- MacDonald, B.T.; Tamai, K.; He, X. Wnt/β-catenin signaling: Components, mechanisms, and diseases. Dev. Cell 2009, 17, 9–26. [Google Scholar] [CrossRef]
- Bao, J.; Zheng, J.J.; Wu, D. The structural basis of DKK-mediated inhibition of Wnt/LRP signaling. Sci. Signal. 2012, 5, pe22. [Google Scholar] [CrossRef] [PubMed]
- Ter Steege, E.J.; Bakker, E.R.M. The role of R-spondin proteins in cancer biology. Oncogene 2021, 40, 6469–6478. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, Q.; Xiao, J.; Niu, C.; Li, Y.; Zhang, X.; Zhou, Z.; Shu, G.; Yin, G. Wnt/β-catenin signalling: Function, biological mechanisms, and therapeutic opportunities. Signal Transduct. Target. Ther. 2022, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Qin, K.; Yu, M.; Fan, J.; Wang, H.; Zhao, P.; Zhao, G.; Zeng, W.; Chen, C.; Wang, Y.; Wang, A.; et al. Canonical and noncanonical Wnt signaling: Multilayered mediators, signaling mechanisms and major signaling crosstalk. Genes Dis. 2024, 11, 103–134. [Google Scholar] [CrossRef] [PubMed]
- He, T.C.; Sparks, A.B.; Rago, C.; Hermeking, H.; Zawel, L.; da Costa, L.T.; Morin, P.J.; Vogelstein, B.; Kinzler, K.W. Identification of c-MYC as a target of the APC pathway. Science 1998, 281, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Shtutman, M.; Zhurinsky, J.; Simcha, I.; Albanese, C.; D’Amico, M.; Pestell, R.; Ben-Ze’ev, A. The cyclin D1 gene is a target of the β-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA 1999, 96, 5522–5527. [Google Scholar] [CrossRef]
- Jho, E.H.; Zhang, T.; Domon, C.; Joo, C.K.; Freund, J.N.; Costantini, F. Wnt/β-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 2002, 22, 1172–1183. [Google Scholar] [CrossRef]
- Liao, J.Y.; Yang, B.; Shi, C.P.; Deng, W.X.; Deng, J.S.; Cen, M.F.; Zheng, B.Q.; Zhan, Z.L.; Liang, Q.L.; Wang, J.E.; et al. RBPWorld for exploring functions and disease associations of RNA-binding proteins across species. Nucleic Acids Res. 2025, 53, D220–D232. [Google Scholar] [CrossRef]
- Lunde, B.M.; Moore, C.; Varani, G. RNA-binding proteins: Modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007, 8, 479–490. [Google Scholar] [CrossRef]
- Sherwood, A.V.; Henkin, T.M. Riboswitch-Mediated Gene Regulation: Novel RNA Architectures Dictate Gene Expression Responses. Annu. Rev. Microbiol. 2016, 70, 361–374. [Google Scholar] [CrossRef]
- Qin, H.; Ni, H.; Liu, Y.; Yuan, Y.; Xi, T.; Li, X.; Zheng, L. RNA-binding proteins in tumor progression. J. Hematol. Oncol. 2020, 13, 90. [Google Scholar] [CrossRef]
- Noubissi, F.K.; Elcheva, I.; Bhatia, N.; Shakoori, A.; Ougolkov, A.; Liu, J.; Minamoto, T.; Ross, J.; Fuchs, S.Y.; Spiegelman, V.S. CRD-BP mediates stabilization of betaTrCP1 and c-myc mRNA in response to β-catenin signalling. Nature 2006, 441, 898–901. [Google Scholar] [CrossRef]
- Fang, F.; Guo, C.; Zheng, W.; Wang, Q.; Zhou, L. Exosome-Mediated Transfer of miR-1323 from Cancer-Associated Fibroblasts Confers Radioresistance of C33A Cells by Targeting PABPN1 and Activating Wnt/β-Catenin Signaling Pathway in Cervical Cancer. Reprod. Sci. 2022, 29, 1809–1821. [Google Scholar] [CrossRef]
- Liu, L.; Christodoulou-Vafeiadou, E.; Rao, J.N.; Zou, T.; Xiao, L.; Chung, H.K.; Yang, H.; Gorospe, M.; Kontoyiannis, D.; Wang, J.Y. RNA-binding protein HuR promotes growth of small intestinal mucosa by activating the Wnt signaling pathway. Mol. Biol. Cell 2014, 25, 3308–3318. [Google Scholar] [CrossRef]
- Huai, Y.; Chen, Z.; Deng, X.; Wang, X.; Mao, W.; Miao, Z.; Li, Y.; Li, H.; Lin, X.; Qian, A. An integrated genome-wide analysis identifies HUR/ELAVL1 as a positive regulator of osteogenesis through enhancing the β-catenin signaling activity. Genes Dis. 2023, 10, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Spears, E.; Neufeld, K.L. Novel double-negative feedback loop between adenomatous polyposis coli and Musashi1 in colon epithelia. J. Biol. Chem. 2011, 286, 4946–4950. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Yousefi, M.; Nakauka-Ddamba, A.; Li, F.; Vandivier, L.; Parada, K.; Woo, D.H.; Wang, S.; Naqvi, A.S.; Rao, S.; et al. The Msi Family of RNA-Binding Proteins Function Redundantly as Intestinal Oncoproteins. Cell Rep. 2015, 13, 2440–2455. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, M.; Sevim, O.; Goff, A.; Raynor, M.; Park, H.; Mancini, E.J.; Nguyen, D.T.T.; Chevassut, T.; Blair, A.; Castellano, L.; et al. β-Catenin interacts with canonical RBPs including MSI2 to associate with a Wnt signalling mRNA network in myeloid leukaemia cells. Oncogene 2025, 44, 2490–2503. [Google Scholar] [CrossRef]
- Zhang, S.; Lv, C.; Niu, Y.; Li, C.; Li, X.; Shang, Y.; Zhang, Y.; Zhang, Y.; Zhang, Y.; Zeng, Y. RBM3 suppresses stemness remodeling of prostate cancer in bone microenvironment by modulating N6-methyladenosine on CTNNB1 mRNA. Cell Death Dis. 2023, 14, 91. [Google Scholar] [CrossRef]
- Vanharanta, S.; Marney, C.B.; Shu, W.; Valiente, M.; Zou, Y.; Mele, A.; Darnell, R.B.; Massagué, J. Loss of the multifunctional RNA-binding protein RBM47 as a source of selectable metastatic traits in breast cancer. eLife 2014, 3, e02734. [Google Scholar] [CrossRef]
- Shen, D.J.; Jiang, Y.H.; Li, J.Q.; Xu, L.W.; Tao, K.Y. The RNA-binding protein RBM47 inhibits non-small cell lung carcinoma metastasis through modulation of AXIN1 mRNA stability and Wnt/β-catentin signaling. Surg. Oncol. 2020, 34, 31–39. [Google Scholar] [CrossRef]
- Bikkavilli, R.K.; Malbon, C.C. Dishevelled-KSRP complex regulates Wnt signaling through post-transcriptional stabilization of β-catenin mRNA. J. Cell Sci. 2010, 123, 1352–1362. [Google Scholar] [CrossRef]
- Luo, Y.; Shan, G.; Guo, W.; Smrt, R.D.; Johnson, E.B.; Li, X.; Pfeiffer, R.L.; Szulwach, K.E.; Duan, R.; Barkho, B.Z.; et al. Fragile X mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells. PLoS Genet. 2010, 6, e1000898. [Google Scholar] [CrossRef] [PubMed]
- Pedini, G.; Buccarelli, M.; Bianchi, F.; Pacini, L.; Cencelli, G.; D’Alessandris, Q.G.; Martini, M.; Giannetti, S.; Sasso, F.; Melocchi, V.; et al. FMRP modulates the Wnt signalling pathway in glioblastoma. Cell Death Dis. 2022, 13, 719. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, X.; Sun, C.; Shi, C.; Hua, D.; Yu, L.; Wen, Y.; Hua, F.; Wang, Q.; Zhou, Q.; et al. Quaking-5 suppresses aggressiveness of lung cancer cells through inhibiting β-catenin signaling pathway. Oncotarget 2017, 8, 82174–82184. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Fu, H.; Zhang, J.; Lu, X.; Yu, F.; Jin, L.; Bai, L.; Huang, B.; Shen, L.; Feng, Y.; et al. RNA-binding protein quaking, a critical regulator of colon epithelial differentiation and a suppressor of colon cancer. Gastroenterology 2010, 138, 231–240.e231–e235. [Google Scholar] [CrossRef]
- Cheng, L.T.; Tan, G.Y.T.; Chang, F.P.; Wang, C.K.; Chou, Y.C.; Hsu, P.H.; Hwang-Verslues, W.W. Core clock gene BMAL1 and RNA-binding protein MEX3A collaboratively regulate Lgr5 expression in intestinal crypt cells. Sci. Rep. 2023, 13, 17597. [Google Scholar] [CrossRef]
- Wang, Y.; Liang, Q.; Lei, K.; Zhu, Q.; Zeng, D.; Liu, Y.; Lu, Y.; Kang, T.; Tang, N.; Huang, L.; et al. Targeting MEX3A attenuates metastasis of breast cancer via β-catenin signaling pathway inhibition. Cancer Lett. 2021, 521, 50–63. [Google Scholar] [CrossRef]
- Jiang, X.; Liu, B.; Nie, Z.; Duan, L.; Xiong, Q.; Jin, Z.; Yang, C.; Chen, Y. The role of m6A modification in the biological functions and diseases. Signal Transduct. Target. Ther. 2021, 6, 74. [Google Scholar] [CrossRef]
- Nielsen, F.C.; Nielsen, J.; Kristensen, M.A.; Koch, G.; Christiansen, J. Cytoplasmic trafficking of IGF-II mRNA-binding protein by conserved KH domains. J. Cell Sci. 2002, 115, 2087–2097. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295, Correction in Nat. Cell Biol. 2018, 20, 1098. [Google Scholar] [CrossRef]
- Duan, M.; Liu, H.; Xu, S.; Yang, Z.; Zhang, F.; Wang, G.; Wang, Y.; Zhao, S.; Jiang, X. IGF2BPs as novel m6A readers: Diverse roles in regulating cancer cell biological functions, hypoxia adaptation, metabolism, and immunosuppressive tumor microenvironment. Genes Dis. 2024, 11, 890–920. [Google Scholar] [CrossRef]
- Singh, V.; Walter, V.; Elcheva, I.; Imamura Kawasawa, Y.; Spiegelman, V.S. Global role of IGF2BP1 in controlling the expression of Wnt/β-catenin-regulated genes in colorectal cancer cells. Front. Cell Dev. Biol. 2023, 11, 1236356. [Google Scholar] [CrossRef] [PubMed]
- Elcheva, I.; Goswami, S.; Noubissi, F.K.; Spiegelman, V.S. CRD-BP protects the coding region of betaTrCP1 mRNA from miR-183-mediated degradation. Mol. Cell 2009, 35, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Manieri, N.A.; Drylewicz, M.R.; Miyoshi, H.; Stappenbeck, T.S. Igf2bp1 is required for full induction of Ptgs2 mRNA in colonic mesenchymal stem cells in mice. Gastroenterology 2012, 143, 110–121.e10. [Google Scholar] [CrossRef] [PubMed]
- Noubissi, F.K.; Goswami, S.; Sanek, N.A.; Kawakami, K.; Minamoto, T.; Moser, A.; Grinblat, Y.; Spiegelman, V.S. Wnt signaling stimulates transcriptional outcome of the Hedgehog pathway by stabilizing GLI1 mRNA. Cancer Res. 2009, 69, 8572–8578. [Google Scholar] [CrossRef]
- Noubissi, F.K.; Nikiforov, M.A.; Colburn, N.; Spiegelman, V.S. Transcriptional Regulation of CRD-BP by c-myc: Implications for c-myc Functions. Genes Cancer 2010, 1, 1074–1082. [Google Scholar] [CrossRef]
- Leeds, P.; Kren, B.T.; Boylan, J.M.; Betz, N.A.; Steer, C.J.; Gruppuso, P.A.; Ross, J. Developmental regulation of CRD-BP, an RNA-binding protein that stabilizes c-myc mRNA in vitro. Oncogene 1997, 14, 1279–1286. [Google Scholar] [CrossRef]
- Singh, V.; Gowda, C.P.; Singh, V.; Ganapathy, A.S.; Karamchandani, D.M.; Eshelman, M.A.; Yochum, G.S.; Nighot, P.; Spiegelman, V.S. The mRNA-binding protein IGF2BP1 maintains intestinal barrier function by up-regulating occludin expression. J. Biol. Chem. 2020, 295, 8602–8612. [Google Scholar] [CrossRef]
- Lv, D.; Ding, S.; Zhong, L.; Tu, J.; Li, H.; Yao, H.; Zou, Y.; Zeng, Z.; Liao, Y.; Wan, X.; et al. M6A demethylase FTO-mediated downregulation of DACT1 mRNA stability promotes Wnt signaling to facilitate osteosarcoma progression. Oncogene 2022, 41, 1727–1741. [Google Scholar] [CrossRef]
- Huang, Q.; Guo, H.; Wang, S.; Ma, Y.; Chen, H.; Li, H.; Li, J.; Li, X.; Yang, F.; Qiu, M.; et al. A novel circular RNA, circXPO1, promotes lung adenocarcinoma progression by interacting with IGF2BP1. Cell Death Dis. 2020, 11, 1031. [Google Scholar] [CrossRef]
- Singh, V.; Singh, A.; Liu, A.J.; Fuchs, S.Y.; Sharma, A.K.; Spiegelman, V.S. RNA Binding Proteins as Potential Therapeutic Targets in Colorectal Cancer. Cancers 2024, 16, 3502. [Google Scholar] [CrossRef]
- Wallis, N.; Gershon, T.; Shaaby, S.; Oberman, F.; Grunewald, M.; Duran, D.; Singh, A.; Vainer, G.; Spiegelman, V.S.; Sharma, A.K.; et al. AVJ16 inhibits lung carcinoma by targeting IGF2BP1. Oncogene 2025, 44, 3239–3254. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Singh, V.; Wallis, N.; Abis, G.; Oberman, F.; Wood, T.; Dhamdhere, M.; Gershon, T.; Ramos, A.; Yisraeli, J.; et al. Development of a specific and potent IGF2BP1 inhibitor: A promising therapeutic agent for IGF2BP1-expressing cancers. Eur. J. Med. Chem. 2024, 263, 115940. [Google Scholar] [CrossRef] [PubMed]
- Shang, F.F.; Lu, Q.; Lin, T.; Pu, M.; Xiao, R.; Liu, W.; Deng, H.; Guo, H.; Quan, Z.S.; Ding, C.; et al. Discovery of Triazolyl Derivatives of Cucurbitacin B Targeting IGF2BP1 against Non-Small Cell Lung Cancer. J. Med. Chem. 2023, 66, 12931–12949. [Google Scholar] [CrossRef] [PubMed]
- Wallis, N.; Oberman, F.; Shurrush, K.; Germain, N.; Greenwald, G.; Gershon, T.; Pearl, T.; Abis, G.; Singh, V.; Singh, A.; et al. Small molecule inhibitor of Igf2bp1 represses Kras and a pro-oncogenic phenotype in cancer cells. RNA Biol. 2022, 19, 26–43. [Google Scholar] [CrossRef]
- Mahapatra, L.; Andruska, N.; Mao, C.; Le, J.; Shapiro, D.J. A Novel IMP1 Inhibitor, BTYNB, Targets c-Myc and Inhibits Melanoma and Ovarian Cancer Cell Proliferation. Transl. Oncol. 2017, 10, 818–827. [Google Scholar] [CrossRef]
- Ma, W.J.; Cheng, S.; Campbell, C.; Wright, A.; Furneaux, H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 1996, 271, 8144–8151. [Google Scholar] [CrossRef]
- Pascale, A.; Govoni, S. The complex world of post-transcriptional mechanisms: Is their deregulation a common link for diseases? Focus on ELAV-like RNA-binding proteins. Cell. Mol. Life Sci. 2012, 69, 501–517. [Google Scholar] [CrossRef]
- Srikantan, S.; Gorospe, M. HuR function in disease. Front. Biosci. (Landmark Ed.) 2012, 17, 189–205. [Google Scholar] [CrossRef]
- Wu, X.; Xu, L. The RNA-binding protein HuR in human cancer: A friend or foe? Adv. Drug Deliv. Rev. 2022, 184, 114179. [Google Scholar] [CrossRef]
- Fan, X.C.; Steitz, J.A. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. USA 1998, 95, 15293–15298. [Google Scholar] [CrossRef]
- Peng, S.S.; Chen, C.Y.; Xu, N.; Shyu, A.B. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998, 17, 3461–3470. [Google Scholar] [CrossRef]
- Wang, J.; Guo, Y.; Chu, H.; Guan, Y.; Bi, J.; Wang, B. Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis. Int. J. Mol. Sci. 2013, 14, 10015–10041. [Google Scholar] [CrossRef] [PubMed]
- Good, P.J. A conserved family of elav-like genes in vertebrates. Proc. Natl. Acad. Sci. USA 1995, 92, 4557–4561. [Google Scholar] [CrossRef] [PubMed]
- Doller, A.; Pfeilschifter, J.; Eberhardt, W. Signalling pathways regulating nucleo-cytoplasmic shuttling of the mRNA-binding protein HuR. Cell. Signal. 2008, 20, 2165–2173. [Google Scholar] [CrossRef] [PubMed]
- Gallouzi, I.E.; Brennan, C.M.; Steitz, J.A. Protein ligands mediate the CRM1-dependent export of HuR in response to heat shock. RNA 2001, 7, 1348–1361, Erratum in RNA 2003, 9, 1410. [Google Scholar] [CrossRef]
- Güttinger, S.; Mühlhäusser, P.; Koller-Eichhorn, R.; Brennecke, J.; Kutay, U. Transportin2 functions as importin and mediates nuclear import of HuR. Proc. Natl. Acad. Sci. USA 2004, 101, 2918–2923. [Google Scholar] [CrossRef]
- Koyama, M.; Matsuura, Y. Mechanistic insights from the recent structures of the CRM1 nuclear export complex and its disassembly intermediate. Biophysics 2012, 8, 145–150. [Google Scholar] [CrossRef]
- Natalizio, B.J.; Wente, S.R. Postage for the messenger: Designating routes for nuclear mRNA export. Trends Cell Biol. 2013, 23, 365–373. [Google Scholar] [CrossRef]
- Finan, J.M.; Sutton, T.L.; Dixon, D.A.; Brody, J.R. Targeting the RNA-Binding Protein HuR in Cancer. Cancer Res. 2023, 83, 3507–3516. [Google Scholar] [CrossRef]
- Lebedeva, S.; Jens, M.; Theil, K.; Schwanhäusser, B.; Selbach, M.; Landthaler, M.; Rajewsky, N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell 2011, 43, 340–352. [Google Scholar] [CrossRef]
- Uren, P.J.; Burns, S.C.; Ruan, J.; Singh, K.K.; Smith, A.D.; Penalva, L.O. Genomic analyses of the RNA-binding protein Hu antigen R (HuR) identify a complex network of target genes and novel characteristics of its binding sites. J. Biol. Chem. 2011, 286, 37063–37066. [Google Scholar] [CrossRef]
- Liu, Z.; Li, B.; Hu, H.; Li, X.; Zhang, X. Potential of RNA-binding protein human antigen R as a driver of osteogenic differentiation in osteoporosis. J. Orthop. Surg. Res. 2022, 17, 234. [Google Scholar] [CrossRef]
- López de Silanes, I.; Fan, J.; Yang, X.; Zonderman, A.B.; Potapova, O.; Pizer, E.S.; Gorospe, M. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene 2003, 22, 7146–7154. [Google Scholar] [CrossRef] [PubMed]
- Ale-Agha, N.; Galban, S.; Sobieroy, C.; Abdelmohsen, K.; Gorospe, M.; Sies, H.; Klotz, L.O. HuR regulates gap junctional intercellular communication by controlling β-catenin levels and adherens junction integrity. Hepatology 2009, 50, 1567–1576. [Google Scholar] [CrossRef]
- Chou, S.D.; Murshid, A.; Eguchi, T.; Gong, J.; Calderwood, S.K. HSF1 regulation of β-catenin in mammary cancer cells through control of HuR/elavL1 expression. Oncogene 2015, 34, 2178–2188. [Google Scholar] [CrossRef] [PubMed]
- Chai, W.; Liu, R.; Li, F.; Zhang, Z.; Lei, B. Long noncoding RNA TSLNC8 enhances pancreatic cancer aggressiveness by regulating CTNNB1 expression via association with HuR. Hum. Cell 2021, 34, 165–176. [Google Scholar] [CrossRef]
- Lee, H.K.; Jeong, S. β-Catenin stabilizes cyclooxygenase-2 mRNA by interacting with AU-rich elements of 3′-UTR. Nucleic Acids Res. 2006, 34, 5705–5714. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.; Kwak, H.; Lee, H.K.; Hyun, S.; Jeong, S. β-Catenin recognizes a specific RNA motif in the cyclooxygenase-2 mRNA 3′-UTR and interacts with HuR in colon cancer cells. Nucleic Acids Res. 2012, 40, 6863–6872. [Google Scholar] [CrossRef]
- Kim, I.; Hur, J.; Jeong, S. HuR represses Wnt/β-catenin-mediated transcriptional activity by promoting cytoplasmic localization of β-catenin. Biochem. Biophys. Res. Commun. 2015, 457, 65–70. [Google Scholar] [CrossRef]
- Chu, P.C.; Chuang, H.C.; Kulp, S.K.; Chen, C.S. The mRNA-stabilizing factor HuR protein is targeted by β-TrCP protein for degradation in response to glycolysis inhibition. J. Biol. Chem. 2012, 287, 43639–43650. [Google Scholar] [CrossRef]
- Nakamura, M.; Okano, H.; Blendy, J.A.; Montell, C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron 1994, 13, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, S.; Imai, T.; Hamaguchi, K.; Okabe, M.; Aruga, J.; Nakajima, K.; Yasutomi, D.; Nagata, T.; Kurihara, Y.; Uesugi, S.; et al. Mouse-Musashi-1, a neural RNA-binding protein highly enriched in the mammalian CNS stem cell. Dev. Biol. 1996, 176, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Chen, H.; Cao, S.; Guo, J.; Liu, Z.; Long, S. RNA-binding MSI proteins and their related cancers: A medicinal chemistry perspective. Bioorg. Chem. 2024, 143, 107044. [Google Scholar] [CrossRef] [PubMed]
- Nagata, T.; Kanno, R.; Kurihara, Y.; Uesugi, S.; Imai, T.; Sakakibara, S.; Okano, H.; Katahira, M. Structure, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi1. J. Mol. Biol. 1999, 287, 315–330. [Google Scholar] [CrossRef]
- Iwaoka, R.; Nagata, T.; Tsuda, K.; Imai, T.; Okano, H.; Kobayashi, N.; Katahira, M. Backbone and side chain assignments of the second RNA-binding domain of Musashi-1 in its free form and in complex with 5-mer RNA. Biomol. NMR Assign. 2017, 11, 265–268. [Google Scholar] [CrossRef]
- Kawahara, H.; Okada, Y.; Imai, T.; Iwanami, A.; Mischel, P.S.; Okano, H. Musashi1 cooperates in abnormal cell lineage protein 28 (Lin28)-mediated let-7 family microRNA biogenesis in early neural differentiation. J. Biol. Chem. 2011, 286, 16121–16130. [Google Scholar] [CrossRef]
- Chen, H.Y.; Wang, M.L.; Laurent, B.; Hsu, C.H.; Chen, M.T.; Lin, L.T.; Shen, J.; Chang, W.C.; Hsu, J.; Hung, M.C.; et al. Musashi-1 promotes stress-induced tumor progression through recruitment of AGO2. Theranostics 2020, 10, 201–217. [Google Scholar] [CrossRef]
- Bley, N.; Hmedat, A.; Müller, S.; Rolnik, R.; Rausch, A.; Lederer, M.; Hüttelmaier, S. Musashi-1-A Stemness RBP for Cancer Therapy? Biology 2021, 10, 407. [Google Scholar] [CrossRef]
- Darai, N.; Mahalapbutr, P.; Wolschann, P.; Lee, V.S.; Wolfinger, M.T.; Rungrotmongkol, T. Theoretical studies on RNA recognition by Musashi 1 RNA-binding protein. Sci. Rep. 2022, 12, 12137. [Google Scholar] [CrossRef]
- Horisawa, K.; Imai, T.; Okano, H.; Yanagawa, H. The Musashi family RNA-binding proteins in stem cells. Biomol. Concepts 2010, 1, 59–66. [Google Scholar] [CrossRef]
- Kawahara, H.; Imai, T.; Imataka, H.; Tsujimoto, M.; Matsumoto, K.; Okano, H. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J. Cell Biol. 2008, 181, 639–653. [Google Scholar] [CrossRef] [PubMed]
- de Sousa Abreu, R.; Sanchez-Diaz, P.C.; Vogel, C.; Burns, S.C.; Ko, D.; Burton, T.L.; Vo, D.T.; Chennasamudaram, S.; Le, S.Y.; Shapiro, B.A.; et al. Genomic analyses of musashi1 downstream targets show a strong association with cancer-related processes. J. Biol. Chem. 2009, 284, 12125–12135. [Google Scholar] [CrossRef] [PubMed]
- Cragle, C.; MacNicol, A.M. Musashi protein-directed translational activation of target mRNAs is mediated by the poly(A) polymerase, germ line development defective-2. J. Biol. Chem. 2014, 289, 14239–14251. [Google Scholar] [CrossRef] [PubMed]
- Rezza, A.; Skah, S.; Roche, C.; Nadjar, J.; Samarut, J.; Plateroti, M. The overexpression of the putative gut stem cell marker Musashi-1 induces tumorigenesis through Wnt and Notch activation. J. Cell Sci. 2010, 123, 3256–3265. [Google Scholar] [CrossRef]
- Orzechowska-Licari, E.J.; Bialkowska, A.B.; Yang, V.W. Sonic Hedgehog and WNT Signaling Regulate a Positive Feedback Loop Between Intestinal Epithelial and Stromal Cells to Promote Epithelial Regeneration. Cell. Mol. Gastroenterol. Hepatol. 2023, 16, 607–642. [Google Scholar] [CrossRef]
- Wang, X.Y.; Yin, Y.; Yuan, H.; Sakamaki, T.; Okano, H.; Glazer, R.I. Musashi1 modulates mammary progenitor cell expansion through proliferin-mediated activation of the Wnt and Notch pathways. Mol. Cell. Biol. 2008, 28, 3589–3599. [Google Scholar] [CrossRef]
- Li, J.; Yan, K.; Yang, Y.; Li, H.; Wang, Z.; Xu, X. Musashi-1 positively regulates growth and proliferation of hepatoma cells in vitro. J. South. Med. Univ. 2019, 39, 1436–1442. [Google Scholar] [CrossRef]
- Liu, Q.; Zhou, C.; Zhang, B. Upregulation of musashi1 increases malignancy of hepatocellular carcinoma via the Wnt/β-catenin signaling pathway and predicts a poor prognosis. BMC Gastroenterol. 2019, 19, 230. [Google Scholar] [CrossRef]
- Gong, P.; Wang, Y.; Gao, Y.; Gao, M.; Liu, L.; Qu, P.; Jin, X.; Gao, Q. Msi1 promotes tumor progression by epithelial-to-mesenchymal transition in cervical cancer. Hum. Pathol. 2017, 65, 53–61. [Google Scholar] [CrossRef]
- Zhang, H.; Guo, R.; Li, Z.; Ma, R.; Xu, S.; Yin, L.; Zhu, H.; Huang, Z.; Xing, C.; Yang, Y.; et al. MSI2 mediates WNT/β-Catenin pathway function in hematopoietic stem cells. Leukemia 2025, 39, 265–270. [Google Scholar] [CrossRef]
- Li, Z.; Guo, Q.; Zhang, J.; Fu, Z.; Wang, Y.; Wang, T.; Tang, J. The RNA-Binding Motif Protein Family in Cancer: Friend or Foe? Front. Oncol. 2021, 11, 757135. [Google Scholar] [CrossRef]
- Wu, Z.; Guo, L.; Wan, L.; Xu, K.; Luo, L.; Wen, Z. Comprehensive bioinformatics analysis of a RBM family-based prognostic signature with experiment validation in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2023, 149, 11891–11905. [Google Scholar] [CrossRef]
- Query, C.C.; Bentley, R.C.; Keene, J.D. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein. Cell 1989, 57, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Birney, E.; Kumar, S.; Krainer, A.R. Analysis of the RNA-recognition motif and RS and RGG domains: Conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993, 21, 5803–5816. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, L.C.; Rintala-Maki, N.D.; White, R.D.; Morin, C.D. RNA binding motif (RBM) proteins: A novel family of apoptosis modulators? J. Cell. Biochem. 2005, 94, 5–24. [Google Scholar] [CrossRef] [PubMed]
- Inoue, A.; Takahashi, K.P.; Kimura, M.; Watanabe, T.; Morisawa, S. Molecular cloning of a RNA binding protein, S1-1. Nucleic Acids Res. 1996, 24, 2990–2997. [Google Scholar] [CrossRef]
- Ryan, J.C.; Morey, J.S.; Ramsdell, J.S.; Van Dolah, F.M. Acute phase gene expression in mice exposed to the marine neurotoxin domoic acid. Neuroscience 2005, 136, 1121–1132. [Google Scholar] [CrossRef]
- Wellmann, S.; Truss, M.; Bruder, E.; Tornillo, L.; Zelmer, A.; Seeger, K.; Bührer, C. The RNA-binding protein RBM3 is required for cell proliferation and protects against serum deprivation-induced cell death. Pediatr. Res. 2010, 67, 35–41. [Google Scholar] [CrossRef]
- Zhou, R.B.; Lu, X.L.; Zhang, C.Y.; Yin, D.C. RNA binding motif protein 3: A potential biomarker in cancer and therapeutic target in neuroprotection. Oncotarget 2017, 8, 22235–22250. [Google Scholar] [CrossRef]
- Sureban, S.M.; Ramalingam, S.; Natarajan, G.; May, R.; Subramaniam, D.; Bishnupuri, K.S.; Morrison, A.R.; Dieckgraefe, B.K.; Brackett, D.J.; Postier, R.G.; et al. Translation regulatory factor RBM3 is a proto-oncogene that prevents mitotic catastrophe. Oncogene 2008, 27, 4544–4556. [Google Scholar] [CrossRef]
- Derry, J.M.; Kerns, J.A.; Francke, U. RBM3, a novel human gene in Xp11.23 with a putative RNA-binding domain. Hum. Mol. Genet. 1995, 4, 2307–2311. [Google Scholar] [CrossRef]
- Danno, S.; Nishiyama, H.; Higashitsuji, H.; Yokoi, H.; Xue, J.H.; Itoh, K.; Matsuda, T.; Fujita, J. Increased transcript level of RBM3, a member of the glycine-rich RNA-binding protein family, in human cells in response to cold stress. Biochem. Biophys. Res. Commun. 1997, 236, 804–807. [Google Scholar] [CrossRef] [PubMed]
- Godin, K.S.; Varani, G. How arginine-rich domains coordinate mRNA maturation events. RNA Biol. 2007, 4, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Smart, F.; Aschrafi, A.; Atkins, A.; Owens, G.C.; Pilotte, J.; Cunningham, B.A.; Vanderklish, P.W. Two isoforms of the cold-inducible mRNA-binding protein RBM3 localize to dendrites and promote translation. J. Neurochem. 2007, 101, 1367–1379. [Google Scholar] [CrossRef] [PubMed]
- Gonatopoulos-Pournatzis, T.; Cowling, V.H. Cap-binding complex (CBC). Biochem. J. 2014, 457, 231–242. [Google Scholar] [CrossRef]
- Zhan, X.; Qian, B.; Cao, F.; Wu, W.; Yang, L.; Guan, Q.; Gu, X.; Wang, P.; Okusolubo, T.A.; Dunn, S.L.; et al. An Arabidopsis PWI and RRM motif-containing protein is critical for pre-mRNA splicing and ABA responses. Nat. Commun. 2015, 6, 8139. [Google Scholar] [CrossRef]
- Guan, R.; El-Rass, S.; Spillane, D.; Lam, S.; Wang, Y.; Wu, J.; Chen, Z.; Wang, A.; Jia, Z.; Keating, A.; et al. rbm47, a novel RNA binding protein, regulates zebrafish head development. Dev. Dyn. 2013, 242, 1395–1404. [Google Scholar] [CrossRef]
- Fossat, N.; Tourle, K.; Radziewic, T.; Barratt, K.; Liebhold, D.; Studdert, J.B.; Power, M.; Jones, V.; Loebel, D.A.; Tam, P.P. C to U RNA editing mediated by APOBEC1 requires RNA-binding protein RBM47. EMBO Rep. 2014, 15, 903–910. [Google Scholar] [CrossRef]
- Sakurai, T.; Isogaya, K.; Sakai, S.; Morikawa, M.; Morishita, Y.; Ehata, S.; Miyazono, K.; Koinuma, D. RNA-binding motif protein 47 inhibits Nrf2 activity to suppress tumor growth in lung adenocarcinoma. Oncogene 2016, 35, 5000–5009, Erratum in Oncogene 2017, 36, 5083. https://doi.org/10.1038/onc.2017.191. [Google Scholar] [CrossRef]
- Kim, Y.E.; Won, M.; Lee, S.G.; Park, C.; Song, C.H.; Kim, K.K. RBM47-regulated alternative splicing of TJP1 promotes actin stress fiber assembly during epithelial-to-mesenchymal transition. Oncogene 2019, 38, 6521–6536. [Google Scholar] [CrossRef]
- Shivalingappa, P.K.M.; Sharma, V.; Shiras, A.; Bapat, S.A. RNA binding motif 47 (RBM47): Emerging roles in vertebrate development, RNA editing and cancer. Mol. Cell. Biochem. 2021, 476, 4493–4505. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zhang, F.; Zhang, Y.; Wang, X.; Xing, C.; Guo, J.; Zhang, H.; Suo, Z.; Li, Y.; Wang, J.; et al. Post-transcriptional regulator Rbm47 elevates IL-10 production and promotes the immunosuppression of B cells. Cell. Mol. Immunol. 2019, 16, 580–589. [Google Scholar] [CrossRef] [PubMed]
- Venugopal, A.; Subramaniam, D.; Balmaceda, J.; Roy, B.; Dixon, D.A.; Umar, S.; Weir, S.J.; Anant, S. RNA binding protein RBM3 increases β-catenin signaling to increase stem cell characteristics in colorectal cancer cells. Mol. Carcinog. 2016, 55, 1503–1516. [Google Scholar] [CrossRef]
- Soleymanjahi, S.; Blanc, V.; Molitor, E.A.; Alvarado, D.M.; Xie, Y.; Gazit, V.; Brown, J.W.; Byrnes, K.; Liu, T.C.; Mills, J.C.; et al. RBM47 regulates intestinal injury and tumorigenesis by modifying proliferation, oxidative response, and inflammatory pathways. JCI Insight 2023, 8, e161118. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Zhang, Y.; Zhou, T.; Qiao, Q.; Shan, J.; Chen, Y.; Jiang, W.; Wang, Y.; Liu, S.; Wang, Y.; et al. RBM10 C761Y mutation induced oncogenic ASPM isoforms and regulated β-catenin signaling in cholangiocarcinoma. J. Exp. Clin. Cancer Res. 2024, 43, 104. [Google Scholar] [CrossRef]
- Cao, Y.; Di, X.; Zhang, Q.; Li, R.; Wang, K. RBM10 Regulates Tumor Apoptosis, Proliferation, and Metastasis. Front. Oncol. 2021, 11, 603932. [Google Scholar] [CrossRef]
- Cao, Y.; Geng, J.; Wang, X.; Meng, Q.; Xu, S.; Lang, Y.; Zhou, Y.; Qi, L.; Wang, Z.; Wei, Z.; et al. RNA-binding motif protein 10 represses tumor progression through the Wnt/β-catenin pathway in lung adenocarcinoma. Int. J. Biol. Sci. 2022, 18, 124–139. [Google Scholar] [CrossRef]
- Zhu, L.; Xi, P.W.; Li, X.X.; Sun, X.; Zhou, W.B.; Xia, T.S.; Shi, L.; Hu, Y.; Ding, Q.; Wei, J.F. The RNA binding protein RBMS3 inhibits the metastasis of breast cancer by regulating Twist1 expression. J. Exp. Clin. Cancer Res. 2019, 38, 105, Erratum in J. Exp. Clin. Cancer Res. 2020, 39, 21. https://doi.org/10.1186/s13046-019-1509-0. [Google Scholar] [CrossRef]
- Liu, J.; Shu, G.; Wu, A.; Zhang, X.; Zhou, Z.; Alvero, A.B.; Mor, G.; Yin, G. TWIST1 induces proteasomal degradation of β-catenin during the differentiation of ovarian cancer stem-like cells. Sci. Rep. 2022, 12, 15650. [Google Scholar] [CrossRef]
- Palzer, K.A.; Bolduan, V.; Käfer, R.; Kleinert, H.; Bros, M.; Pautz, A. The Role of KH-Type Splicing Regulatory Protein (KSRP) for Immune Functions and Tumorigenesis. Cells 2022, 11, 1482. [Google Scholar] [CrossRef]
- Avigan, M.I.; Strober, B.; Levens, D. A far upstream element stimulates c-myc expression in undifferentiated leukemia cells. J. Biol. Chem. 1990, 265, 18538–18545. [Google Scholar] [CrossRef] [PubMed]
- Davis-Smyth, T.; Duncan, R.C.; Zheng, T.; Michelotti, G.; Levens, D. The far upstream element-binding proteins comprise an ancient family of single-strand DNA-binding transactivators. J. Biol. Chem. 1996, 271, 31679–31687. [Google Scholar] [CrossRef] [PubMed]
- Min, H.; Turck, C.W.; Nikolic, J.M.; Black, D.L. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997, 11, 1023–1036. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-C.; Ho, K.-H.; Hua, K.-T.; Chien, M.-H. Roles of K(H)SRP in modulating gene transcription throughout cancer progression: Insights from cellular studies to clinical perspectives. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2024, 1879, 189202. [Google Scholar] [CrossRef]
- García-Mayoral, M.F.; Hollingworth, D.; Masino, L.; Díaz-Moreno, I.; Kelly, G.; Gherzi, R.; Chou, C.F.; Chen, C.Y.; Ramos, A. The structure of the C-terminal KH domains of KSRP reveals a noncanonical motif important for mRNA degradation. Structure 2007, 15, 485–498. [Google Scholar] [CrossRef]
- García-Mayoral, M.F.; Díaz-Moreno, I.; Hollingworth, D.; Ramos, A. The sequence selectivity of KSRP explains its flexibility in the recognition of the RNA targets. Nucleic Acids Res. 2008, 36, 5290–5296. [Google Scholar] [CrossRef]
- Gherzi, R.; Chen, C.Y.; Trabucchi, M.; Ramos, A.; Briata, P. The role of KSRP in mRNA decay and microRNA precursor maturation. Wiley Interdiscip. Rev. RNA 2010, 1, 230–239. [Google Scholar] [CrossRef]
- Hall, M.P.; Huang, S.; Black, D.L. Differentiation-induced colocalization of the KH-type splicing regulatory protein with polypyrimidine tract binding protein and the c-src pre-mRNA. Mol. Biol. Cell 2004, 15, 774–786. [Google Scholar] [CrossRef]
- Liu, J.; Kouzine, F.; Nie, Z.; Chung, H.J.; Elisha-Feil, Z.; Weber, A.; Zhao, K.; Levens, D. The FUSE/FBP/FIR/TFIIH system is a molecular machine programming a pulse of c-myc expression. EMBO J. 2006, 25, 2119–2130. [Google Scholar] [CrossRef]
- Chen, C.Y.; Gherzi, R.; Ong, S.E.; Chan, E.L.; Raijmakers, R.; Pruijn, G.J.; Stoecklin, G.; Moroni, C.; Mann, M.; Karin, M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 2001, 107, 451–464. [Google Scholar] [CrossRef]
- Gherzi, R.; Lee, K.Y.; Briata, P.; Wegmüller, D.; Moroni, C.; Karin, M.; Chen, C.Y. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell 2004, 14, 571–583. [Google Scholar] [CrossRef]
- Trabucchi, M.; Briata, P.; Garcia-Mayoral, M.; Haase, A.D.; Filipowicz, W.; Ramos, A.; Gherzi, R.; Rosenfeld, M.G. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 2009, 459, 1010–1014. [Google Scholar] [CrossRef]
- Tong, L.; Luo, Y.; Wei, T.; Guo, L.; Wang, H.; Zhu, W.; Zhang, J. KH-type splicing regulatory protein (KHSRP) contributes to tumorigenesis by promoting miR-26a maturation in small cell lung cancer. Mol. Cell. Biochem. 2016, 422, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Chien, M.H.; Lee, W.J.; Yang, Y.C.; Li, Y.L.; Chen, B.R.; Cheng, T.Y.; Yang, P.W.; Wang, M.Y.; Jan, Y.H.; Lin, Y.K.; et al. KSRP suppresses cell invasion and metastasis through miR-23a-mediated EGR3 mRNA degradation in non-small cell lung cancer. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Masuda, K.; Hamada, J.; Shoda, K.; Naruto, T.; Hamada, S.; Miyakami, Y.; Kohmoto, T.; Watanabe, M.; Takahashi, R.; et al. KH-type splicing regulatory protein is involved in esophageal squamous cell carcinoma progression. Oncotarget 2017, 8, 101130–101145. [Google Scholar] [CrossRef]
- Pan, R.; Cai, W.; Sun, J.; Yu, C.; Li, P.; Zheng, M. Inhibition of KHSRP sensitizes colorectal cancer to 5-fluoruracil through miR-501-5p-mediated ERRFI1 mRNA degradation. J. Cell. Physiol. 2020, 235, 1576–1587. [Google Scholar] [CrossRef] [PubMed]
- Nie, E.; Jin, X.; Miao, F.; Yu, T.; Zhi, T.; Shi, Z.; Wang, Y.; Zhang, J.; Xie, M.; You, Y. TGF-β1 modulates temozolomide resistance in glioblastoma via altered microRNA processing and elevated MGMT. Neuro Oncol. 2021, 23, 435–446. [Google Scholar] [CrossRef]
- Yang, Y.C.; Lin, Y.W.; Lee, W.J.; Lai, F.R.; Ho, K.H.; Chu, C.Y.; Hua, K.T.; Chen, J.Q.; Tung, M.C.; Hsiao, M.; et al. The RNA-binding protein KSRP aggravates malignant progression of clear cell renal cell carcinoma through transcriptional inhibition and post-transcriptional destabilization of the NEDD4L ubiquitin ligase. J. Biomed. Sci. 2023, 30, 68. [Google Scholar] [CrossRef]
- Bikkavilli, R.K.; Zerayesus, S.A.; Van Scoyk, M.; Wilson, L.; Wu, P.Y.; Baskaran, A.; Tang, K.; Raheem, S.; Samuelson, B.A.; Reddy, N.M.; et al. K-homology splicing regulatory protein (KSRP) promotes post-transcriptional destabilization of Spry4 transcripts in non-small cell lung cancer. J. Biol. Chem. 2017, 292, 7423–7434. [Google Scholar] [CrossRef]
- Taniuchi, K.; Ogasawara, M. KHSRP-bound small nucleolar RNAs associate with promotion of cell invasiveness and metastasis of pancreatic cancer. Oncotarget 2020, 11, 131–147, Correction in Oncotarget 2023, 14, 104. https://doi.org/10.18632/oncotarget.28361. [Google Scholar] [CrossRef]
- Huang, J.; Sachdeva, M.; Xu, E.; Robinson, T.J.; Luo, L.; Ma, Y.; Williams, N.T.; Lopez, O.; Cervia, L.D.; Yuan, F.; et al. The Long Noncoding RNA NEAT1 Promotes Sarcoma Metastasis by Regulating RNA Splicing Pathways. Mol. Cancer Res. 2020, 18, 1534–1544. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zhang, X.; Feng, X.; Fan, X.; Jin, Z. The crosstalk between microRNAs and the Wnt/β-catenin signaling pathway in cancer. Oncotarget 2017, 8, 14089–14106. [Google Scholar] [CrossRef] [PubMed]
- Trabucchi, M.; Briata, P.; Filipowicz, W.; Ramos, A.; Gherzi, R.; Rosenfeld, M.G. KSRP promotes the maturation of a group of miRNA precursors. Adv. Exp. Med. Biol. 2010, 700, 36–42. [Google Scholar] [PubMed]
- Yang, Y.; Wang, Y.; Jia, H.; Li, B.; Xing, D.; Li, J.J. MicroRNA-1 Modulates Chondrocyte Phenotype by Regulating FZD7 of Wnt/β-Catenin Signaling Pathway. Cartilage 2021, 13, 1019S–1029S. [Google Scholar] [CrossRef]
- Li, N.; Yang, L.; Sun, Y.; Wu, X. MicroRNA-16 inhibits migration and invasion via regulation of the Wnt/β-catenin signaling pathway in ovarian cancer. Oncol. Lett. 2019, 17, 2631–2638. [Google Scholar] [CrossRef]
- Sun, J.; Yan, P.; Chen, Y.; Chen, Y.; Yang, J.; Xu, G.; Mao, H.; Qiu, Y. MicroRNA-26b inhibits cell proliferation and cytokine secretion in human RASF cells via the Wnt/GSK-3β/β-catenin pathway. Diagn. Pathol. 2015, 10, 72. [Google Scholar] [CrossRef]
- Ma, J.; Song, Y.; Mabrouk, I.; Zhou, Y.; Liu, Q.; Yu, J.; Li, X.; Xue, G.; Wang, J.; Yu, Z.; et al. miR-140-y targets TCF4 to regulate the Wnt signaling pathway and promote embryonic feather follicle development in Hungarian white goose. Poult. Sci. 2024, 103, 103508. [Google Scholar] [CrossRef]
- Jiang, Q.; He, M.; Ma, M.T.; Wu, H.Z.; Yu, Z.J.; Guan, S.; Jiang, L.Y.; Wang, Y.; Zheng, D.D.; Jin, F.; et al. MicroRNA-148a inhibits breast cancer migration and invasion by directly targeting WNT-1. Oncol. Rep. 2016, 35, 1425–1432, Erratum in Oncol. Rep. 2022, 47, 106. https://doi.org/10.3892/or.2022.8317. [Google Scholar] [CrossRef]
- Banerjee, A.; Chawla-Sarkar, M.; Mukherjee, A. Rotavirus-Mediated Suppression of miRNA-192 Family and miRNA-181a Activates Wnt/β-Catenin Signaling Pathway: An In Vitro Study. Viruses 2022, 14, 558. [Google Scholar] [CrossRef]
- Ai, G.; Meng, M.; Wang, L.; Shao, X.; Li, Y.; Cheng, J.; Tong, X.; Cheng, Z. microRNA-196a promotes osteogenic differentiation and inhibit adipogenic differentiation of adipose stem cells via regulating β-catenin pathway. Am. J. Transl. Res. 2019, 11, 3081–3091. [Google Scholar]
- Song, J.; Gao, L.; Yang, G.; Tang, S.; Xie, H.; Wang, Y.; Wang, J.; Zhang, Y.; Jin, J.; Gou, Y.; et al. MiR-199a regulates cell proliferation and survival by targeting FZD7. PLoS ONE 2014, 9, e110074. [Google Scholar] [CrossRef]
- Zhou, F.; Cao, W.; Xu, R.; Zhang, J.; Yu, T.; Xu, X.; Zhi, T.; Yin, J.; Cao, S.; Liu, N.; et al. MicroRNA-206 attenuates glioma cell proliferation, migration, and invasion by blocking the WNT/β-catenin pathway via direct targeting of Frizzled 7 mRNA. Am. J. Transl. Res. 2019, 11, 4584–4601. [Google Scholar] [PubMed]
- Wang, Y.; Zhao, Y.; Herbst, A.; Kalinski, T.; Qin, J.; Wang, X.; Jiang, Z.; Benedix, F.; Franke, S.; Wartman, T.; et al. miR-221 Mediates Chemoresistance of Esophageal Adenocarcinoma by Direct Targeting of DKK2 Expression. Ann. Surg. 2016, 264, 804–814. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Tang, H.; Li, N.; Chen, H.; Chen, X.; Gu, L.; Zhang, L.; Tian, G.; Tao, D. MicroRNA-361-3p Inhibit the Progression of Lymphoma by the Wnt/β-Catenin Signaling Pathway. Cancer Manag. Res. 2020, 12, 12375–12384. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.H.; Ho, J.C.; Li, S.C.; Cheng, Y.W.; Yang, Y.C.; Chen, J.F.; Hsu, C.Y.; Nakano, T.; Wang, F.S.; Yang, M.Y.; et al. Upregulation of miR-941 in Circulating CD14+ Monocytes Enhances Osteoclast Activation via WNT16 Inhibition in Patients with Psoriatic Arthritis. Int. J. Mol. Sci. 2020, 21, 4301. [Google Scholar] [CrossRef]
- Sampson, V.B.; Rong, N.H.; Han, J.; Yang, Q.; Aris, V.; Soteropoulos, P.; Petrelli, N.J.; Dunn, S.P.; Krueger, L.J. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007, 67, 9762–9770. [Google Scholar] [CrossRef]
- Nicastro, G.; García-Mayoral, M.F.; Hollingworth, D.; Kelly, G.; Martin, S.R.; Briata, P.; Gherzi, R.; Ramos, A. Noncanonical G recognition mediates KSRP regulation of let-7 biogenesis. Nat. Struct. Mol. Biol. 2012, 19, 1282–1286. [Google Scholar] [CrossRef]
- Briata, P.; Ilengo, C.; Corte, G.; Moroni, C.; Rosenfeld, M.G.; Chen, C.Y.; Gherzi, R. The Wnt/β-catenin→Pitx2 pathway controls the turnover of Pitx2 and other unstable mRNAs. Mol. Cell 2003, 12, 1201–1211. [Google Scholar] [CrossRef]
- Basu, M.; Roy, S.S. Wnt/β-catenin pathway is regulated by PITX2 homeodomain protein and thus contributes to the proliferation of human ovarian adenocarcinoma cell, SKOV-3. J. Biol. Chem. 2013, 288, 4355–4367. [Google Scholar] [CrossRef]
- Laggerbauer, B.; Ostareck, D.; Keidel, E.M.; Ostareck-Lederer, A.; Fischer, U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum. Mol. Genet. 2001, 10, 329–338. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Y.; Ku, L.; Wilkinson, K.D.; Warren, S.T.; Feng, Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2001, 29, 2276–2283. [Google Scholar] [CrossRef] [PubMed]
- Huber, K.M.; Gallagher, S.M.; Warren, S.T.; Bear, M.F. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc. Natl. Acad. Sci. USA 2002, 99, 7746–7750. [Google Scholar] [CrossRef] [PubMed]
- Sidorov, M.S.; Auerbach, B.D.; Bear, M.F. Fragile X mental retardation protein and synaptic plasticity. Mol. Brain 2013, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Richter, J.D.; Zhao, X. The molecular biology of FMRP: New insights into fragile X syndrome. Nat. Rev. Neurosci. 2021, 22, 209–222. [Google Scholar] [CrossRef]
- Zhao, X.; Bhattacharyya, A. Human Models Are Needed for Studying Human Neurodevelopmental Disorders. Am. J. Hum. Genet. 2018, 103, 829–857. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, T.; Han, J. Targeting FMRP: A new window for cancer immunotherapy. MedComm (2020) 2023, 4, e233. [Google Scholar] [CrossRef]
- Ashley, C.T., Jr.; Wilkinson, K.D.; Reines, D.; Warren, S.T. FMR1 protein: Conserved RNP family domains and selective RNA binding. Science 1993, 262, 563–566. [Google Scholar] [CrossRef]
- Siomi, H.; Siomi, M.C.; Nussbaum, R.L.; Dreyfuss, G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell 1993, 74, 291–298. [Google Scholar] [CrossRef]
- Adinolfi, S.; Ramos, A.; Martin, S.R.; Dal Piaz, F.; Pucci, P.; Bardoni, B.; Mandel, J.L.; Pastore, A. The N-terminus of the fragile X mental retardation protein contains a novel domain involved in dimerization and RNA binding. Biochemistry 2003, 42, 10437–10444. [Google Scholar] [CrossRef]
- Maurer-Stroh, S.; Dickens, N.J.; Hughes-Davies, L.; Kouzarides, T.; Eisenhaber, F.; Ponting, C.P. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 2003, 28, 69–74. [Google Scholar] [CrossRef]
- Lu, R.; Wang, G.G. Tudor: A versatile family of histone methylation ‘readers’. Trends Biochem. Sci. 2013, 38, 546–555. [Google Scholar] [CrossRef] [PubMed]
- Myrick, L.K.; Hashimoto, H.; Cheng, X.; Warren, S.T. Human FMRP contains an integral tandem Agenet (Tudor) and KH motif in the amino terminal domain. Hum. Mol. Genet. 2015, 24, 1733–1740. [Google Scholar] [CrossRef] [PubMed]
- Eberhart, D.E.; Malter, H.E.; Feng, Y.; Warren, S.T. The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum. Mol. Genet. 1996, 5, 1083–1091. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Chen, Z.; Fu, Y.; He, Q.; Jiang, L.; Zheng, J.; Gao, Y.; Mei, P.; Chen, Z.; Ren, X. The amino-terminal structure of human fragile X mental retardation protein obtained using precipitant-immobilized imprinted polymers. Nat. Commun. 2015, 6, 6634. [Google Scholar] [CrossRef]
- Edens, B.M.; Vissers, C.; Su, J.; Arumugam, S.; Xu, Z.; Shi, H.; Miller, N.; Rojas Ringeling, F.; Ming, G.L.; He, C.; et al. FMRP Modulates Neural Differentiation through m6A-Dependent mRNA Nuclear Export. Cell Rep. 2019, 28, 845–854.e5. [Google Scholar] [CrossRef]
- Zhou, X.; Guo, C.J.; Wang, R.; Li, Y.L.; Zhang, T.; Qiu, Z.; Gao, S.; Liu, J.L.; Gao, Y. FMR1 KH0-KH1 domains coordinate m6A binding and phase separation in Fragile X syndrome. Exp. Cell Res. 2025, 450, 114664. [Google Scholar] [CrossRef]
- Zhang, F.; Kang, Y.; Wang, M.; Li, Y.; Xu, T.; Yang, W.; Song, H.; Wu, H.; Shu, Q.; Jin, P. Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum. Mol. Genet. 2018, 27, 3936–3950. [Google Scholar] [CrossRef]
- Darnell, J.C.; Van Driesche, S.J.; Zhang, C.; Hung, K.Y.; Mele, A.; Fraser, C.E.; Stone, E.F.; Chen, C.; Fak, J.J.; Chi, S.W.; et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 2011, 146, 247–261. [Google Scholar] [CrossRef]
- Zeng, Q.; Saghafinia, S.; Chryplewicz, A.; Fournier, N.; Christe, L.; Xie, Y.Q.; Guillot, J.; Yucel, S.; Li, P.; Galván, J.A.; et al. Aberrant hyperexpression of the RNA binding protein FMRP in tumors mediates immune evasion. Science 2022, 378, eabl7207. [Google Scholar] [CrossRef]
- Cao, J.; Wang, J.; Shi, B.; Ma, X.; Wu, W.; Wang, N. DDX5-Targeting Fragile X Mental Retardation Protein Regulates the Wnt/β-catenin Signaling Pathway to Promote Epithelial Mesenchymal Transition in Breast Cancer. J. Sichuan Univ. (Med. Sci.) 2024, 55, 1138–1149. [Google Scholar] [CrossRef]
- Jia, Y.; Jia, R.; Chen, Y.; Lin, X.; Aishan, N.; Li, H.; Wang, L.; Zhang, X.; Ruan, J. The role of RNA binding proteins in cancer biology: A focus on FMRP. Genes Dis. 2025, 12, 101493. [Google Scholar] [CrossRef] [PubMed]
- Clark, E.L.; Hadjimichael, C.; Temperley, R.; Barnard, A.; Fuller-Pace, F.V.; Robson, C.N. p68/DdX5 supports β-catenin & RNAP II during androgen receptor mediated transcription in prostate cancer. PLoS ONE 2013, 8, e54150. [Google Scholar] [CrossRef]
- Marfull-Oromí, P.; Onishi, K.; Han, X.; Yates, J.R., 3rd; Zou, Y. The Fragile X Messenger Ribonucleoprotein 1 Participates in Axon Guidance Mediated by the Wnt/Planar Cell Polarity Pathway. Neuroscience 2023, 508, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Chyung, E.; LeBlanc, H.F.; Fallon, J.R.; Akins, M.R. Fragile X granules are a family of axonal ribonucleoprotein particles with circuit-dependent protein composition and mRNA cargos. J. Comp. Neurol. 2018, 526, 96–108. [Google Scholar] [CrossRef]
- Casingal, C.R.; Kikkawa, T.; Inada, H.; Sasaki, Y.; Osumi, N. Identification of FMRP target mRNAs in the developmental brain: FMRP might coordinate Ras/MAPK, Wnt/β-catenin, and mTOR signaling during corticogenesis. Mol. Brain 2020, 13, 167. [Google Scholar] [CrossRef]
- Forrest, M.P.; Hill, M.J.; Kavanagh, D.H.; Tansey, K.E.; Waite, A.J.; Blake, D.J. The Psychiatric Risk Gene Transcription Factor 4 (TCF4) Regulates Neurodevelopmental Pathways Associated with Schizophrenia, Autism, and Intellectual Disability. Schizophr. Bull. 2018, 44, 1100–1110. [Google Scholar] [CrossRef]
- Song, C.; Broadie, K. Dysregulation of BMP, Wnt, and Insulin Signaling in Fragile X Syndrome. Front. Cell Dev. Biol. 2022, 10, 934662, Erratum in Front. Cell Dev. Biol. 2024, 12, 1416720. https://doi.org/10.3389/fcell.2024.1416720. [Google Scholar] [CrossRef]
- Ehyai, S.; Miyake, T.; Williams, D.; Vinayak, J.; Bayfield, M.A.; McDermott, J.C. FMRP recruitment of β-catenin to the translation pre-initiation complex represses translation. EMBO Rep. 2018, 19, e45536. [Google Scholar] [CrossRef]
- Friedman, S.H.; Dani, N.; Rushton, E.; Broadie, K. Fragile X mental retardation protein regulates trans-synaptic signaling in Drosophila. Dis. Model. Mech. 2013, 6, 1400–1413. [Google Scholar] [CrossRef]
- Neumann, D.P.; Goodall, G.J.; Gregory, P.A. The Quaking RNA-binding proteins as regulators of cell differentiation. Wiley Interdiscip. Rev. RNA 2022, 13, e1724. [Google Scholar] [CrossRef]
- Sidman, R.L.; Dickie, M.M.; Appel, S.H. Mutant Mice (Quaking and Jimpy) with Deficient Myelination in the Central Nervous System. Science 1964, 144, 309–311. [Google Scholar] [CrossRef]
- Ebersole, T.A.; Chen, Q.; Justice, M.J.; Artzt, K. The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins. Nat. Genet. 1996, 12, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Zorn, A.M.; Krieg, P.A. The KH domain protein encoded by quaking functions as a dimer and is essential for notochord development in Xenopus embryos. Genes Dev. 1997, 11, 2176–2190. [Google Scholar] [CrossRef]
- Chen, T.; Richard, S. Structure-function analysis of Qk1: A lethal point mutation in mouse quaking prevents homodimerization. Mol. Cell. Biol. 1998, 18, 4863–4871. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Luyten, I.; Bottomley, M.J.; Messias, A.C.; Houngninou-Molango, S.; Sprangers, R.; Zanier, K.; Krämer, A.; Sattler, M. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science 2001, 294, 1098–1102. [Google Scholar] [CrossRef] [PubMed]
- Daubner, G.M.; Brümmer, A.; Tocchini, C.; Gerhardy, S.; Ciosk, R.; Zavolan, M.; Allain, F.H. Structural and functional implications of the QUA2 domain on RNA recognition by GLD-1. Nucleic Acids Res. 2014, 42, 8092–8105. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Furuta, T.; Mitsunaga, K.; Ebersole, T.A.; Shichiri, M.; Wu, J.; Artzt, K.; Yamamura, K.; Abe, K. Genomic organization and expression analysis of the mouse qkI locus. Mamm. Genome 1999, 10, 662–669. [Google Scholar] [CrossRef]
- Darbelli, L.; Richard, S. Emerging functions of the Quaking RNA-binding proteins and link to human diseases. Wiley Interdiscip. Rev. RNA 2016, 7, 399–412. [Google Scholar] [CrossRef]
- Zhu, W.; Yang, W.; Sun, G.; Huang, J. RNA-binding protein quaking: A multifunctional regulator in tumour progression. Ann. Med. 2025, 57, 2443046. [Google Scholar] [CrossRef]
- Galarneau, A.; Richard, S. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat. Struct. Mol. Biol. 2005, 12, 691–698. [Google Scholar] [CrossRef]
- Feng, Y.; Bankston, A. The star family member QKI and cell signaling. Adv. Exp. Med. Biol. 2010, 693, 25–36. [Google Scholar] [PubMed]
- Min, J.K.; Park, H.S.; Lee, Y.B.; Kim, J.G.; Kim, J.I.; Park, J.B. Cross-Talk between Wnt Signaling and Src Tyrosine Kinase. Biomedicines 2022, 10, 1112. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, R.; Tsusaka, T.; Mitsunaga, H.; Maehata, T.; Hoshino, S. The STAR protein QKI-7 recruits PAPD4 to regulate post-transcriptional polyadenylation of target mRNAs. Nucleic Acids Res. 2016, 44, 2475–2490. [Google Scholar] [CrossRef] [PubMed]
- de Bruin, R.G.; van der Veer, E.P.; Prins, J.; Lee, D.H.; Dane, M.J.; Zhang, H.; Roeten, M.K.; Bijkerk, R.; de Boer, H.C.; Rabelink, T.J.; et al. The RNA-binding protein quaking maintains endothelial barrier function and affects VE-cadherin and β-catenin protein expression. Sci. Rep. 2016, 6, 21643. [Google Scholar] [CrossRef]
- Zhu, Z.; Wei, D.; Li, X.; Wang, F.; Yan, F.; Xing, Z.; Yan, Z.; Lu, H.; Zhai, D.; Ye, Z.; et al. RNA-binding protein QKI regulates contact inhibition via Yes-associate protein in ccRCC. Acta Biochim. Biophys. Sin. 2019, 51, 9–19. [Google Scholar] [CrossRef]
- Zhu, Y.J.; Zheng, B.; Luo, G.J.; Ma, X.K.; Lu, X.Y.; Lin, X.M.; Yang, S.; Zhao, Q.; Wu, T.; Li, Z.X.; et al. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics 2019, 9, 3526–3540. [Google Scholar] [CrossRef]
- Yan, Z.; Ruan, B.; Wang, S.; Du, T.; Shao, X.; Chen, G.; Wang, L.; Zhai, D.; Zhu, S.; Lu, Z.; et al. RNA-binding Protein QKI Inhibits Osteogenic Differentiation Via Suppressing Wnt Pathway. Arch. Med. Res. 2023, 54, 102853. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, S.; Chen, Z.; Gu, X.; Zhang, G.; Zhang, H.; Yuan, W. N7-methyladenosine-induced SLC7A7 serves as a prognostic biomarker in pan-cancer and promotes CRC progression in colorectal cancer. Sci. Rep. 2024, 14, 30755. [Google Scholar] [CrossRef]
- Bufalieri, F.; Caimano, M.; Lospinoso Severini, L.; Basili, I.; Paglia, F.; Sampirisi, L.; Loricchio, E.; Petroni, M.; Canettieri, G.; Santoro, A.; et al. The RNA-Binding Ubiquitin Ligase MEX3A Affects Glioblastoma Tumorigenesis by Inducing Ubiquitylation and Degradation of RIG-I. Cancers 2020, 12, 321. [Google Scholar] [CrossRef]
- Wei, L.; Wang, B.; Hu, L.; Xu, Y.; Li, Z.; Shen, Y.; Huang, H. MEX3A is upregulated in esophageal squamous cell carcinoma (ESCC) and promotes development and progression of ESCC through targeting CDK6. Aging 2020, 12, 21091–21113. [Google Scholar] [CrossRef]
- Qiu, Y.; Meng, M.; Cao, C.; Zhang, J.; Cheng, X.; Huang, Y.; Cao, H.; Li, Y.; Tian, D.; Huang, Y.; et al. RNA-binding protein MEX3A controls G1/S transition via regulating the RB/E2F pathway in clear cell renal cell carcinoma. Mol. Ther. Nucleic Acids 2022, 27, 241–255. [Google Scholar] [CrossRef]
- Wang, C.K.; Chen, T.J.; Tan, G.Y.T.; Chang, F.P.; Sridharan, S.; Yu, C.A.; Chang, Y.H.; Chen, Y.J.; Cheng, L.T.; Hwang-Verslues, W.W. MEX3A Mediates p53 Degradation to Suppress Ferroptosis and Facilitate Ovarian Cancer Tumorigenesis. Cancer Res. 2023, 83, 251–263. [Google Scholar] [CrossRef]
- Draper, B.W.; Mello, C.C.; Bowerman, B.; Hardin, J.; Priess, J.R. MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell 1996, 87, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Buchet-Poyau, K.; Courchet, J.; Le Hir, H.; Séraphin, B.; Scoazec, J.Y.; Duret, L.; Domon-Dell, C.; Freund, J.N.; Billaud, M. Identification and characterization of human Mex-3 proteins, a novel family of evolutionarily conserved RNA-binding proteins differentially localized to processing bodies. Nucleic Acids Res. 2007, 35, 1289–1300. [Google Scholar] [CrossRef]
- Hahn, W.C.; Bader, J.S.; Braun, T.P.; Califano, A.; Clemons, P.A.; Druker, B.J.; Ewald, A.J.; Fu, H.; Jagu, S.; Kemp, C.J.; et al. An expanded universe of cancer targets. Cell 2021, 184, 1142–1155. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liang, J.; Wang, J.; Han, J.; Li, S.; Huang, K.; Liu, C. Mex3a promotes oncogenesis through the RAP1/MAPK signaling pathway in colorectal cancer and is inhibited by hsa-miR-6887-3p. Cancer Commun. 2021, 41, 472–491. [Google Scholar] [CrossRef] [PubMed]
- Gan, T.; Wang, Y.; Xie, M.; Wang, Q.; Zhao, S.; Wang, P.; Shi, Q.; Qian, X.; Miao, F.; Shen, Z.; et al. MEX3A Impairs DNA Mismatch Repair Signaling and Mediates Acquired Temozolomide Resistance in Glioblastoma. Cancer Res. 2022, 82, 4234–4246. [Google Scholar] [CrossRef]
- Xu, J.; Chen, S.; Hao, T.; Liu, G.; Zhang, K.; Zhang, C.; He, Y. MEX3A promotes colorectal cancer migration, invasion and EMT via regulating the Wnt/β-catenin signaling pathway. J. Cancer Res. Clin. Oncol. 2024, 150, 319. [Google Scholar] [CrossRef]
- Lederer, M.; Müller, S.; Glaß, M.; Bley, N.; Ihling, C.; Sinz, A.; Hüttelmaier, S. Oncogenic Potential of the Dual-Function Protein MEX3A. Biology 2021, 10, 415. [Google Scholar] [CrossRef]
- Helms, M.W.; Jahn-Hofmann, K.; Gnerlich, F.; Metz-Weidmann, C.; Braun, M.; Dietert, G.; Scherer, P.; Grandien, K.; Theilhaber, J.; Cao, H.; et al. Utility of the RIG-I Agonist Triphosphate RNA for Melanoma Therapy. Mol. Cancer Ther. 2019, 18, 2343–2356. [Google Scholar] [CrossRef]
- Fornerod, M.; Ohno, M.; Yoshida, M.; Mattaj, I.W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 1997, 90, 1051–1060. [Google Scholar] [CrossRef]
- Oda, T.; Yamazumi, Y.; Hiroko, T.; Kamiya, A.; Kiriya, S.; Suyama, S.; Shiozaki-Sato, Y.; Akiyama, T. Mex-3B induces apoptosis by inhibiting miR-92a access to the Bim-3′UTR. Oncogene 2018, 37, 5233–5247. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Li, H.; Han, J.; Jiang, J.; Wang, J.; Li, Y.; Feng, Z.; Zhao, R.; Sun, Z.; Lv, B.; et al. Mex3a interacts with LAMA2 to promote lung adenocarcinoma metastasis via PI3K/AKT pathway. Cell Death Dis. 2020, 11, 614. [Google Scholar] [CrossRef] [PubMed]
- Panzeri, V.; Manni, I.; Capone, A.; Naro, C.; Sacconi, A.; Di Agostino, S.; de Latouliere, L.; Montori, A.; Pilozzi, E.; Piaggio, G.; et al. The RNA-binding protein MEX3A is a prognostic factor and regulator of resistance to gemcitabine in pancreatic ductal adenocarcinoma. Mol. Oncol. 2021, 15, 579–595. [Google Scholar] [CrossRef] [PubMed]
- Barriga, F.M.; Montagni, E.; Mana, M.; Mendez-Lago, M.; Hernando-Momblona, X.; Sevillano, M.; Guillaumet-Adkins, A.; Rodriguez-Esteban, G.; Buczacki, S.J.A.; Gut, M.; et al. Mex3a Marks a Slowly Dividing Subpopulation of Lgr5+ Intestinal Stem Cells. Cell Stem Cell 2017, 20, 801–816.e7. [Google Scholar] [CrossRef]
- Pereira, B.; Amaral, A.L.; Dias, A.; Mendes, N.; Muncan, V.; Silva, A.R.; Thibert, C.; Radu, A.G.; David, L.; Máximo, V.; et al. MEX3A regulates Lgr5+ stem cell maintenance in the developing intestinal epithelium. EMBO Rep. 2020, 21, e48938. [Google Scholar] [CrossRef]
- Guo, B.; Chatterjee, S.; Li, L.; Kim, J.M.; Lee, J.; Yechoor, V.K.; Minze, L.J.; Hsueh, W.; Ma, K. The clock gene, brain and muscle Arnt-like 1, regulates adipogenesis via Wnt signaling pathway. FASEB J. 2012, 26, 3453–3463. [Google Scholar] [CrossRef]
- Yang, X.; Li, G.; Tian, Y.; Wang, X.; Xu, J.; Liu, R.; Deng, M.; Shao, C.; Pan, Y.; Wu, X.; et al. Identifying the E2F3-MEX3A-KLF4 signaling axis that sustains cancer cells in undifferentiated and proliferative state. Theranostics 2022, 12, 6865–6882. [Google Scholar] [CrossRef]
- Yang, P.; Zhang, P.; Zhang, S. RNA-Binding Protein MEX3A Interacting with DVL3 Stabilizes Wnt/β-Catenin Signaling in Endometrial Carcinoma. Int. J. Mol. Sci. 2022, 24, 592. [Google Scholar] [CrossRef]
- Ji, P.X.; Zhang, P.; Zhou, H.L.; Yu, H.; Fu, Y. MEX3A promotes cell proliferation by regulating the RORA/β-catenin pathway in hepatocellular carcinoma. World J. Gastrointest. Oncol. 2025, 17, 102084. [Google Scholar] [CrossRef]

| RBP | Targets | Regulation | Target Region | Model Cell Line | References |
|---|---|---|---|---|---|
| IGF2BP1 | βTRCP | ↑ S | CDS | HCT116 | [20] |
| GSK3β | ↑ S | undetermined | C33AA | [21] | |
| ELAVL1 | LRP6 | ↑ S, ↑ T | 3′UTR | IEC-6 | [22] |
| CTNNB1 | ↑ S, ↑ T | undetermined | BMSCs | [23] | |
| MSI1 | APC | ↓ T | 3′UTR | NIH3T3 | [24] |
| HCT116, HEK293 | [25] | ||||
| CTNNB1 | ↓ T | 3′UTR | HCT116, HEK293, Mouse Epithelium (in vitro) | [25] | |
| MSI2 | LEF1 | ↓ S | undetermined | K562 | [26] |
| RBM3 | CTNNB1 | ↓ S | 3′UTR | PC-3 | [27] |
| RBM47 | DKK1 | ↑ S | 3′UTR | 231-BrM2 | [28] |
| AXIN1 | ↑ S | 3′UTR | H1299 and A549 | [29] | |
| KSRP | CTNNB1 | ↓ S | 3′UTR | F9 | [30] |
| FMRP | GSK3β | ↓ T | 3′UTR | aNPCs (mouse-derived) | [31] |
| CTNNB1 | ↑ S | undetermined | GSCs (patient-derived), T98G | [32] | |
| QKI 5/6/7 | CTNNB1 | ↓ T | 3′UTR | HT29 | [33,34] |
| MEX3A | LGR5 | ↑ S | CDS, 3′UTR | mIECs | [35] |
| DKK1 | ↓ S, ↓ T | 3′UTR | BT549, MDA-MB-468, SUM159PT | [36] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Czap, M.S.; Singh, V.; Spiegelman, V.S. RNA-Binding Proteins: Modulators of Canonical Wnt Signaling Pathway. Int. J. Mol. Sci. 2026, 27, 205. https://doi.org/10.3390/ijms27010205
Czap MS, Singh V, Spiegelman VS. RNA-Binding Proteins: Modulators of Canonical Wnt Signaling Pathway. International Journal of Molecular Sciences. 2026; 27(1):205. https://doi.org/10.3390/ijms27010205
Chicago/Turabian StyleCzap, Michael S., Vikash Singh, and Vladimir S. Spiegelman. 2026. "RNA-Binding Proteins: Modulators of Canonical Wnt Signaling Pathway" International Journal of Molecular Sciences 27, no. 1: 205. https://doi.org/10.3390/ijms27010205
APA StyleCzap, M. S., Singh, V., & Spiegelman, V. S. (2026). RNA-Binding Proteins: Modulators of Canonical Wnt Signaling Pathway. International Journal of Molecular Sciences, 27(1), 205. https://doi.org/10.3390/ijms27010205







