Abstract
Missense mutations in the BCR::ABL1 kinase domain are found in approximately 12–80% of patients with chronic myeloid leukemia (CML). Clinically significant mutations include T315I, M244V, Y253H/F, E255K/V, V299L, and F359V. The aim of this study was to create a diagnostic system for rapid and inexpensive detection of the above mutations. We used genomic DNA and RNA from peripheral blood and bone marrow cells of 57 patients with a Ph-positive CML diagnosis established in the chronic phase. We have developed a method to detect mutations in the BCR::ABL1 gene based on allele-specific real-time polymerase chain reaction (AS-PCR). In parallel, we analyzed the RNA sequence of the protein kinase domain of the same samples by next-generation sequencing (NGS) covering the points of putative mutations. In this work, we compared the results obtained by both methods for mutation detection and variant allele frequency (VAF) estimation of mutated vs. normal alleles. The sensitivity and specificity of our diagnostic system were also evaluated. It was found that AS-PCR gives reliable results at VAF up to 0.01%. AS-PCR has high sensitivity and may serve as an alternative for the more time-consuming NGS in some cases, as well as for monitoring CML treatment and for analyzing archival material.
1. Introduction
Missense mutations in the BCR::ABL1 kinase domain associated with resistance to tyrosine kinase inhibitor (TKI) therapy are found in approximately 12–35% of patients with therapy failure in the chronic phase and in 50–80% of patients in the acceleration and blast crisis phases [1,2]. Such a wide range of reported mutation frequencies is associated not only with the stage of disease and individual patient characteristics but also with the sensitivity and specificity of diagnostic methods [3,4,5].
The majority of mutations responsible for the failure of TKI therapy affect nine amino acids of the protein kinase domain encoded by the chimeric BCR::ABL1 gene [6]. One of the most common and clinically significant mutations is the pan-resistant T315I mutation, which causes resistance to both 1st generation TKI (imatinib) and 2nd generation TKIs (nilotinib, dasatinib, and bosutinib) [7,8]. According to the literature, the detection frequency of the T315I mutation varies from 2% to 20% [9]. In addition to T315I, the most frequent mutations are M244V, Y253H/F, E255K/V, V299L, and F359V, conferring different sensitivity to certain TKIs [10,11,12,13].
In the presence of combined mutations, the tumor clone becomes more resistant to the effects of 1st and 2nd generation TKIs. In such cases, 3rd generation TKIs (ponatinib) and the allosteric TKI asciminib may be effective [14].
Thus, early detection of mutations leading to the emergence of TKI resistance provides physicians with a tool for selecting individualized therapy.
Currently, next-generation sequencing (NGS) is the primary method for detecting 47 missense mutations [15]. Despite its versatility, this technique can be time-consuming and costly. To address the issue of detecting clinically significant mutations, we propose the use of sensitive, fast, and inexpensive allele-specific polymerase chain reaction (AS-PCR) [15,16,17]. Thus, the goal of this study was to develop a diagnostic system for detecting clinically significant mutations in the chimeric BCR::ABL1 gene, such as T315I, M244V, Y253H, V299L, and F359V, as an alternative to NGS in certain cases. The system is based on AS-PCR and can use both cDNA and genomic DNA, facilitating the analysis of archived materials.
2. Results
We developed a system of primers for the rapid detection of the most frequent clinically significant mutations of the chimeric BCR::ABL1 gene conferring resistance to TKI therapy (Table 1). Primers for quantitative TaqMan PCR were selected by analyzing the ABL1 gene sequence in the vicinity of mutation points [18]. The last nucleotide at the 3′ end of each primer corresponded to a normal or mutant nucleotide in the ABL1 gene sequence. The penultimate nucleotide of the 3′ end of each primer was modified for greater specificity of the AS-PCR.

Table 1.
Primers and fluorescent probes used for AS-PCR detection of mutations in the chimeric BCR::ABL1 gene.
The sensitivity of the proposed system was evaluated by serial dilutions of the patient’s mutated DNA with the DNA of a healthy donor (Figure 1 and Figure 2). Figure 1 shows the amplification curves in the experiment with tenfold dilution from the first point using the DNA of a patient with the T315I mutation. The difference in threshold cycles between the amplification curves of the healthy donor DNA obtained in AS-PCR with primers for normal alleles (green curves in Figure 1, marked by arrow 1) and primers for detection of mutant alleles (red curves in Figure 1, marked by arrow 1) is equal to 15 cycles. Threshold cycles of DNA amplification curves with primer for mutation detection at a 105-fold dilution of patient DNA (blue curves in Figure 1, marked by dotted arrow 2.5) coincide with the threshold cycle of the corresponding curve of healthy donor DNA (red curves in Figure 1, marked by arrow 1). Thus, at an allele ratio of 0.003% or less, we may report the absence of mutant alleles. The threshold cycles of DNA amplification curves corresponding to a 104-fold dilution of the DNA of the patient with the mutation (blue curves in Figure 1, marked by the dotted arrow 2.4) reproducibly indicate the presence of the mutation. Consequently, at an allele ratio greater than 0.01%, we can confidently assert the presence of a mutation. Such sensitivity of T315I mutation detection is in full agreement with what is required for clinical diagnosis [19]. Similar results were obtained for other mutations.
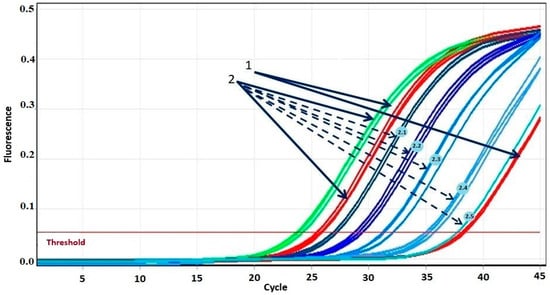
Figure 1.
Amplification curves of real-time PCR of DNA from a patient with T315I mutation in different dilutions. 1—DNA amplification curves of a healthy donor during real-time PCR. 2—DNA amplification curves of a patient with the T315I mutation during real-time PCR. Solid arrows indicate DNA amplification curves without dilution with normal (green) and mutant (red) primers. The color gradient from dark blue to light blue indicates five amplification curves of consecutive DNA dilutions during real-time PCR with mutant primers: dashed arrow 2.1 indicates the curve corresponding to dilution 1/3; 2.2—dilution 1/10; 2.3—dilution 1/100; 2.4—dilution 1/1000; 2.5—dilution 1/10,000.
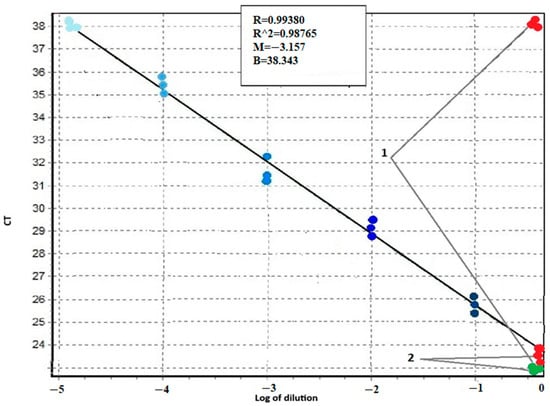
Figure 2.
Calibration curve for determining the sensitivity of AS-PCR with primers detecting the T315I mutation of the BCR::ABL1 chimeric gene. The X-axis is the decimal logarithm of the sample DNA dilution. On the Y-axis is the value of the CT threshold cycle. 1—threshold cycles for AS-PCR DNA of a healthy donor 2—threshold cycles for AS-PCR DNA of a patient with T315I mutation (green dots refer to normal alleles, red—to mutant alleles). The color gradient from dark blue to light blue indicates the points corresponding to the threshold cycles of PCR amplification with mutant primers for consecutive tenfold dilutions of DNA of the patient with the T315I mutation in the DNA of a healthy donor.
According to recent guidelines, NGS performed on cDNA is considered to be the gold standard for detecting mutations that confer resistance to TKI [15]. In our work we compared mutation data obtained on DNA from peripheral blood or bone marrow using the AS-PCR method and the NGS DNA and cDNA mutation data (Table 2).

Table 2.
Comparison of VAF obtained by AS-PCR and NGS mutation studies.
In the case of AS-PCR, the results for VAF estimation were close to those obtained by NGS in the cases with VAF > 5%. VAFs lower than 5% were NGS undetectable. AS-PCR, on the other hand, gave reliable results for VAF up to 0.01%.
3. Discussion
We compared the VAF values of mutations obtained using AS-PCR versus NGS after either nested or single-stage PCR on DNA or cDNA. The VAF calculated from NGS data was significantly higher than the VAF measured by AS-PCR. This result is expected and can be explained by the fact that, in the case of AS-PCR, the VAF represents the ratio of mutated alleles to total alleles of the ABL1 gene in a sample. Whereas in the case of cDNA analysis, VAF refers to the ratio of mutant and normal alleles in BCR::ABL1 chimeric transcripts only. The VAF values obtained from genomic DNA analysis using AS-PCR and NGS methods (Table 2) were similar. However, the sensitivity of NGS in our analysis was 5%, while AS-PCR allowed for the determination of VAFs up to 0.01%.
Thus, the sensitivity of the AS-PCR method, which is more than 10 times higher than the sensitivity of sequencing, allows obtaining reliable results in real time about the presence of mutations at a VAF of up to 0.01%. Similar results are obtained by the NGS cDNA method; however, it is more expensive, labor-intensive, and involves multiple steps—RNA isolation, reverse transcription, subsequent nested PCR, NGS, and subsequent computer processing of the results.
The AS-PCR method is also indispensable for analyzing archival material when the RNA is degraded. In addition, the high sensitivity of the method allows monitoring of mutation during TKI therapy of patients or when changing the drug or detecting a new mutant clone that has emerged [20].
Below we present examples of allele ratio monitoring for different mutations in the chimeric gene of three CML patients with TKI-1 treatment failure (Table 3, Table 4 and Table 5). The row with the maximum allele ratio and BCR::ABL1 transcript level is highlighted in bold.

Table 3.
Allele ratio monitoring of M244V and F359V BCR::ABL1 gene mutations for patient A.

Table 4.
Allele ratio monitoring of T315I BCR::ABL1 gene mutation for patient N.

Table 5.
Allele ratio monitoring of T315I BCR::ABL1 gene mutation for patient B.
The results of AS-PCR were confirmed by NGS; besides, it was performed less frequently.
4. Materials and Methods
4.1. Patients
DNA was isolated from peripheral blood and bone marrow cells of 57 patients treated at the National Medical Research Center for Hematology (Moscow, Russia) from 2022 to 2024. All patients had a diagnosis of Ph-positive CML and failure of 1st generation TKI therapy. BCR::ABL1 chimeric transcript levels were at least 1% in all patients. Informed consents from patients to participate in the study were obtained. The study was conducted in accordance with the Declaration of Helsinki and with the approval of the Institutional Ethics Committee (protocol code 185 of 27 February 2025).
Peripheral blood mononuclear cells were obtained after lysis of erythrocytes from 2 to 8 mL of peripheral blood or bone marrow [21]. DNA and RNA isolation from leukocytes was performed using the reagent kit (Interlabservice, Moscow, Russia). Reverse transcription of isolated RNA to obtain cDNA was performed using a reagent kit (Interlabservice, Russia).
Quantitation of BCR::ABL1 transcript level was performed using the Rotor-Gene amplifier (QIAGEN, Hilden, Germany) and the AmpliSense® Leukemia Quant M-bcr-FRT reagent kit (Interlabservice, Russia).
4.2. Next-Generation Sequencing
Missense mutations were measured in the BCR::ABL1 chimeric transcript and in the ABL1 gene (in cDNA and DNA, respectively). The target BCR::ABL1 region was amplified using either one-step PCR of sample DNA or “nested” PCR as described by van Dongen J. [22]. A panel of primers was created for the amplification of exons 4–9 of the ABL1 gene (Table 6). The conditions for PCR were as follows: preheating—94 °C 300 s; subsequent thermocycling—35 cycles: denaturation 94 °C 30 s, annealing 60 °C 30 s, elongation 72 °C 90 s; final elongation 72 °C 5 min. Quality control of amplification products in both variants was performed by electrophoresis in 2% agarose gel. The amplicons obtained were used to create sequencing libraries using the Nextera XT DNA Library Preparation Kit and Nextera XT Index v2 (Illumina, San Diego, CA, USA). Sequencing was performed on a MiSeq genetic analyzer (Illumina, USA), and bioinformatic analysis was performed using the open-source software Trimmomatic (version 0.39) [23], BWA (version 0.7.17-r1188) [24], SAMtools (version 1.10) [25], Vardict (version 1.8.2) [26], and Annovar (release date 2020-06-08) [27]. Sequencing results were compared with the reference sequence NM_005157. The probable pathogenicity of detected mutations was analyzed using the Franklin by Genoox online database [28]. In the case of cDNA sequencing of the chimeric BCR::ABL1 gene, the ratio of the number of reads with the mutation to the total number of reads determined the proportion of the mutated transcript. In the case of ABL1 gene DNA sequencing, the ratio of the number of reads with a mutation to the total number of reads determined the variant allele frequency (VAF).

Table 6.
Primers used for PCR amplification of exons 4–9 of the ABL1 gene.
4.3. AS-PCR
All primers and fluorescent TaqMan probes were synthesized by «Syntol™» (Moscow, Russia). The sequences are summarized in Table 1. To detect each target mutation, two almost identical PCR reactions were performed, differing only by one primer for amplification of the normal or mutant allele. The conditions for AS-PCR were as follows: preheating—94 °C 300 s; subsequent thermocycling—45 cycles: denaturation at 94 °C 20 s, annealing and elongation at 65 °C 50 s. The amount of each primer—10 pmol per reaction, each probe—5 pmol; reaction volume—25 μL. PCR buffer, MgCl2 dNTP, and Taq polymerase (Syntol, Moscow, Russia) were used according to the manufacturer’s instructions. AS-PCR was performed on a CFX96 C1000 Touch instrument (Bio-Rad Laboratories, Hercules, CA, USA).
4.4. Allele Ratio and VAF in AS-PCR Assays
Allele ratio (AR)—the ratio of the number of alleles carrying a mutation in the ABL1 gene to the number of ABL1 gene alleles without mutations was calculated using the formula: AR = (2CABL1:2CABL1mt) × 100%, where CABL1 is the PCR threshold cycle for the normal gene and CABL1mt is the threshold cycle for the ABL1 gene with a certain mutation.
ΔCT was the difference between the threshold cycle (Ct) value for the mutant allele and the Ct value for the normal allele.
Variant allele frequency (VAF)—i.e., the ratio of the proportion of alleles carrying the mutation in the ABL1 gene to all alleles of the ABL1 gene both with and without the mutation—was calculated using the formula: VAF = 100% × AR: (100% + AR), where AR is the allele ratio.
5. Conclusions
The developed diagnostic system based on allele-specific polymerase chain reaction with primers for detection of T315I, M244V, Y253H, V299L, and F359V mutations in the chimeric BCR::ABL1 is a sensitive, specific, and simple method. The sensitivity of AS-PCR allows the detection of leukemia clones with mutation levels up to 0.01%. This analysis is useful prior to choosing farther-targeted TKI drugs. The detection of low-percentage mutant tumor clones is also of great value, particularly when, against the background of TKI therapy, there is a selection of tumor clones with a mutation that is resistant to this therapy. This is especially relevant in the case of the panresistant T315I mutation, which can sometimes be detected even at the onset of CML in certain patients. Moreover, the abundance of this leukemia clone can increase or decrease, which is impossible to predict without continuous monitoring of the presence of mutations. It should also be noted that we are not proposing to replace sequencing with AS-PCR. In all PCR negative cases, samples will still be sent for NGS. However, in the majority of cases, critical information for making clinical decisions will be available faster and at a lower cost.
AS-PCR has another significant advantage over NGS: faster results, lower costs, and a simple research protocol. The AS-PCR method for diagnosing BCR::ABL1 mutations can be applied in any laboratory worldwide, as the reaction requires only allele-specific primers and probes, as well as the creation of optimal PCR conditions. We propose to use our diagnostic system for primary screening of resistant CML cases and for monitoring TKI treatment of patients. This system allows us to work with both DNA and RNA. It also provides the opportunity to work with archival material when RNA degrades.
Author Contributions
Conceptualization, I.F.; investigation, A.S. (Anastasia Skripkina), B.B., and E.S.; methodology, A.S. (Anastasia Skripkina) and I.F.; project administration, A.S. (Andrey Sudarikov); resources, E.C. and E.K.; supervision, A.T.; writing—original draft, A.S. (Anastasia Skripkina); writing—review and editing, A.S. (Andrey Sudarikov). All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of the National Medical Research Center for Hematology (protocol code 185, 27 February 2025).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Braun, T.P.; Eide, C.A.; Druker, B.J. Response and Resistance to BCR-ABL1-Targeted Therapies. Cancer Cell 2020, 37, 530–542. [Google Scholar] [CrossRef]
- Goranova-Marinova, V.; Linev, A.; Ivanov, H.J.; Zheljazkov, I.; Stoyanova, V.; Grudeva-Popova, Z. Clinical characteristics, disease evolution and survival in patients with chronic myeloid leukemia, BCR-ABL1 (+) and T315I mutation. Folia Med. 2021, 63, 670–675. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, S.; Kim, H.J.; Kim, Y.-K.; Kwak, J.-Y.; Yhim, H.-Y.; Kim, S.-H.; Do, Y.R.; Oh, S.; Lee, S.-E.; et al. Comparison of Frequency and Sensitivity of BCR-ABL1 Kinase Domain Mutations in Asian and White Patients With Imatinib-resistant Chronic-Phase Chronic Myeloid Leukemia. Clin. Lymphoma Myeloma Leuk. 2018, 18, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Chahardouli, B.; Zaker, F.; Mousavi, S.A.; Kazemi, A.; Ostadali, M.; Nadali, F.; Rostami, S.; Alimoghaddam, K.; Ghavamzade, A. Evaluation of T315I mutation frequency in chronic myeloid leukemia patients after imatinib resistance. Hematology 2013, 18, 158–162. [Google Scholar] [CrossRef]
- Sun, J.; Hu, R.; Han, M.; Tan, Y.; Xie, M.; Gao, S.; Hu, J.F. Mechanisms underlying therapeutic resistance of tyrosine kinase inhibitors in chronic myeloid leukemia. Int. J. Biol. Sci. 2024, 20, 175–181. [Google Scholar] [CrossRef]
- Jabbour, E.; Parikh, S.A.; Kantarjian, H.; Cortes, J. Chronic myeloid leukemia: Mechanisms of resistance and treatment. Hematol. Oncol. Clin. N. Am. 2011, 25, 981–995. [Google Scholar] [CrossRef]
- Cortes, J.; Jabbour, E.; Kantarjian, H.; Yin, C.C.; Shan, J.; O’Brien, S.; Garcia-Manero, G.; Giles, F.; Breeden, M.; Reeves, N.; et al. Dynamics of BCR-ABL kinase domain mutations in chronic myeloid leukemia after sequential treatment with multiple tyrosine kinase inhibitors. Blood 2007, 110, 4005–4011. [Google Scholar] [CrossRef] [PubMed]
- Soverini, S. Resistance mutations in CML and how we approach them. Hematol. Am. Soc. Hematol. Educ. Program. 2023, 2023, 469–475. [Google Scholar] [CrossRef]
- Turkina, A.G.; Lomaia, E.G.; Morozova, E.V.; Vinogradova, O.Y.; Mitina, T.A.; Shatokhin, Y.V.; Ovsyannikova, E.G.; Vlasova, Y.Y.; Kulikov, S.M.; Chelysheva, E.Y. Evolution of therapeutic approaches in patients with chronic myeloid leukemia and T315I mutation. Oncohematology 2024, 19, 93–107. [Google Scholar] [CrossRef]
- Soverini, S.; Branford, S.; Nicolini, F.E.; Talpaz, M.; Deininger, M.W.N.; Martinelli, G.; Müller, M.C.; Radich, J.P.; Shah, N.P. Implications of BCR-ABL1 kinase domain-mediated resistance in chronic myeloid leukemia. Leuk. Res. 2014, 38, 10–20. [Google Scholar] [CrossRef]
- Kockerols, C.; Valk, P.J.M.; Blijlevens, N.M.A.; Cornelissen, J.J.; Dinmohamed, A.G.; Geelen, I.; Hoogendoorn, M.; Janssen, J.J.W.M.; Daenen, L.G.M.; Reijden, B.A.v.d. BCR::ABL1 kinase domain mutation testing and clinical outcome in a nationwide chronic myeloid leukemia patient population. Eur. J. Haematol. 2023, 111, 938–945. [Google Scholar] [CrossRef]
- Wang, S.; Qiao, C.; Zhu, Y.; Shen, W.; He, G.M.; Li, J. The third-time chronic myeloid leukemia in lymphoblastic crisis with ABL1 kinase mutation induced by decitabine, dexamethason combined with nilotinib and dasatinib. J. Transl. Int. Med. 2016, 4, 182–184. [Google Scholar] [CrossRef] [PubMed]
- Chelysheva, E.Y.; Shukhov, O.A.; Lazareva, O.V.; Turkina, A.V.; Turkina, A.G. Mutations of the kinase domain of the BCR-ABL gene in chronic myeloleukemia. Clin. Oncohematol. 2012, 1, 13–21. [Google Scholar]
- Zabriskie, M.S.; Eide, C.A.; Tantravahi, S.K.; Vellore, N.A.; Estrada, J.; Nicolini, F.E.; Khoury, H.J.; Larson, R.A.; Konopleva, M.; Cortes, J.E.; et al. BCR::ABL1 compound mutations combining key kinase domain positions confer clinical resistance to ponatinib in Ph chromosome-positive leukemia. Cancer Cell 2014, 26, 428–442. [Google Scholar] [CrossRef] [PubMed]
- Soverini, S.; Bernardi, S.; Galimberti, S. Molecular Testing in CML between Old and New Methods: Are We at a Turning Point? J. Clin. Med. 2020, 9, 3865. [Google Scholar] [CrossRef]
- Polivkova, V.; Benesova, A.; Zizkova, H.; Koblihova, J.; Curik, N.; Motlova, E.; Klamova, H.; Salek, C.; Polakova, K.M. Sensitivity and reliability of DNA-based mutation analysis by allele-specific digital PCR to follow resistant BCR-ABL1-positive cells. Leukemia 2021, 35, 2419–2423. [Google Scholar] [CrossRef]
- Branford, S.; Rudzki, Z.; Parkinson, I.; Grigg, A.; Taylor, K.; Seymour, J.F.; Durrant, S.; Browett, P.; Anthony, P.; Arthur, C. Real-time quantitative PCR analysis can be used as a primary screen to identify patients with CML treated with imatinib who have BCR-ABL kinase domain mutations. Blood 2004, 104, 2926–2932. [Google Scholar] [CrossRef]
- Dyer, S.C.; Austine-Orimoloye, O.; Azov, A.G.; Barba, M.; Barnes, I.; Barrera-Enriquez, V.P.; Becker, A.; Bennett, R.; Beracochea, M.; Berry, A.; et al. Ensembl. Nucleic Acids Res. 2025, 53, D948–D957. [Google Scholar] [CrossRef]
- Arechavaleta, G.M.; Scholl, V.; Pérez, V.; Bittencourt, R.; Moellmann, A.; Hassan, R.; Seuánez, H.N.; Dobbin, J.; Martinez, L.; Renault, I.Z.; et al. Rapid and sensitive allele-specific (AS)-RT-PCR assay for detection of T315I mutation in chronic myeloid leukemia patients treated with tyrosine-kinase inhibitors. Clin. Exp. Med. 2011, 11, 55–59. [Google Scholar] [CrossRef]
- Innes, A.J.; Hayden, C.; Orovboni, V.; Bittencourt, R.; Moellmann, A.; Hassan, R.; Seuánez, H.N.; Dobbin, J.; Martinez, L.; Renault, I.Z. Impact of BCR::ABL1 single nucleotide variants on asciminib efficacy. Leukemia 2024, 38, 2443–2455. [Google Scholar] [CrossRef]
- Sidorova, Y.V.; Sorokina, T.V.; Biderman, B.V.; Nikulina, E.E.; Kisilichina, D.G.; Naumova, E.V.; Pochtar, M.E.; Lugovskaya, S.A.; Ivanova, V.L.; Kovaleva, L.G.; et al. Determination of minimal residual disease in patients with B-cell chronic lympholeukemia by patient-specific PCR. Klin. Lab. Diagn. 2011, 12, 22–35. [Google Scholar]
- van Dongen, J.; Macintyre, E.; Gabert, J.; Delabesse, E.; Rossi, V.; Saglio, G.; Gottardi, E.; Rambaldi, A.; Dotti, G.; Griesinger, F.; et al. Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease. Report of the BIOMED-1 Concerted Action: Investigation of minimal residual disease in acute leukemia. Leukemia 1999, 13, 1901–1928. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Lai, Z.; Markovets, A.; Ahdesmaki, M.; Chapman, B.; Hofmann, O.; McEwen, R.; Johnson, J.; Dougherty, B.; Barrett, J.C.; Dry, J.R. VarDict: A novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016, 44, e108. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Franklin. Available online: https://franklin.genoox.com/clinical-db/home (accessed on 4 September 2024).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).