The Nucleolus: A Central Hub for Ribosome Biogenesis and Cellular Regulatory Signals
Abstract
1. Introduction
2. Structural and Functional Organization of the Nucleolus
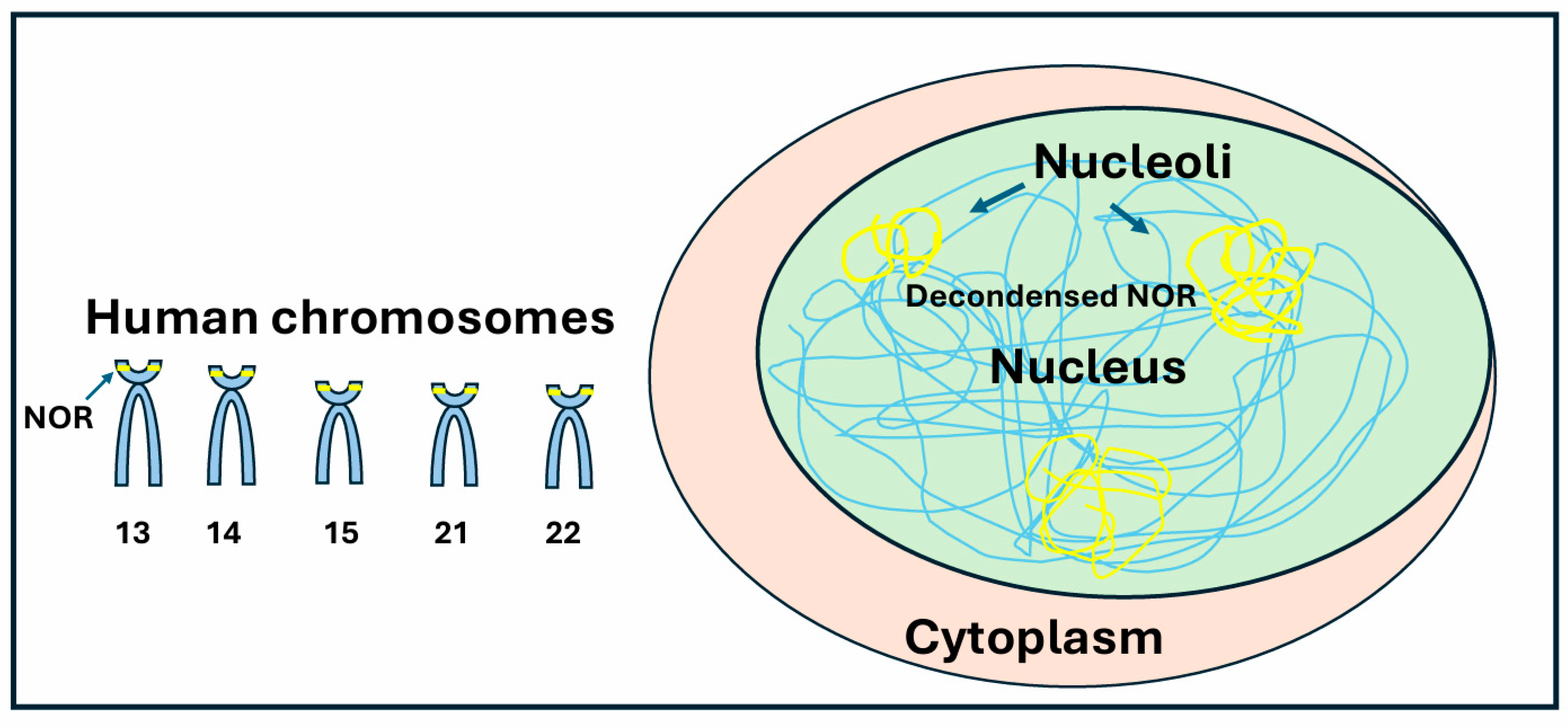
2.1. Nucleolus Organizer Regions
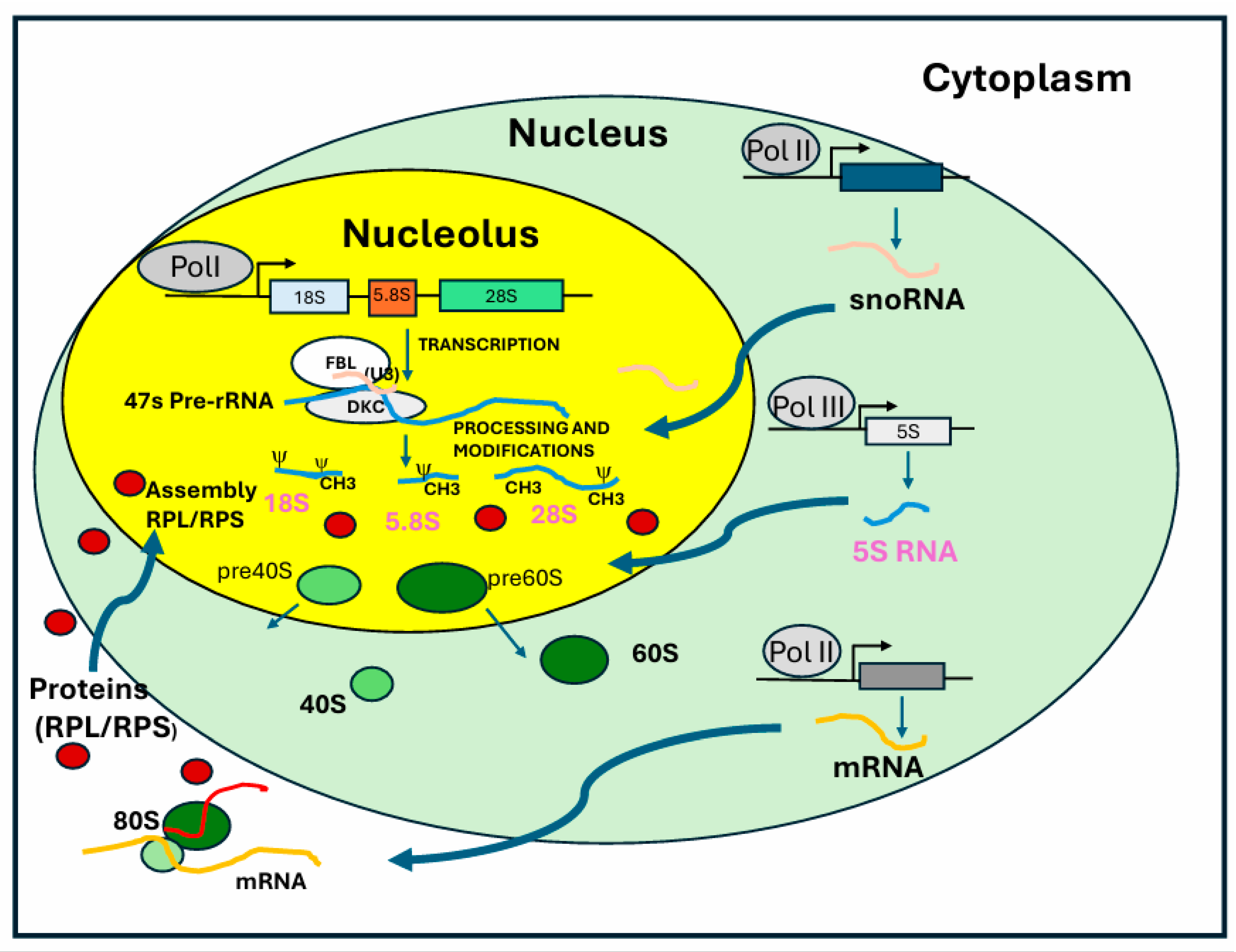
2.2. Processing of Precursor rRNA
2.3. Nucleolar Compartments
3. Regulation of Ribosomal RNA Transcription
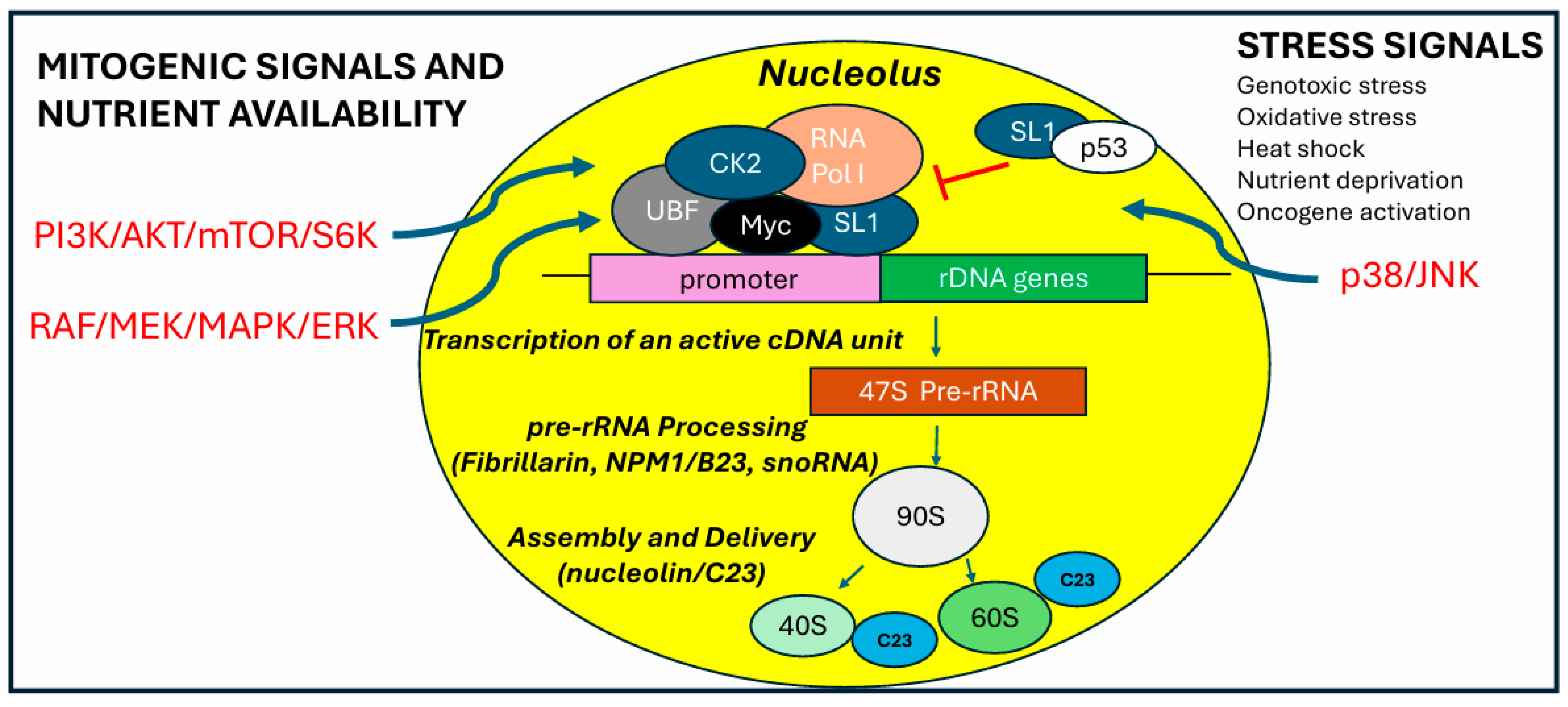
3.1. RNA Polymerase I Activity
3.2. RNA Polymerase I and the Proto-Oncogene Myc
3.3. RNA Polymerase I and the Tumor Suppressor p53
4. Ribosomal DNA and Epigenetic Modifications
4.1. UBF Orchestrates Nucleolar Architecture
4.2. rRNA Transcription and Transcription Termination Factor 1
5. Stress-Induced Transcription of Noncoding RNA from the Ribosomal Intergenic Spacer
5.1. Long Noncoding RNA (lncRNA)
5.2. Ribosomal Intergenic Spacer (rIGS RNAs)
6. RNA Polymerase I and Cell Signaling
6.1. RNA Polymerase I and mTOR Signaling
6.2. RNA Polymerase I Activity and MAPK Signaling
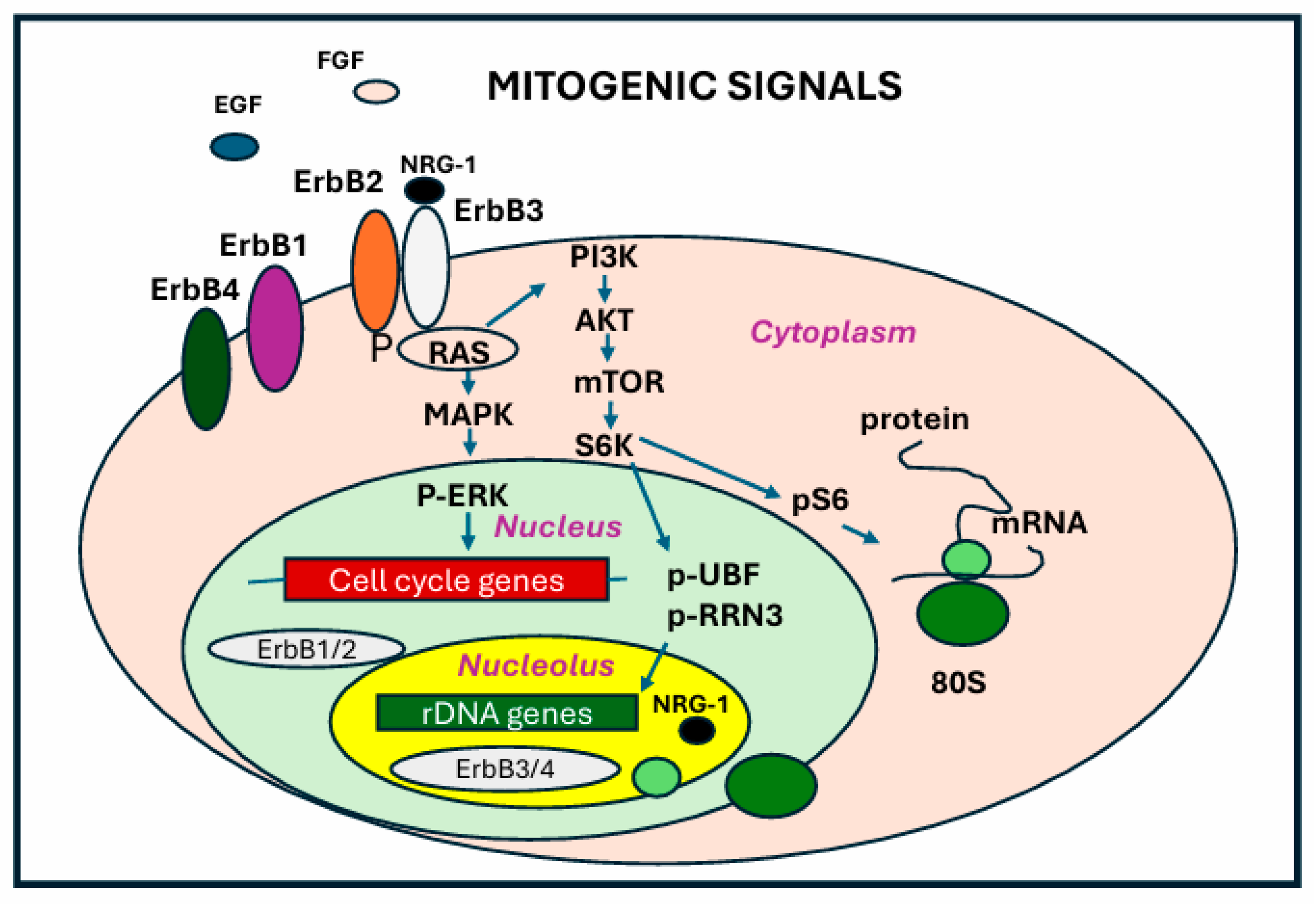
6.3. ErbB Receptors in the Nucleolus
7. The Nucleolus in Cancer: A Hidden Driver
7.1. Fibrillarin and Ribosome Heterogeneity
7.2. Nucleolus and Epithelial–Mesenchymal Transition (EMT)
7.2.1. Snail and Signaling Network
7.2.2. USP36-Snail Nucleolar Axis
7.2.3. RPL11-c-Myc-Snail Axis
8. Conclusions
Funding
Conflicts of Interest
Abbreviations
| Erb | Receptor tyrosine-protein kinase |
| UBF | Upstream binding factor |
| ARF | Alternate Reading Frame protein |
| EGR1 | Early growth response 1 |
| ActD | Actinomycin D |
| ErbB3 | v-erb-b erythroblastic leukemia viral oncogene homolog |
| SL-1 | Selectivity factor 1 |
| mTOR | Mammalian target of rapamycin |
| IGS | Intergenic spacers |
| PTEN | Phosphatase and tensin homolog |
| pRB | Retinoblastoma protein |
| NORs | Nucleolus organizer regions |
| FC | Fibrillar centers |
| DFC | Dense fibrillar components |
| GC | Granular component |
| rRNA | Ribosomal RNA |
| rDNA | Ribosomal DNA |
| NPM | Nucleophosmin |
| B23 | Nucleophosmin |
| TTF-1 | Transcription termination factor I |
| FBL | Fibrillarin |
| C23 | Nucleolin |
| CK2 | Protein kinase 2 |
References
- Hurt, E.; Cheng, J.; Baβler, J.; Iwasa, J.; Beckmann, R. SnapShot: Eukaryotic ribosome biogenesis I. Cell 2023, 186, 2282–2282.e1. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Xu, C.; Li, G.; Tong, T. Nucleolar TRF2 attenuated nucleolus stress-induced HCC cell-cycle arrest by altering rRNA synthesis. Cell Death Dis. 2018, 9, 518. [Google Scholar] [CrossRef] [PubMed]
- Shore, D.; Albert, B. Ribosome biogenesis and the cellular energy economy. Curr. Biol. 2022, 32, R611–R617. [Google Scholar] [CrossRef] [PubMed]
- Dubois, M.L.; Boisvert, F.M. The Nucleolus: Structure and Function. Funct. Nucl. 2016, 23, 29–49. [Google Scholar] [CrossRef]
- Lewis, J.D.; Tollervey, D. Like attracts like: Getting RNA processing together in the nucleus. Science 2000, 288, 1385–1389. [Google Scholar] [CrossRef] [PubMed]
- Panichnantakul, P.; Lisbeth, C.; Aguilar, L.C.; Evan Daynard, E.; Guest, M.; Peters, C.; Vogel, J.; Oeffinger, M. Protein UFMylation regulates early events of ribosomal DNA double-stranded break response. Cell Rep 2024. [Google Scholar] [CrossRef]
- Lesage, E.; Clouaire, T.; Legube, G. Repair of DNA double-strand breaks in RNAPI- and RNAPII-transcribed loci. DNA Repair 2021, 104, 103139. [Google Scholar] [CrossRef]
- Larsen, D.H.; Hari, F.; Clapperton, J.A.; Gwerder, M.; Gutsche, K.; Altmeyer, M.; Jungmichel, S.; Toledo, L.I.; Fink, D.; Rask, M.B.; et al. The NBS1-Treacle complex controls ribosomal RNA transcription in response to DNA damage. Nat. Cell Biol. 2014, 16, 792–803. [Google Scholar] [CrossRef]
- Wang, R.N.; Li, L.; Zhou, J.; Ran, J. Multifaceted roles of UFMylation in health and disease. Acta Pharmacol. Sin. 2025, 46, 805–815. [Google Scholar] [CrossRef]
- Nelson, J.O.; Watase, G.J.; Warsinger-Pepe, N.; Yamashita, Y.M. Mechanisms of rDNA Copy Number Maintenance. Trends Genet. 2019, 35, 734–742. [Google Scholar] [CrossRef]
- Lindström, M.S.; Jurada, D.; Bursac, S.; Orsolic, I.; Bartek, J.; Volarevic, S. Nucleolus as an emerging hub in maintance of genome stability and cancer pathogenesis. Oncogene 2018, 37, 2351–2366. [Google Scholar] [CrossRef] [PubMed]
- Ataei, L.; Zhang, J.; Monis, S.; Giemza, K.; Mittal, K.; Yang, J.; Shimomura, M.; McStay, B.; Wilson, M.D.; Ramalho-Santos, M. LINE1 elements at distal junctions of rDNA repeats regulate nucleolar organization in human embryonic stem cells. Genes Dev. 2025, 39, 280–298. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Lemos, B. The long-range interaction map of ribosomal DNA arrays. PLoS Genet. 2018, 14, e1007258. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, A.; Lukowiak, A.; Jády, B.E.; Dragon, F.; Kiss, T.; Terns, R.M.; Terns, M.P. Nucleolar localization signals of box H/ACA small nucleolar RNAs. EMBO J. 1999, 18, 5120–5130. [Google Scholar] [CrossRef]
- Dunbar, D.A.; Wormsley, S.; Lowe, T.M.; Baserga, S.J. Fibrillarin-associated Box C/D Small Nucleolar RNAs in Trypanosoma brucei: Sequence conservation and implications for 2′-o-ribose methylation of rRNA. J. Biol. Chem. 2000, 275, 14767–14776. [Google Scholar] [CrossRef]
- Rothé, B.; Manival, X.; Rolland, N.; Charron, C.; Senty-Ségault, V.; Branlant, C.; Charpentier, B. Implication of the box C/D snoRNP assembly factor Rsa1p in U3 snoRNP assembly. Nucleic Acids Res. 2017, 45, 7455–7473. [Google Scholar] [CrossRef]
- Sklias, A.; Cruciani, S.; Marchand, V.; Spagnuolo, M.; Lavergne, G.; Bourguignon, V.; Brambilla, A.; Dreos, R.; Marygold, S.J.; Novoa, E.M.; et al. Comprehensive map of ribosomal 2′-O-methylation and C/D box snoRNAs in Drosophila melanogaster. Nucleic Acids Res. 2024, 52, 2848–2864. [Google Scholar] [CrossRef]
- Kugler, J.E.; Deng, T.; Bustin, M. The HMGN family of chromatin-binding proteins: Dynamic modulators of epigenetic processes. Biochim. Biophys. Acta. 2012, 1819, 652–656. [Google Scholar] [CrossRef]
- Castillo Duque de Estrada, N.M.; Thoms, M.; Flemming, D.; Hammaren, H.M.; Buschauer, R.; Ameismeier, M.; Baßler, J.; Beck, M.; Beckmann, R.; Hurt, E. Structure of nascent 5S RNPs at the crossroad between ribosome assembly and MDM2-p53 pathways. Nat. Struct. Mol. Biol. 2023, 30, 1119–1131. [Google Scholar] [CrossRef]
- Tsekrekou, M.; Stratigi, K.; Chatzinikolaou, G. The Nucleolus: In Genome Maintenance and Repair. Int. J. Mol. Sci. 2017, 18, 1411. [Google Scholar] [CrossRef]
- Lafontaine, D.L.J.; Riback, J.A.; Bascetin, R.; Brangwynne, C.P. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 2021, 22, 165–182. [Google Scholar] [CrossRef] [PubMed]
- Moore, H.M.; Bai, B.; Boisvert, F.M.; Latonen, L.; Rantanen, V.; Simpson, J.C.; Pepperkok, R.; Lamond, A.I.; Laiho, M. Quantitative proteomics and dynamic imaging of the nucleolus reveal distinct responses to UV and ionizing radiation. Mol. Cell Proteom. 2011, 10, M111.009241. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.S.; Lam, Y.W.; Leung, A.K.; Ong, S.E.; Lyon, C.E.; Lamond, A.I.; Mann, M. Nucleolar proteome dynamics. Nature 2005, 433, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Aitken, S.; Semple, C.A. The circadian dynamics of small nucleolar RNA in the mouse liver. J. R. Soc. Interface 2017, 14, 20170034. [Google Scholar] [CrossRef]
- Voit, R.; Grummt, I. Phosphorylation of UBF at serine 388 is required for interaction with RNA polymerase I and activation of rDNA transcription. Proc. Natl. Acad. Sci. USA 2001, 98, 13631–13636. [Google Scholar] [CrossRef]
- Cavanaugh, A.H.; Hempel, W.M.; Taylor, L.J.; Rogalsky, V.; Todorov, G.; Rothblum, L.I. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature 1995, 374, 177–180. [Google Scholar] [CrossRef]
- O’Mahony, D.J.; Xie, W.Q.; Smith, S.D.; Singer, H.A.; Rothblum, L.I. Differential phosphorylation and localization of the transcription factor UBF in vivo in response to serum deprivation. In vitro dephosphorylation of UBF reduces its transactivation properties. J. Biol. Chem. 1992, 267, 35–38. [Google Scholar] [CrossRef] [PubMed]
- Pitts, S.; Laiho, M. Regulation of RNA Polymerase I Stability and Function. Cancers 2022, 14, 5776. [Google Scholar] [CrossRef]
- Panov, K.I.; Friedrich, J.K.; Russell, J.; Zomerdijk, J.C. UBF activates RNA polymerase I transcription by stimulating promoter escape. EMBO J. 2006, 25, 3310–3322. [Google Scholar] [CrossRef]
- Gorski, J.J.; Pathak, S.; Panov, K.; Kasciukovic, T.; Panova, T.; Russell, J.; Zomerdijk, J.C. A novel TBP-associated factor of SL1 functions in RNA polymerase I transcription. EMBO J. 2007, 26, 1560–1568. [Google Scholar] [CrossRef]
- Sadian, Y.; Baudin, F.; Tafur, L.; Murciano, B.; Wetzel, R.; Weis, F.; Müller, C.W. Molecular insight into RNA polymerase I promoter recognition and promoter melting. Nat. Commun. 2019, 10, 5543. [Google Scholar] [CrossRef] [PubMed]
- Peyroche, G.; Milkereit, P.; Bischler, N.; Tschochner, H.; Schultz, P.; Sentenac, A.; Carles, C.; Riva, M. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J. 2000, 19, 5473–5482. [Google Scholar] [CrossRef] [PubMed]
- Stepanchick, A.; Zhi, H.; Cavanaugh, A.H.; Rothblum, K.; Schneider, D.A.; Rothblum, L.I. DNA binding by the ribosomal DNA transcription factor rrn3 is essential for ribosomal DNA transcription. J. Biol. Chem. 2013, 288, 9135–9144. [Google Scholar] [CrossRef] [PubMed]
- Pilsl, M.; Crucifix, C.; Papai, G.; Krupp, F.; Steinbauer, R.; Griesenbeck, J.; Milkereit, P.; Tschochner, H.; Schultz, P. Structure of the initiation-competent RNA polymerase I and its implication for transcription. Nat. Commun. 2016, 7, 12126. [Google Scholar] [CrossRef]
- Schwank, K.; Schmid, C.; Fremter, T.; Milkereit, P.; Griesenbeck, J.; Tschochner, H. RNA polymerase I (Pol I) lobe-binding subunit Rpa12.2 promotes RNA cleavage and proofreading. J. Biol. Chem. 2022, 298, 101862. [Google Scholar] [CrossRef]
- Ruan, W.; Lehmann, E.; Thomm, M.; Kostrewa, D.; Cramer, P. Evolution of two modes of intrinsic RNA polymerase transcript cleavage. J. Biol. Chem. 2011, 286, 18701–18707. [Google Scholar] [CrossRef]
- Hoppe, S.; Bierhoff, H.; Cado, I.; Weber, A.; Tiebe, M.; Grummt, I.; Voit, R. AMP-activated protein kinase adapts rRNA synthesis to cellular energy supply. Proc. Natl. Acad. Sci. USA 2009, 106, 17781–17786. [Google Scholar] [CrossRef]
- Meraner, J.; Lechner, M.; Loidl, A.; Goralik-Schramel, M.; Voit, R.; Grummt, I.; Loidl, P. Acetylation of UBF changes during the cell cycle and regulates the interaction of UBF with RNA polymerase I. Nucleic Acids Res. 2006, 34, 1798–1806. [Google Scholar] [CrossRef]
- Arabi, A.; Wu, S.; Ridderstrale, K.; Bierhoff, H.; Shiue, C.; Fatyol, K.; Fahlén, S.; Hydbring, P.; Söderberg, O.; Grummt, I.; et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 2005, 7, 303–310. [Google Scholar] [CrossRef]
- van Riggelen, J.; Yetil, A.; Felsher, D. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer. 2010, 10, 301–309. [Google Scholar] [CrossRef]
- Shiue, C.-N.; Nematollahi-Mahani, A.; Wright, A.P. Wright, Myc-induced anchorage of the rDNA IGS region to nucleolar matrix modulates growth-stimulated changes in higher-order rDNA architecture. Nucleic Acids Res. 2014, 42, 5505–5517. [Google Scholar] [CrossRef]
- Grandori, C.; Mac, J.; Siëbelt, F.; Ayer, D.E.; Eisenman, R.N. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 1996, 15, 4344–4357. [Google Scholar] [CrossRef] [PubMed]
- Destefanis, F.; Manara, V.; Bellosta, P. Myc as a Regulator of Ribosome Biogenesis and Cell Competition: A Link to Cancer. Int. J. Mol. Sci. 2020, 21, 4037. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.; Park, H.; Ro, S.W. c-Myc-driven Hepatocarcinogenesis. Anticancer. Res. 2021, 41, 4937–4946. [Google Scholar] [CrossRef] [PubMed]
- Boone, D.N.; Qi, Y.; Li, Z.; Hann, S.R. Egr1 mediates p53-independent c-Myc-induced apoptosis via a non-canonical ARF-dependent transcriptional mechanism. Proc. Natl. Acad. Sci. USA 2011, 108, 632–637. [Google Scholar] [CrossRef]
- Ayrault, O.; Andrique, L.; Fauvin, D.; Eymin, B.; Gazzeri, S.; Séité, P. Human tumor suppressor p14ARF negatively regulates rRNA transcription and inhibits UBF1 transcription factor phosphorylation. Oncogene 2026, 25, 7577–7586. [Google Scholar] [CrossRef]
- Xirodimas, D.P.; Chisholm, J.; Desterro, J.M.; Lane, D.P.; Hay, R.T. P14ARF promotes accumulation of SUMO-1 conjugated (H)Mdm2. FEBS Lett. 2002, 528, 207–211. [Google Scholar] [CrossRef]
- Chen, L.; Chen, J. MDM2-ARF complex regulates p53 sumoylation. Oncogene 2003, 22, 5348–5357. [Google Scholar] [CrossRef]
- Ponti, D.; Bellenchi, G.C.; Puca, R.; Bastianelli, D.; Maroder, M.; Ragona, G.; Roussel, P.; Thiry, M.; Mercola, D.; Calogero, A. The transcription factor EGR1 localizes to the nucleolus and is linked to suppression of ribosomal precursor synthesis. PLoS ONE 2014, 9, e96037. [Google Scholar] [CrossRef]
- Ponti, D.; Bastianelli, D.; Rosa, P.; Pacini, L.; Ibrahim, M.; Rendina, E.A.; Ragona, G.; Calogero, A. The expression of B23 and EGR1 proteins is functionally linked in tumor cells under stress conditions. BMC Cell Biol. 2015, 16, 27. [Google Scholar] [CrossRef]
- Xie, W.; Ling, T.; Zhou, Y.; Feng, W.; Zhu, Q.; Stunnenberg, H.G.; Grummt, I.; Tao, W. The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proc. Natl. Acad. Sci. USA 2012, 109, 8161–8166. [Google Scholar] [CrossRef] [PubMed]
- Salifou, K.; Ray, S.; Verrier, L.; Aguirrebengoa, M.; Trouche, D.; Panov, K.I.; Vandromme, M. The histone demethylase JMJD2A/KDM4A links ribosomal RNA transcription to nutrients and growth factors availability. Nat. Commun. 2016, 7, 10174. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Lemos, B. Ribosomal DNA harbors an evolutionarily conserved clock of biological aging. Genome Res. 2019, 29, 325–333. [Google Scholar] [CrossRef]
- Moss, T.; LeDoux, M.S.; Crane-Robinson, C. HMG-boxes, ribosomopathies and neurodegenerative disease. Front. Genet. 2023, 14, 1225832. [Google Scholar] [CrossRef]
- Sanij, E.; Hannan, R.D. The role of UBF in regulating the structure and dynamics of transcriptionally active rDNA chromatin. Epigenetics 2009, 4, 374–382. [Google Scholar] [CrossRef]
- Cerqueira, A.V.; Lemos, B. Ribosomal DNA and the Nucleolus as Keystones of Nuclear Architecture, Organization, and Function. Trends Genet. 2019, 35, 710–723. [Google Scholar] [CrossRef]
- Mais, C.; Wright, J.E.; Prieto, J.L.; Raggett, S.L.; McStay, B. UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery. Genes Dev. 2005, 19, 50–64. [Google Scholar] [CrossRef]
- Dousset, T.; Wang, C.; Verheggen, C.; Chen, D.; Hernandez-Verdun, D.; Huang, S. Initiation of nucleolar assembly is independent of RNA polymerase I transcription. Mol. Biol. Cell 2000, 11, 2705–2717. [Google Scholar] [CrossRef]
- Ueshima, S.; Nagata, K.; Okuwaki, M. Internal Associations of the Acidic Region of Upstream Binding Factor Control Its Nucleolar Localization. Mol. Cell Biol. 2017, 37, e00218-17. [Google Scholar] [CrossRef]
- Chen, J.; Teo, B.H.D.; Cai, Y.; Wee, S.Y.K.; Lu, J. The linker histone H1.2 is a novel component of the nucleolar organizer regions. J. Biol. Chem. 2018, 293, 2358–2369. [Google Scholar] [CrossRef]
- Hamdane, N.; Stefanovsky, V.Y.; Tremblay, M.G.; Németh, A.; Paquet, E.; Lessard, F.; Sanij, E.; Hannan, R.; Moss, T. Conditional inactivation of Upstream Binding Factor reveals its epigenetic functions and the existence of a somatic nucleolar precursor body. PLoS Genet. 2014, 10, e1004505. [Google Scholar] [CrossRef] [PubMed]
- Santoro, R. The silence of the ribosomal RNA genes. Cell. Mol. Life Sci. 2005, 62, 2067–2079. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, V.; Baptiste, B.A.; Okur, M.N.; Bohr, V.A. Current and emerging roles of Cockayne syndrome group B (CSB) protein. Nucleic Acids Res. 2021, 49, 2418–2434. [Google Scholar] [CrossRef]
- Akamatsu, Y.; Kobayashi, T. The Human RNA Polymerase I Transcription Terminator Complex Acts as a Replication Fork Barrier That Coordinates the Progress of Replication with rRNA Transcription Activity. Mol. Cell Biol. 2015, 35, 1871–1881. [Google Scholar] [CrossRef]
- Németh, A.; Strohner, R.; Grummt, I.; Längst, G. The chromatin remodeling complex NoRC and TTF-I cooperate in the regulation of the mammalian rRNA genes in vivo. Nucleic Acids Res. 2004, 32, 4091–4099. [Google Scholar] [CrossRef]
- Gavrilova, A.A.; Neklesova, M.V.; Zagryadskaya, Y.A.; Kuznetsova, I.M.; Turoverov, K.K.; Fonin, A.V. Stress-Induced Evolution of the Nucleolus: The Role of Ribosomal Intergenic Spacer (rIGS) Transcripts. Biomolecules 2024, 14, 1333. [Google Scholar] [CrossRef]
- Audas, T.E.; Jacob, M.D.; Lee, S. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol. Cell 2012, 45, 147–157. [Google Scholar] [CrossRef]
- Bierhoff, H.; Dammert, M.A.; Brocks, D.; Dambacher, S.; Schotta, G.; Grummt, I. Quiescence-induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol. Cell 2014, 54, 675–682. [Google Scholar] [CrossRef]
- Alberti, S.; Carra, S. Nucleolus: A Liquid Droplet Compartment for Misbehaving Proteins. Curr. Biol. 2019, 29, R930–R932. [Google Scholar] [CrossRef]
- Zhao, Z.; Sentürk, N.; Song, C.; Grummt, I. lncRNA PAPAS tethered to the rDNA enhancer recruits hypophosphorylated CHD4/NuRD to repress rRNA synthesis at elevated temperatures. Genes Dev. 2018, 32, 836–848. [Google Scholar] [CrossRef]
- Zhao, Z.; Dammert, M.A.; Grummt, I.; Bierhoff, H. lncRNA-Induced Nucleosome Repositioning Reinforces Transcriptional Repression of rRNA Genes upon Hypotonic Stress. Cell Rep. 2016, 14, 1876–1882. [Google Scholar] [CrossRef] [PubMed]
- Audas, T.E.; Audas, D.E.; Jacob, M.D.; Ho, J.J.; Khacho, M.; Wang, M.; Perera, J.K.; Gardiner, C.; Bennett, C.A.; Head, T.; et al. Adaptation to Stressors by Systemic Protein Amyloidogenesis. Dev. Cell 2016, 39, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Bierhoff, H.; Postepska-Igielska, A.; Grummt, I. Noisy silence: Non-coding RNA and heterochromatin formation at repetitive elements. Epigenetics 2014, 9, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Vydzhak, O.; Luke, B.; Schindler, N. Non-coding RNAs at the Eukaryotic rDNALocus: RNA–DNAHybrids Beyond. J. Mol. Biol. 2020, 432, 4287–4304. [Google Scholar] [CrossRef]
- Stefanovsky, V.; Langlois, F.; Gagnon-Kugler, T.; Rothblum, L.I.; Moss, T. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and r-chromatin remodeling. Mol. Cell 2006, 21, 629–639. [Google Scholar] [CrossRef]
- Hannan, K.M.; Brandenburger, Y.; Jenkins, A.; Sharkey, K.; Cavanaugh, A.; Rothblum, L.; Moss, T.; Poortinga, G.; McArthur, G.A.; Pearson, R.B.; et al. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol. Cell Biol. 2003, 23, 8862–8877. [Google Scholar] [CrossRef]
- Lin, C.Y.; Navarro, S.; Reddy, S.; Comai, L. CK2-mediated stimulation of Pol I transcription by stabilization of UBF-SL1 interaction. Nucleic Acids Res. 2006, 34, 4752–4766. [Google Scholar] [CrossRef]
- Blattner, C.; Jennebach, S.; Herzog, F.; Mayer, A.; Cheung, A.C.; Witte, G.; Lorenzen, K.; Hopfner, K.P.; Heck, A.J.; Aebersold, R.; et al. Molecular basis of Rrn3-regulated RNA polymerase I initiation and cell growth. Genes Dev. 2011, 25, 2093–2105. [Google Scholar] [CrossRef]
- Lavoie, H.; Gagnon, J.; Therrien, M. ERK signalling: A master regulator of cell behaviour, life and fate. Nat. Rev. Mol. Cell Biol. 2020, 21, 607–632. [Google Scholar] [CrossRef]
- Mayer, C.; Bierhoff, H.; Grummt, I. The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes Dev. 2005, 19, 933–941. [Google Scholar] [CrossRef]
- Andrique, L.; Fauvin, D.; Maassarani, M.; Colasson, H.; Vannier, B.; Séité, P. ErbB3 80kDa, a nuclear variant of the ErbB3 receptor, binds to the Cyclin D1 promoter to activate cell proliferation but is negatively controlled by p14ARF. Cell Signal 2012, 24, 1074–1085. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z. ErbB Receptors and Cancer. Methods Mol. Biol. 2017, 1652, 3–35. [Google Scholar] [CrossRef] [PubMed]
- Kruspig, B.; Monteverde, T.; Neidler, S.; Hock, A.; Kerr, E.; Nixon, C.; Clark, W.; Hedley, A.; Laing, S.; Coffelt, S.B.; et al. The Erbb network facilitates KRAS-driven lung tumorigeneis. Sci. Transl. Med. 2018, 10, eaao2565. [Google Scholar] [CrossRef]
- Sahu, A.; Verma, S.; Varma, M.; Yadav, M.K. Impact of ErbB Receptors and Anticancer Drugs against Breast Cancer: A Review. Curr. Pharm. Biotechnol. 2022, 23, 787–802. [Google Scholar] [CrossRef]
- Wang, Y.N.; Yamaguchi, H.; Hsu, J.M.; Hung, M.C. Nuclear trafficking of the epidermal growth factor receptor family membrane proteins. Oncogene 2010, 29, 3997–4006. [Google Scholar] [CrossRef]
- Giri, D.K.; Ali-Seyed, M.; Li, L.Y.; Lee, D.F.; Ling, P.; Bartholomeusz, G.; Wang, S.C.; Hung, M.C. Endosomal transport of ErbB-2: Mechanism for nuclear entry of the cell surface receptor. Mol. Cell Biol. 2005, 25, 11005–11018. [Google Scholar] [CrossRef]
- Ni, C.Y.; Murphy, M.P.; Golde, T.E.; Carpenter, G. gamma -Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science 2001, 294, 2179–2181. [Google Scholar] [CrossRef]
- Reif, R.; Adawy, A.; Vartak, N.; Schröder, J.; Günther, G.; Ghallab, A.; Schmidt, M.; Schormann, W.; Hengstler, J.G. Activated ErbB3 Translocates to the Nucleus via Clathrin-independent Endocytosis, Which Is Associated with Proliferating Cells. J. Biol. Chem. 2016, 291, 3837–3847. [Google Scholar] [CrossRef]
- Tagliaferro, M.; Rosa, P.; Bellenchi, G.C.; Bastianelli, D.; Trotta, R.; Tito, C.; Fazi, F.; Calogero, A.; Ponti, D. Nucleolar localization of the ErbB3 receptor as a new target in glioblastoma. BMC Mol. Cell Biol. 2022, 23, 13. [Google Scholar] [CrossRef]
- Balça-Silva, J.; do Carmo, A.; Tão, H.; Rebelo, O.; Barbosa, M.; Moura-Neto, V.; Sarmento-Ribeiro, A.B.; Lopes, M.C.; Moreira, J.N. Nucleolin is expressed in patient-derived samples and glioblastoma cells, enabling improved intracellular drug delivery and cytotoxicity. Exp. Cell Res. 2018, 370, 68–77. [Google Scholar] [CrossRef]
- González-Arzola, K. The nucleolus: Coordinating stress response and genomic stability. Biochim. Et Biophys. Acta (BBA)-Gene Regul. Mech. 2024, 1867, 195029. [Google Scholar] [CrossRef] [PubMed]
- Derenzini, M.; Trerè, D.; Pession, A.; Montanaro, L.; Sirri, V.; Ochs, R.L. Nucleolar function and size in cancer cells. Am. J. Pathol. 1998, 152, 1291–1297. [Google Scholar] [PubMed] [PubMed Central]
- Montironi, R.; Braccischi, A.; Scarpelli, M.; Matera, G.; Alberti, R. Value of quantitative nucleolar features in the preoperative cytological diagnosis of follicular neoplasias of the thyroid. J. Clin. Pathol. 1991, 44, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Belin, S.; Beghin, A.; Solano-Gonzàlez, E.; Bezin, L.; Brunet-Manquat, S.; Textoris, J.; Prats, A.C.; Mertani, H.C.; Dumontet, C.; Diaz, J.J. Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PLoS ONE 2009, 4, e7147. [Google Scholar] [CrossRef]
- Williamson, D.; Lu, Y.J.; Fang, C.; Pritchard-Jones, K.; Shipley, J. Nascent pre-rRNA overexpression correlates with an adverse prognosis in alveolar rhabdomyosarcoma. Genes Chromosomes Cancer 2006, 45, 839–845. [Google Scholar] [CrossRef]
- Gamel, J.W.; McLean, I.W. Computerized histopathologic assessment of malignant potential. II. A practical method for predicting survival following enucleation for uveal melanoma. Cancer 1983, 52, 1032–1038. [Google Scholar] [CrossRef]
- van Diest, P.J.; Mouriquand, J.; Schipper, N.W.; Baak, J.P. Prognostic value of nucleolar morphometric variables in cytological breast cancer specimens. J. Clin. Pathol. 1990, 43, 157–159. [Google Scholar] [CrossRef]
- Elhamamsy, A.R.; Metge, B.J.; Alsheikh, H.A.; Shevde, L.A.; Samant, R.S. Ribosome Biogenesis: A Central Player in Cancer Metastasis and Therapeutic Resistance. Cancer Res. 2022, 82, 2344–2353. [Google Scholar] [CrossRef]
- Goodpasture, C.; Bloom, S.E. Visualization of nucleolar organizer regions im mammalian chromosomes using silver staining. Chromosoma 1975, 53, 37–50. [Google Scholar] [CrossRef]
- Aspesi, A.; Ellis, S.R. Rare ribosomopathies: Insights into mechanisms of cancer. Nat. Rev. Cancer 2019, 19, 228–238. [Google Scholar] [CrossRef]
- Venturi, G.; Montanaro, L. How Altered Ribosome Production Can Cause or Contribute to Human Disease: The Spectrum of Ribosomopathies. Cells 2020, 9, 2300. [Google Scholar] [CrossRef] [PubMed]
- Mensah, M.A.; Niskanen, H.; Magalhaes, A.P.; Basu, S.; Kircher, M.; Sczakiel, H.L.; Reiter, A.M.V.; Elsner, J.; Meinecke, P.; Biskup, S.; et al. Aberrant phase separation and nucleolar dysfunction in rare genetic diseases. Nature 2023, 614, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Yu, X. NBS1 facilitates preribosomal RNA biogenesis. Proc. Natl. Acad. Sci. USA 2025, 122, e2422029122. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, J.; Lim, C.U. The role of NBS1 in DNA double strand break repair, telomere stability, and cell cycle checkpoint control. Cell Res. 2006, 16, 45–54. [Google Scholar] [CrossRef]
- Chen, H.; Li, Y.; Li, H.; Chen, X.; Fu, H.; Mao, D.; Chen, W.; Lan, L.; Wang, C.; Hu, K.; et al. NBS1 lactylation is required for efficient DNA repair and chemotherapy resistance. Nature 2024, 631, 663–669. [Google Scholar] [CrossRef]
- Babaian, A.; Rothe, K.; Girodat, D.; Minia, I.; Djondovic, S.; Milek, M.; Spencer Miko, S.E.; Wieden, H.J.; Landthaler, M.; Morin, G.B.; et al. Loss of m1acp3Ψ Ribosomal RNA Modification Is a Major Feature of Cancer. Cell Rep. 2020, 31, 107611. [Google Scholar] [CrossRef]
- Li, D.; Wang, J. Ribosome heterogeneity in stem cells and development. J. Cell Biol. 2020, 219, e202001108. [Google Scholar] [CrossRef]
- Xue, M.; Dong, L.; Zhang, H.; Li, Y.; Qiu, K.; Zhao, Z.; Gao, M.; Han, L.; Chan, A.K.N.; Li, W.; et al. METTL16 promotes liver cancer stem cell self-renewal via controlling ribosome biogenesis and mRNA translation. J. Hematol. Oncol. 2024, 17, 7. [Google Scholar] [CrossRef]
- Barros-Silva, D.; Klavert, J.; Jenster, G.; Jerónimo, C.; Lafontaine, D.L.J.; Martens-Uzunova, E.S. The role of OncoSnoRNAs and Ribosomal RNA 2′-O-methylation in Cancer. RNA Biol. 2021, 18, 61–74. [Google Scholar] [CrossRef]
- Iyer-Bierhoff, A.; Krogh, N.; Tessarz, P.; Ruppert, T.; Nielsen, H.; Grummt, I. SIRT7-Dependent Deacetylation of Fibrillarin Controls Histone H2A Methylation and rRNA Synthesis during the Cell Cycle. Cell Rep. 2018, 25, 2946–2954.e5. [Google Scholar] [CrossRef]
- Sun, X.; Gao, C.; Xu, X.; Li, M.; Zhao, X.; Wang, Y.; Wang, Y.; Zhang, S.; Yan, Z.; Liu, X.; et al. FBL promotes cancer cell resistance to DNA damage and BRCA1 transcription via YBX1. EMBO Rep. 2023, 24, e56230. [Google Scholar] [CrossRef] [PubMed]
- Schlatter, S.; Senn, C.; Fussenegger, M. Modulation of translation-initiation in CHO-K1 cells by rapamycin-induced heterodimerization of engineered eIF4G fusion proteins. Biotechnol. Bioeng. 2003, 83, 210–225. [Google Scholar] [CrossRef] [PubMed]
- Erales, J.; Marchand, V.; Panthu, B.; Gillot, S.; Belin, S.; Ghayad, S.E.; Garcia, M.; Laforêts, F.; Marcel, V.; Baudin-Baillieu, A.; et al. Evidence for rRNA 2′-O-methylation plasticity: Control of intrinsic translational capabilities of human ribosomes. Proc. Natl. Acad. Sci. USA 2017, 114, 12934–12939. [Google Scholar] [CrossRef]
- Han, Z.; Zhang, Q.; Zhu, Y.; Chen, J.; Li, W. Ribosomes: An Exciting Avenue in Stem Cell Research. Stem Cells Int. 2020, 2020, 8863539. [Google Scholar] [CrossRef]
- Brombin, A.; Joly, J.S.; Jamen, F. New tricks for an old dog: Ribosome biogenesis contributes to stem cell homeostasis. Curr. Opin. Genet. Dev. 2015, 34, 61–70. [Google Scholar] [CrossRef]
- Aigner, K.; Dampier, B.; Descovich, L.; Mikula, M.; Sultan, A.; Schreiber, M.; Mikulits, W.; Brabletz, T.; Strand, D.; Obrist, P.; et al. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene 2007, 26, 6979–6988. [Google Scholar] [CrossRef]
- Gumbiner, B.M. Regulation of cadherin-mediated adhesion in morphogenesis. Nat. Rev. Mol. Cell Biol. 2005, 6, 622–634. [Google Scholar] [CrossRef]
- Moody, S.E.; Perez, D.; Pan, T.C.; Sarkisian, C.J.; Portocarrero, C.P.; Sterner, C.J.; Notorfrancesco, K.L.; Cardiff, R.D.; Chodosh, L.A. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell 2005, 8, 197–209. [Google Scholar] [CrossRef]
- De Craene, B.; Berx, G. Snail in the frame of malignant tumor recurrence. Breast Cancer Res. 2006, 8, 105. [Google Scholar] [CrossRef]
- Whiteman, E.L.; Liu, C.J.; Fearon, E.R.; Margolis, B. The transcription factor snail represses Crumbs3 expression and disrupts apico-basal polarity complexes. Oncogene 2008, 27, 3875–3879. [Google Scholar] [CrossRef]
- Peinado, H.; Ballestar, E.; Esteller, M.; Cano, A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol. Cell Biol. 2004, 24, 306–319. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Sleeman, J.P. Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006, 7, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Qin, K.; Yu, S.; Liu, Y.; Guo, R.; Guo, S.; Fei, J.; Wang, Y.; Jia, K.; Xu, Z.; Chen, H.; et al. USP36 stabilizes nucleolar Snail1 to promote ribosome biogenesis and cancer cell survival upon ribotoxic stress. Nat. Commun. 2023, 14, 6473. [Google Scholar] [CrossRef]
- Yu, Q.; Zhou, B.P.; Wu, Y. The regulation of snail: On the ubiquitin edge. Cancer Cell Microenviron. 2017, 4, e1567. [Google Scholar] [PubMed] [PubMed Central]
- Wang, R.; Peng, C.; Song, J.; Hua, Y.; Wu, Q.; Deng, L.; Cao, Y.; Zhang, J.; Zhang, L.; Wu, L.; et al. Downregulated RRS1 inhibits invasion and metastasis of BT549 through RPL11-c-Myc-SNAIL axis. Int. J. Oncol. 2022, 60, 33. [Google Scholar] [CrossRef]
- Sun, X.X.; Sears, R.C.; Dai, M.S. Deubiquitinating c-Myc: USP36 steps up in the nucleolus. Cell Cycle 2015, 14, 3786–3793. [Google Scholar] [CrossRef]
- Sun, X.X.; He, X.; Yin, L.; Komada, M.; Sears, R.C.; Dai, M.S. The nucleolar ubiquitin-specific protease USP36 deubiquitinates and stabilizes c-Myc. Proc. Natl. Acad. Sci. USA 2015, 112, 3734–3739. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ponti, D. The Nucleolus: A Central Hub for Ribosome Biogenesis and Cellular Regulatory Signals. Int. J. Mol. Sci. 2025, 26, 4174. https://doi.org/10.3390/ijms26094174
Ponti D. The Nucleolus: A Central Hub for Ribosome Biogenesis and Cellular Regulatory Signals. International Journal of Molecular Sciences. 2025; 26(9):4174. https://doi.org/10.3390/ijms26094174
Chicago/Turabian StylePonti, Donatella. 2025. "The Nucleolus: A Central Hub for Ribosome Biogenesis and Cellular Regulatory Signals" International Journal of Molecular Sciences 26, no. 9: 4174. https://doi.org/10.3390/ijms26094174
APA StylePonti, D. (2025). The Nucleolus: A Central Hub for Ribosome Biogenesis and Cellular Regulatory Signals. International Journal of Molecular Sciences, 26(9), 4174. https://doi.org/10.3390/ijms26094174



