Neanderthal and Denisovan Glutamate Dehydrogenase 2 Evolution and Clinical Significance
Abstract
1. Introduction
2. Results
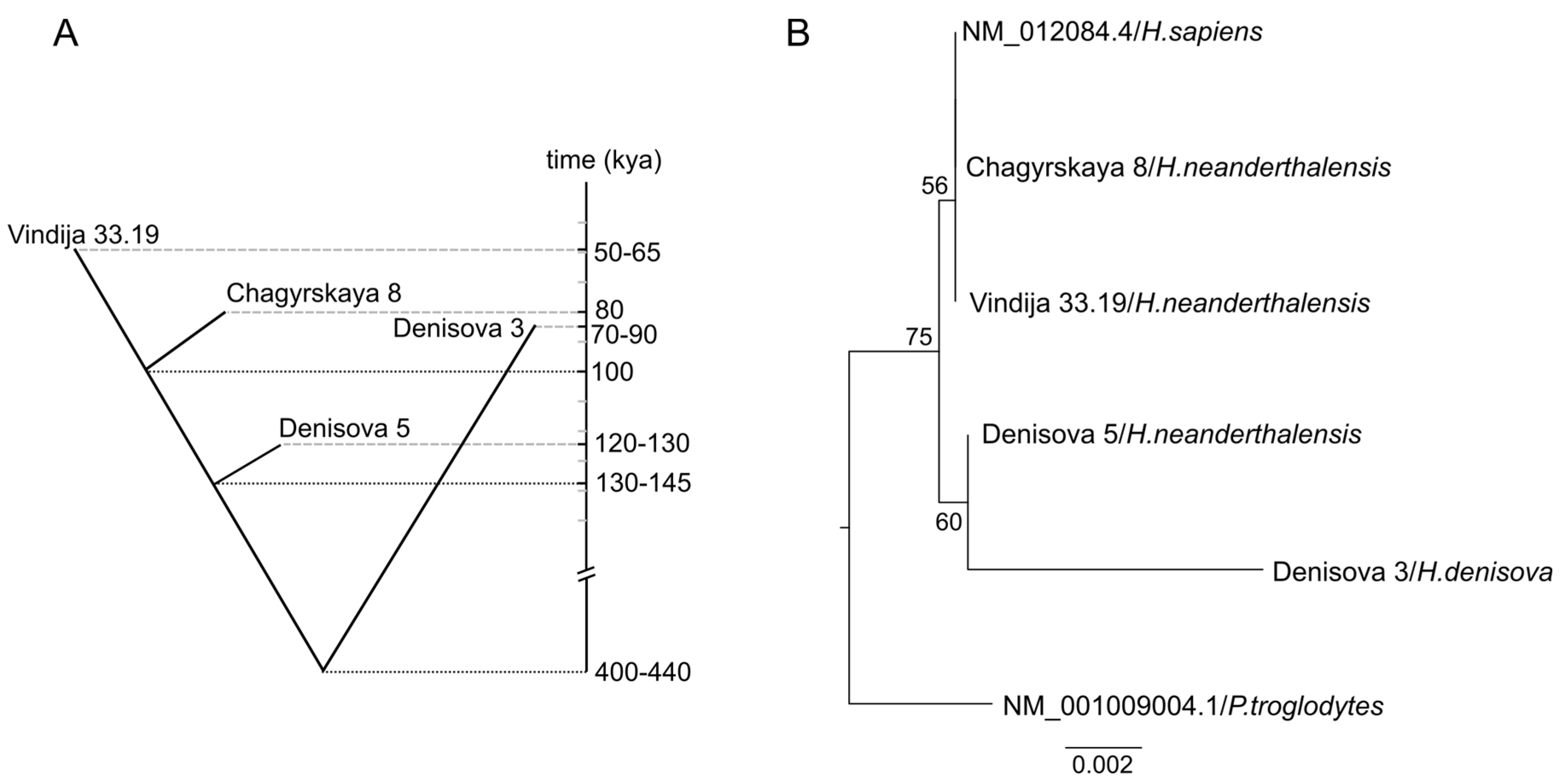
2.1. Phylogenetic Analysis of Neanderthal and Denisovan GLUD2 Sequence
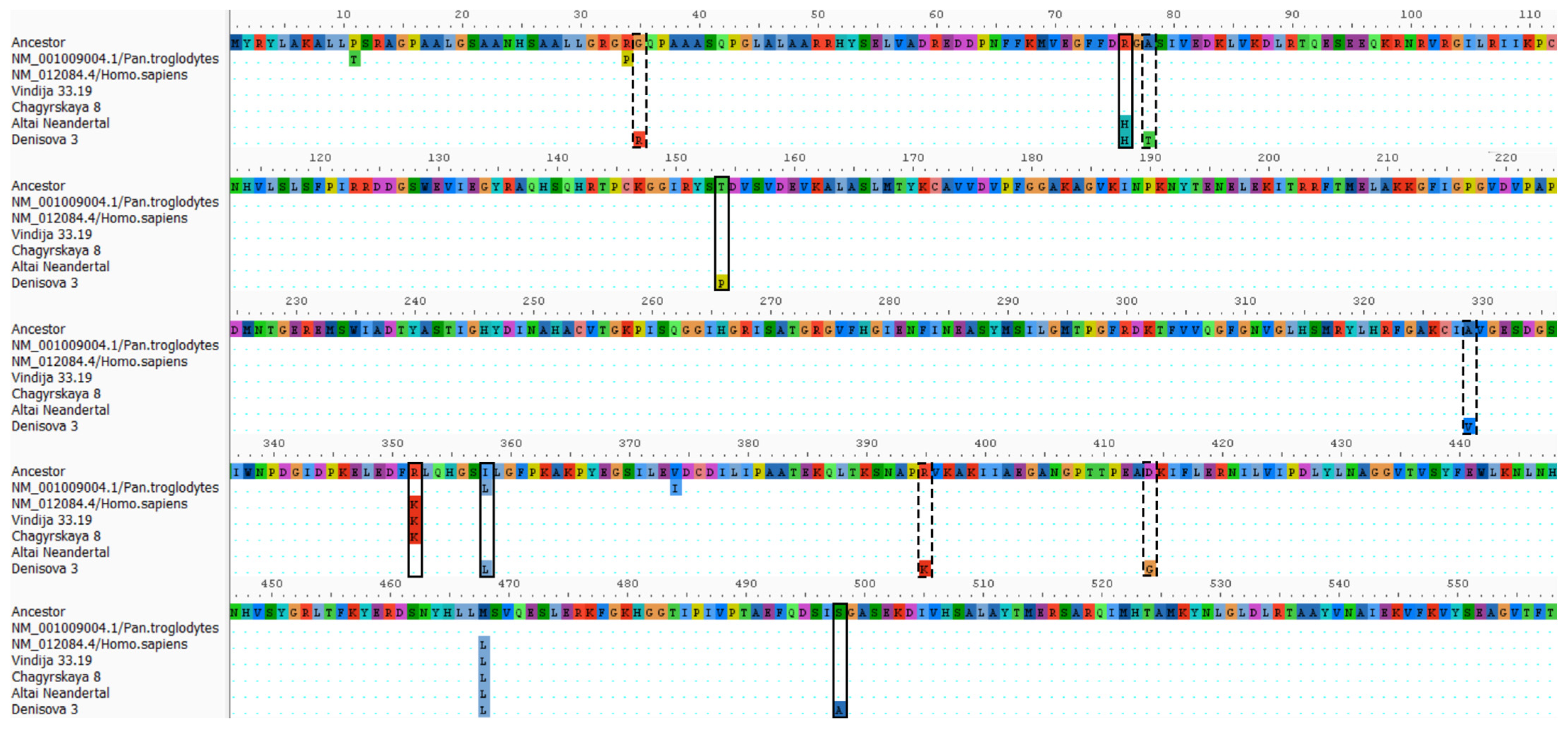
2.2. Substitutions in Denisovan GLUD1 Illustrate the Level of DNA Degradation
2.3. Exclusion of GLUD2 Substitutions Caused by DNA Degradation
2.4. Genetic Insights into GDH2 Evolution from Common Mutations of Altai Neanderthal and Denisovan
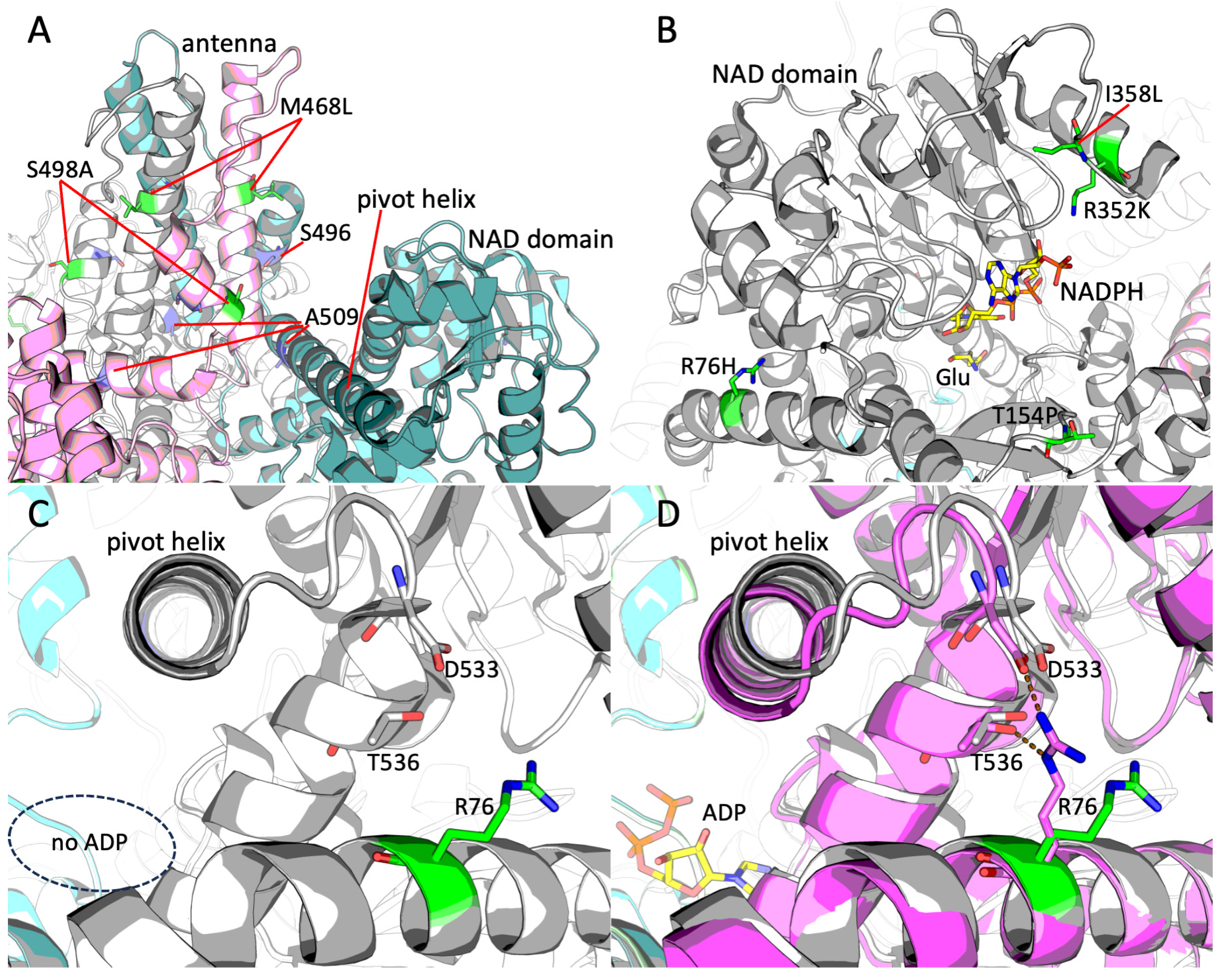
2.5. Structural Analysis of the Novel GDH2 Mutations Found in Altai Neanderthal and Denisovan Genomes
2.6. Potential Clinical Singificance of Neanderthal and Denisovan GDH2 Variants
| Variant | Allele Frequency | Submitted Condition | Structural Data | Database |
|---|---|---|---|---|
| R76H (c.227G>A) | – | Somatic, Colon Adenocarcinoma | N-terminal helix | COSMIC |
| T154P (c.460A>C) | 0.0002231 | – | Entrance to the active site | gnomAD, dbSNP |
| I358L (c.1072A>C) | 8.268 × 10−7 | – | NAD domain | gnomAD |
| L468M (c.1402C>A) | – | – | Antenna, large helix | Ensembl, Uniprot |
| S498A (c.1492T>G) | 0.02618 | Parkinson’s disease, late-onset | Antenna, small helix | gnomAD, ClinVar |
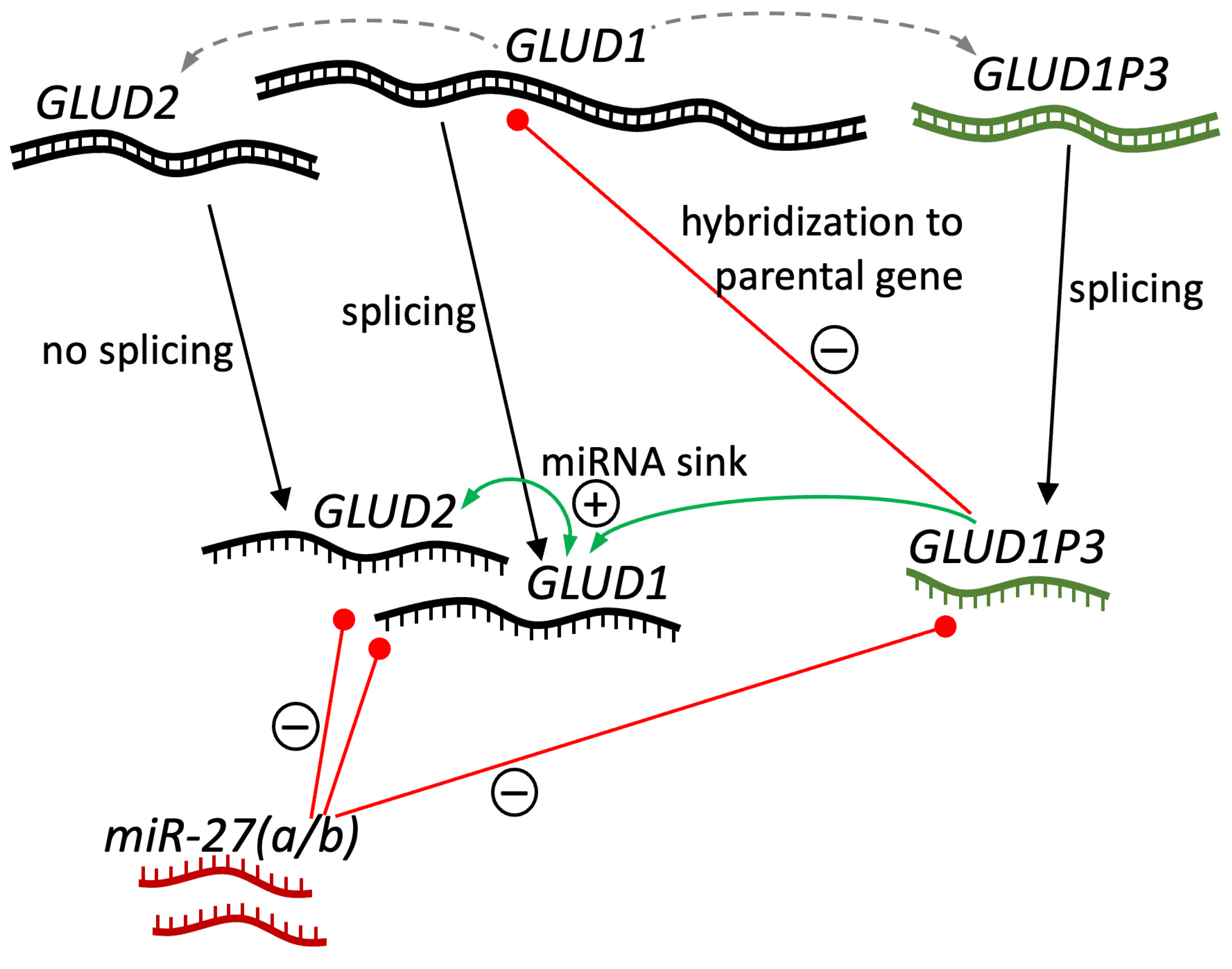
2.7. Availability of GDH Pseudogenes in Neanderthal and Denisovan Genomes and Their Plausible Role
3. Discussion
3.1. Impact of DNA Degradation on the Results
3.2. Potential Role of Gene Flow
3.3. Structural Data on GDH2 Mutations Compared to Other Species
3.4. GDH Pseudogenes and Their Potential Regulatory Role
4. Materials and Methods
4.1. Sequences of GDH Genes and Pseudogenes
4.2. Phylogenetics Analysis
4.3. Structural Analysis of GDH Mutations
4.4. Analysis of Available Neanderthal-like GDH2 Variants and Their Clinical Significance
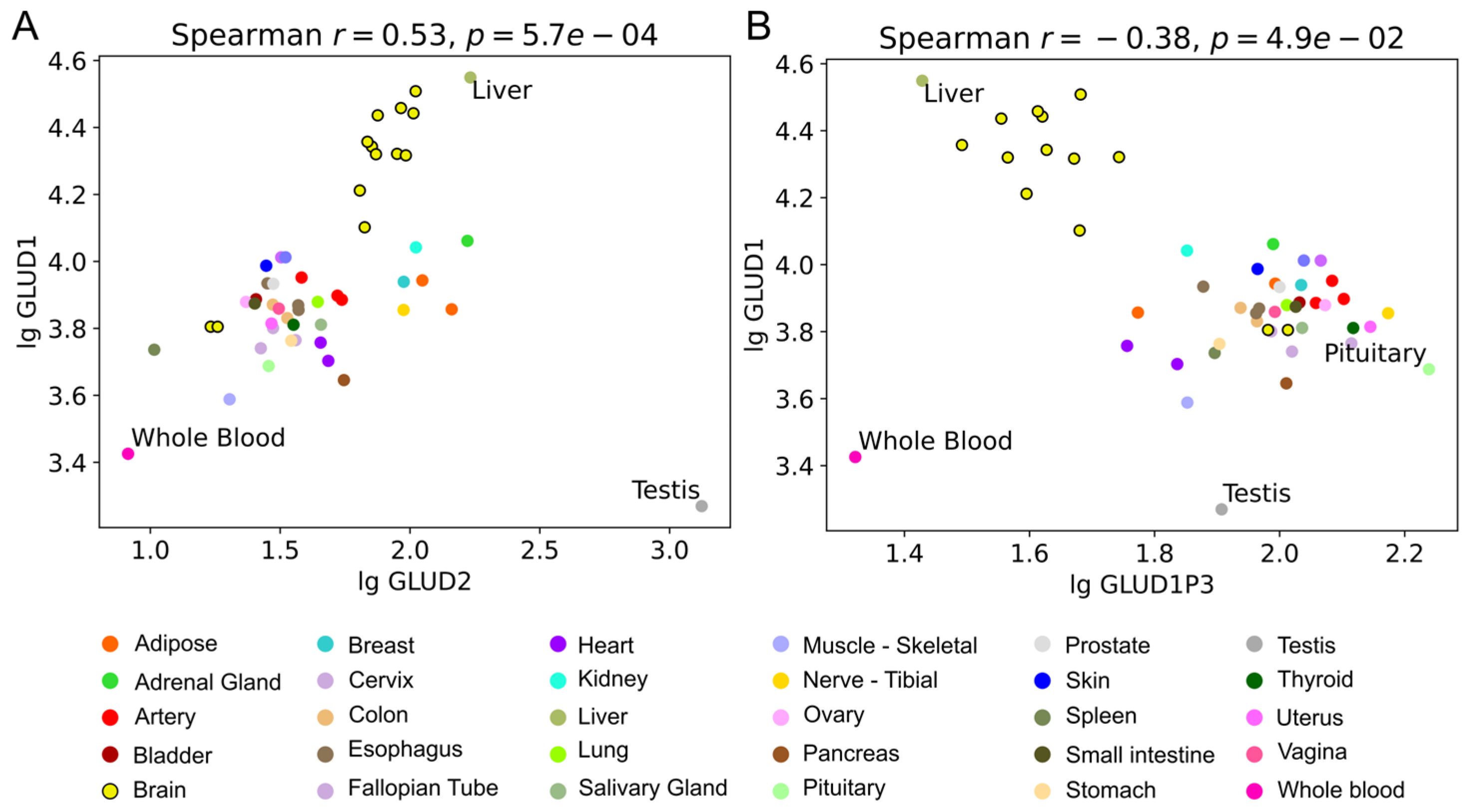
4.5. Correlation Analysis of (Pseudo)Gene Expression
4.6. Sequence Alignments of MiR-27, GLUD1, GLUD2, and GLUD1P3
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GDH | glutamate dehydrogenase |
| ky | 1000 years |
| kya | 1000 years ago |
| TS1 | transition type 2 |
| TS2 | transition type 2 |
| GTEx | Genotype-Tissue Expression |
References
- Shashidharan, P.; Michaelidis, T.M.; Robakis, N.K.; Kresovali, A.; Papamatheakis, J.; Plaitakis, A. Novel human glutamate dehydrogenase expressed in neural and testicular tissues and encoded by an X-linked intronless gene. J. Biol. Chem. 1994, 269, 16971–16976. [Google Scholar] [CrossRef]
- Burki, F.; Kaessmann, H. Birth and adaptive evolution of a hominoid gene that supports high neurotransmitter flux. Nat. Genet. 2004, 36, 1061–1063. [Google Scholar] [CrossRef]
- Zhang, Z.; Carriero, N.; Gerstein, M. Comparative analysis of processed pseudogenes in the mouse and human genomes. Trends Genet. 2004, 20, 62–67. [Google Scholar] [CrossRef]
- Aleshina, Y.A.; Aleshin, V.A. Evolutionary Changes in Primate Glutamate Dehydrogenases 1 and 2 Influence the Protein Regulation by Ligands, Targeting and Posttranslational Modifications. Int. J. Mol. Sci. 2024, 25, 4341. [Google Scholar] [CrossRef]
- Smith, T.J.; Stanley, C.A. Untangling the glutamate dehydrogenase allosteric nightmare. Trends Biochem. Sci. 2008, 33, 557–564. [Google Scholar] [CrossRef]
- Spanaki, C.; Plaitakis, A. The role of glutamate dehydrogenase in mammalian ammonia metabolism. Neurotox. Res. 2012, 21, 117–127. [Google Scholar] [CrossRef]
- Moreno-Sanchez, R.; Marin-Hernandez, A.; Gallardo-Perez, J.C.; Pacheco-Velazquez, S.C.; Robledo-Cadena, D.X.; Padilla-Flores, J.A.; Saavedra, E.; Rodriguez-Enriquez, S. Physiological Role of Glutamate Dehydrogenase in Cancer Cells. Front. Oncol. 2020, 10, 429. [Google Scholar] [CrossRef]
- Munn, E.A. Structure of oligomeric and polymeric forms of ox liver glutamate dehydrogenase examined by electron microscopy. Biochim. Biophys. Acta 1972, 285, 301–313. [Google Scholar] [CrossRef]
- Banerjee, S.; Schmidt, T.; Fang, J.; Stanley, C.A.; Smith, T.J. Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation. Biochemistry 2003, 42, 3446–3456. [Google Scholar] [CrossRef]
- Vedelek, V.; Vedelek, B.; Lorincz, P.; Juhasz, G.; Sinka, R. A comparative analysis of fruit fly and human glutamate dehydrogenases in Drosophila melanogaster sperm development. Front. Cell Dev. Biol. 2023, 11, 1281487. [Google Scholar] [CrossRef]
- Rosso, L.; Marques, A.C.; Reichert, A.S.; Kaessmann, H. Mitochondrial targeting adaptation of the hominoid-specific glutamate dehydrogenase driven by positive Darwinian selection. PLoS Genet 2008, 4, e1000150. [Google Scholar] [CrossRef]
- Kanavouras, K.; Mastorodemos, V.; Borompokas, N.; Spanaki, C.; Plaitakis, A. Properties and molecular evolution of human GLUD2 (neural and testicular tissue-specific) glutamate dehydrogenase. J. Neurosci. Res. 2007, 85, 3398–3406. [Google Scholar] [CrossRef]
- Spanaki, C.; Sidiropoulou, K.; Petraki, Z.; Diskos, K.; Konstantoudaki, X.; Volitaki, E.; Mylonaki, K.; Savvaki, M.; Plaitakis, A. Glutamate-specific gene linked to human brain evolution enhances synaptic plasticity and cognitive processes. iScience 2024, 27, 108821. [Google Scholar] [CrossRef]
- Spanaki, C.; Kotzamani, D.; Plaitakis, A. Widening Spectrum of Cellular and Subcellular Expression of Human GLUD1 and GLUD2 Glutamate Dehydrogenases Suggests Novel Functions. Neurochem. Res. 2016, 42, 92–107. [Google Scholar] [CrossRef]
- Lillepea, K.; Juchnewitsch, A.-G.; Kasak, L.; Valkna, A.; Dutta, A.; Pomm, K.; Poolamets, O.; Nagirnaja, L.; Tamp, E.; Mahyari, E.; et al. Toward clinical exomes in diagnostics and management of male infertility. Am. J. Hum. Genet. 2024, 111, 877–895. [Google Scholar] [CrossRef]
- Consortium, G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Meyer, M.; Kircher, M.; Gansauge, M.-T.; Li, H.; Racimo, F.; Mallick, S.; Schraiber, J.G.; Jay, F.; Prüfer, K.; de Filippo, C.; et al. A High-Coverage Genome Sequence from an Archaic Denisovan Individual. Science 2012, 338, 222–226. [Google Scholar] [CrossRef]
- Prüfer, K.; Racimo, F.; Patterson, N.; Jay, F.; Sankararaman, S.; Sawyer, S.; Heinze, A.; Renaud, G.; Sudmant, P.H.; de Filippo, C.; et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 2013, 505, 43–49. [Google Scholar] [CrossRef]
- Mafessoni, F.; Grote, S.; de Filippo, C.; Slon, V.; Kolobova, K.A.; Viola, B.; Markin, S.V.; Chintalapati, M.; Peyrégne, S.; Skov, L.; et al. A high-coverage Neandertal genome from Chagyrskaya Cave. Proc. Natl. Acad. Sci. USA 2020, 117, 15132–15136. [Google Scholar] [CrossRef]
- Prufer, K.; de Filippo, C.; Grote, S.; Mafessoni, F.; Korlevic, P.; Hajdinjak, M.; Vernot, B.; Skov, L.; Hsieh, P.; Peyregne, S.; et al. A high-coverage Neandertal genome from Vindija Cave in Croatia. Science 2017, 358, 655–658. [Google Scholar] [CrossRef]
- Pearce, E.; Stringer, C.; Dunbar, R.I.M. New insights into differences in brain organization between Neanderthals and anatomically modern humans. Proc. R. Soc. B Biol. Sci. 2013, 280, 168. [Google Scholar] [CrossRef]
- Binladen, J.; Wiuf, C.; Gilbert, M.T.; Bunce, M.; Barnett, R.; Larson, G.; Greenwood, A.D.; Haile, J.; Ho, S.Y.; Hansen, A.J.; et al. Assessing the fidelity of ancient DNA sequences amplified from nuclear genes. Genetics 2006, 172, 733–741. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK, 2000. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Dimovasili, C.; Fadouloglou, V.E.; Kefala, A.; Providaki, M.; Kotsifaki, D.; Kanavouras, K.; Sarrou, I.; Plaitakis, A.; Zaganas, I.; Kokkinidis, M. Crystal structure of glutamate dehydrogenase 2, a positively selected novel human enzyme involved in brain biology and cancer pathophysiology. J. Neurochem. 2021, 157, 802–815. [Google Scholar] [CrossRef]
- Aleshin, V.A.; Bunik, V.I.; Bruch, E.M.; Bellinzoni, M. Structural Basis for the Binding of Allosteric Activators Leucine and ADP to Mammalian Glutamate Dehydrogenase. Int. J. Mol. Sci. 2022, 23, 11306. [Google Scholar] [CrossRef]
- Plaitakis, A.; Latsoudis, H.; Kanavouras, K.; Ritz, B.; Bronstein, J.M.; Skoula, I.; Mastorodemos, V.; Papapetropoulos, S.; Borompokas, N.; Zaganas, I.; et al. Gain-of-function variant in GLUD2 glutamate dehydrogenase modifies Parkinson’s disease onset. Eur. J. Hum. Genet. 2010, 18, 336–341. [Google Scholar] [CrossRef]
- Massilani, D.; Skov, L.; Hajdinjak, M.; Gunchinsuren, B.; Tseveendorj, D.; Yi, S.; Lee, J.; Nagel, S.; Nickel, B.; Devièse, T.; et al. Denisovan ancestry and population history of early East Asians. Science 2020, 370, 579–583. [Google Scholar] [CrossRef]
- Browning, S.R.; Browning, B.L.; Zhou, Y.; Tucci, S.; Akey, J.M. Analysis of Human Sequence Data Reveals Two Pulses of Archaic Denisovan Admixture. Cell 2018, 173, 53–61.e9. [Google Scholar] [CrossRef]
- Karafet, T.M.; Bulayeva, K.B.; Bulayev, O.A.; Gurgenova, F.; Omarova, J.; Yepiskoposyan, L.; Savina, O.V.; Veeramah, K.R.; Hammer, M.F. Extensive genome-wide autozygosity in the population isolates of Daghestan. Eur. J. Hum. Genet. 2015, 23, 1405–1412. [Google Scholar] [CrossRef]
- Landrum, M.J.; Chitipiralla, S.; Brown, G.R.; Chen, C.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; Kaur, K.; Liu, C.; et al. ClinVar: Improvements to accessing data. Nucleic Acids Res. 2020, 48, D835–D844. [Google Scholar] [CrossRef]
- Sondka, Z.; Dhir, N.B.; Carvalho-Silva, D.; Jupe, S.; Madhumita; McLaren, K.; Starkey, M.; Ward, S.; Wilding, J.; Ahmed, M.; et al. COSMIC: A curated database of somatic variants and clinical data for cancer. Nucleic Acids Res. 2024, 52, D1210–D1217. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.; Sirotkin, K. dbSNP—Database for Single Nucleotide Polymorphisms and Other Classes of Minor Genetic Variation. Genome Res. 1999, 9, 677–679. [Google Scholar] [CrossRef]
- Dyer, S.C.; Austine-Orimoloye, O.; Azov, A.G.; Barba, M.; Barnes, I.; Barrera-Enriquez, V.P.; Becker, A.; Bennett, R.; Beracochea, M.; Berry, A.; et al. Ensembl 2025. Nucleic Acids Res. 2025, 53, D948–D957. [Google Scholar] [CrossRef]
- Chen, S.; Francioli, L.C.; Goodrich, J.K.; Collins, R.L.; Kanai, M.; Wang, Q.; Alföldi, J.; Watts, N.A.; Vittal, C.; Gauthier, L.D.; et al. A genomic mutational constraint map using variation in 76,156 human genomes. Nature 2023, 625, 92–100. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Adesina, A.; Ahmad, S.; Bowler-Barnett, E.H.; Bye-A-Jee, H.; Carpentier, D.; Denny, P.; et al. UniProt: The Universal Protein Knowledgebase in 2025. Nucleic Acids Res. 2025, 53, D609–D617. [Google Scholar] [CrossRef]
- Grigorenko, A.P.; Borinskaya, S.A.; Yankovsky, N.K.; Rogaev, E.I. Achievements and peculiarities in studies of ancient DNA and DNA from complicated forensic specimens. Acta Naturae 2009, 1, 58–69. [Google Scholar] [CrossRef]
- Litso, I.; Plaitakis, A.; Fadouloglou, V.E.; Providaki, M.; Kokkinidis, M.; Zaganas, I. Structural Evolution of Primate Glutamate Dehydrogenase 2 as Revealed by In Silico Predictions and Experimentally Determined Structures. Biomolecules 2023, 14, 22. [Google Scholar] [CrossRef]
- Mkrtchyan, G.; Aleshin, V.; Parkhomenko, Y.; Kaehne, T.; Di Salvo, M.L.; Parroni, A.; Contestabile, R.; Vovk, A.; Bettendorff, L.; Bunik, V. Molecular mechanisms of the non-coenzyme action of thiamin in brain: Biochemical, structural and pathway analysis. Sci. Rep. 2015, 5, 12583. [Google Scholar] [CrossRef]
- Mathioudakis, L.; Dimovasili, C.; Bourbouli, M.; Latsoudis, H.; Kokosali, E.; Gouna, G.; Vogiatzi, E.; Basta, M.; Kapetanaki, S.; Panagiotakis, S.; et al. Study of Alzheimer’s disease- and frontotemporal dementia-associated genes in the Cretan Aging Cohort. Neurobiol. Aging 2023, 123, 111–128. [Google Scholar] [CrossRef]
- Milligan, M.J.; Lipovich, L. Pseudogene-derived lncRNAs: Emerging regulators of gene expression. Front. Genet. 2015, 5, 476. [Google Scholar] [CrossRef]
- Barisciano, G.; Colangelo, T.; Rosato, V.; Muccillo, L.; Taddei, M.L.; Ippolito, L.; Chiarugi, P.; Galgani, M.; Bruzzaniti, S.; Matarese, G.; et al. miR-27a is a master regulator of metabolic reprogramming and chemoresistance in colorectal cancer. Br. J. Cancer 2020, 122, 1354–1366. [Google Scholar] [CrossRef]
- Rosato, V.B.G.; Muccillo, L.; Colangelo, T.; Bianchi, F.; Mazzoccoli, G.; Colantuoni, V.; Sabatino, L. miR-27a-3p Is a MasterRegulator of Metabolic Reprogramming in Colorectal Cancer. Available online: https://www.nanaonlus.org/wp-content/uploads/2018/07/Poster-Rosato-Valeria.pdf (accessed on 7 March 2024).
- Grzybowska, E.A. Human intronless genes: Functional groups, associated diseases, evolution, and mRNA processing in absence of splicing. Biochem. Biophys. Res. Commun. 2012, 424, 1–6. [Google Scholar] [CrossRef]
- Guttilla, I.K.; White, B.A. Coordinate Regulation of FOXO1 by miR-27a, miR-96, and miR-182 in Breast Cancer Cells. J. Biol. Chem. 2009, 284, 23204–23216. [Google Scholar] [CrossRef]
- Poliseno, L.; Salmena, L.; Zhang, J.; Carver, B.; Haveman, W.J.; Pandolfi, P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010, 465, 1033–1038. [Google Scholar] [CrossRef]
- Johnsson, P.; Ackley, A.; Vidarsdottir, L.; Lui, W.-O.; Corcoran, M.; Grandér, D.; Morris, K.V. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat. Struct. Mol. Biol. 2013, 20, 440–446. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Chakraborty, R. Molecular evolution and phylogenetics, Masatoshi Nei and Sudhir Kumar. Oxford University Press, Oxford, UK/New York, NY, USA, 2000, xiv+333 pages (hardback) $75; (paperback) $35.00. Mol. Phylogenet. Evol. 2002, 25, 569–570. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Smith, T.F.; Waterman, M.S. Identification of common molecular subsequences. J. Mol. Biol. 1981, 147, 195–197. [Google Scholar] [CrossRef]
| Substitution | Transition Type | Potential Cause |
|---|---|---|
| c.342T>C | TS1 | DNA degradation |
| c.376G>A | TS2 | DNA degradation |
| c.462T>C | TS1 | DNA degradation |
| c.527T>C | TS1 | DNA degradation |
| c.771T>C | TS1 | DNA degradation |
| c.909A>C | none | synonymous mutation |
| c.942A>G | TS1 | DNA degradation |
| c.1175A>G | TS1 | DNA degradation |
| c.1255G>A | TS2 | DNA degradation |
| c.1479G>A | TS2 | DNA degradation |
| Substitution | Transition Type | Potential Cause | Synonymous or Not |
|---|---|---|---|
| c.94C>A | none | mutation | synonymous |
| c.103G>A | TS2 | DNA degradation | G35R |
| c.123G>T | none | mutation | synonymous |
| c.232G>A | TS2 | DNA degradation | A78T |
| c.460A>C | none | mutation | T154P |
| c.582C>T | TS2 | DNA degradation | synonymous |
| c.986C>T | TS2 | DNA degradation | A329V |
| c.1072A>C | none | mutation | I358L |
| c.1184G>A | TS2 | DNA degradation | R395K |
| c.1241A>G | TS1 | DNA degradation | D414G |
| c.1371A>G | TS1 | DNA degradation | synonymous |
| c.1395C>T | TS2 | DNA degradation | synonymous |
| c.1492T>G | none | mutation | S498A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aleshina, Y.A.; Zavileyskiy, L.G.; Aleshin, V.A. Neanderthal and Denisovan Glutamate Dehydrogenase 2 Evolution and Clinical Significance. Int. J. Mol. Sci. 2025, 26, 4322. https://doi.org/10.3390/ijms26094322
Aleshina YA, Zavileyskiy LG, Aleshin VA. Neanderthal and Denisovan Glutamate Dehydrogenase 2 Evolution and Clinical Significance. International Journal of Molecular Sciences. 2025; 26(9):4322. https://doi.org/10.3390/ijms26094322
Chicago/Turabian StyleAleshina, Yulia A., Lev G. Zavileyskiy, and Vasily A. Aleshin. 2025. "Neanderthal and Denisovan Glutamate Dehydrogenase 2 Evolution and Clinical Significance" International Journal of Molecular Sciences 26, no. 9: 4322. https://doi.org/10.3390/ijms26094322
APA StyleAleshina, Y. A., Zavileyskiy, L. G., & Aleshin, V. A. (2025). Neanderthal and Denisovan Glutamate Dehydrogenase 2 Evolution and Clinical Significance. International Journal of Molecular Sciences, 26(9), 4322. https://doi.org/10.3390/ijms26094322






