A Quinoxaline 1,4-Dioxide Activates DNA Repair Systems in Mycobacterium smegmatis: A Transcriptomic Study
Abstract
1. Introduction
2. Results
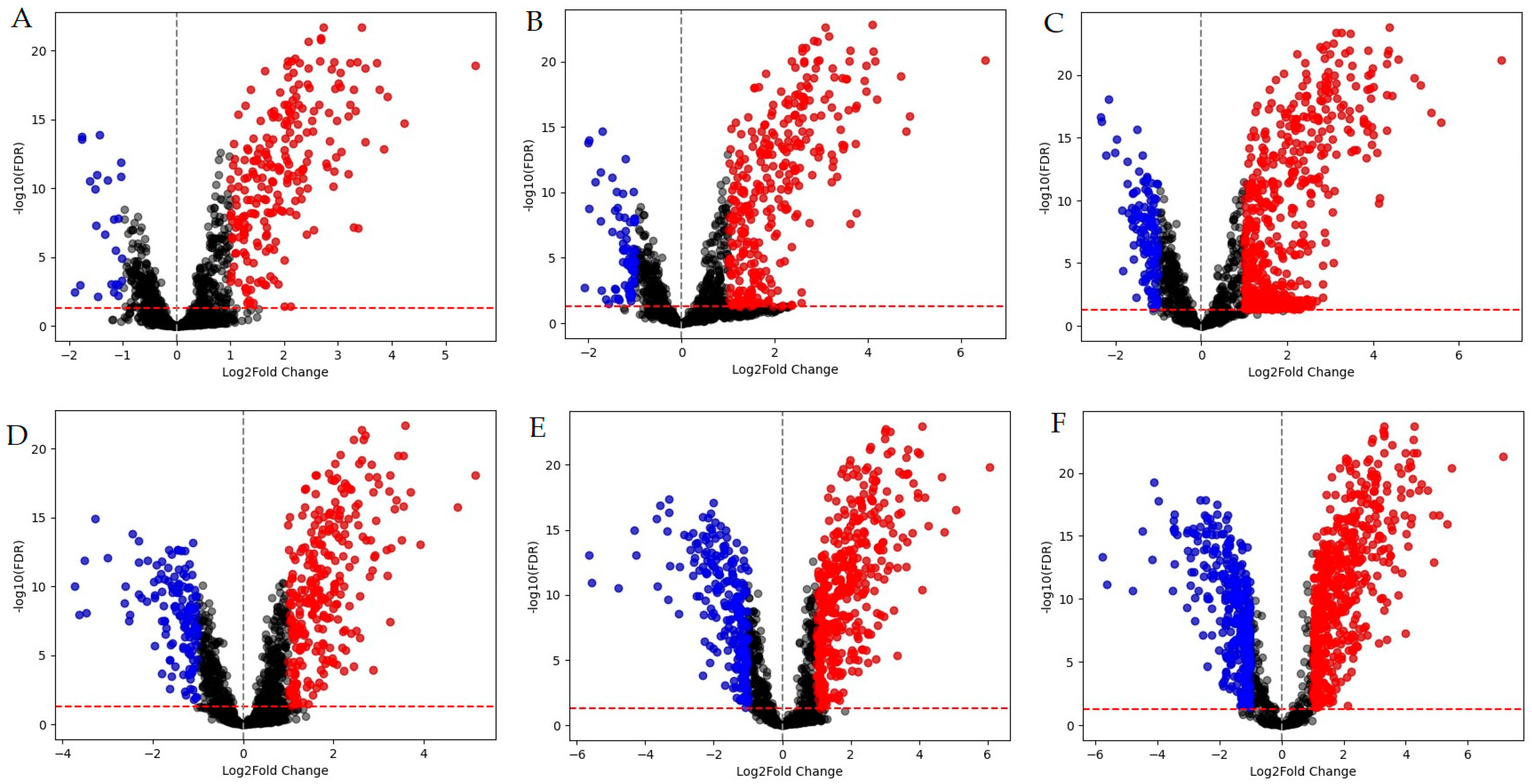
2.1. Whole-Transcriptome Analysis
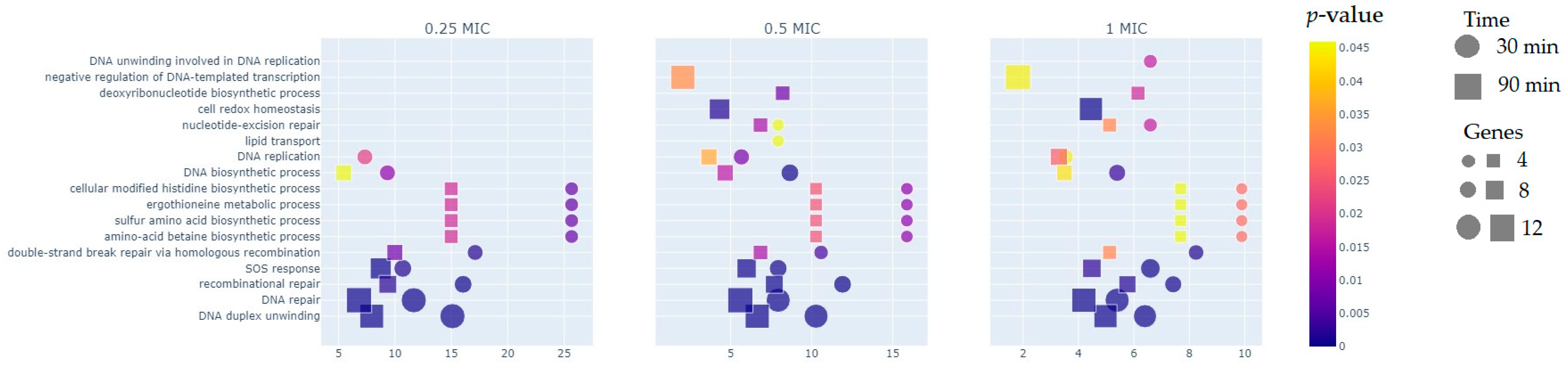
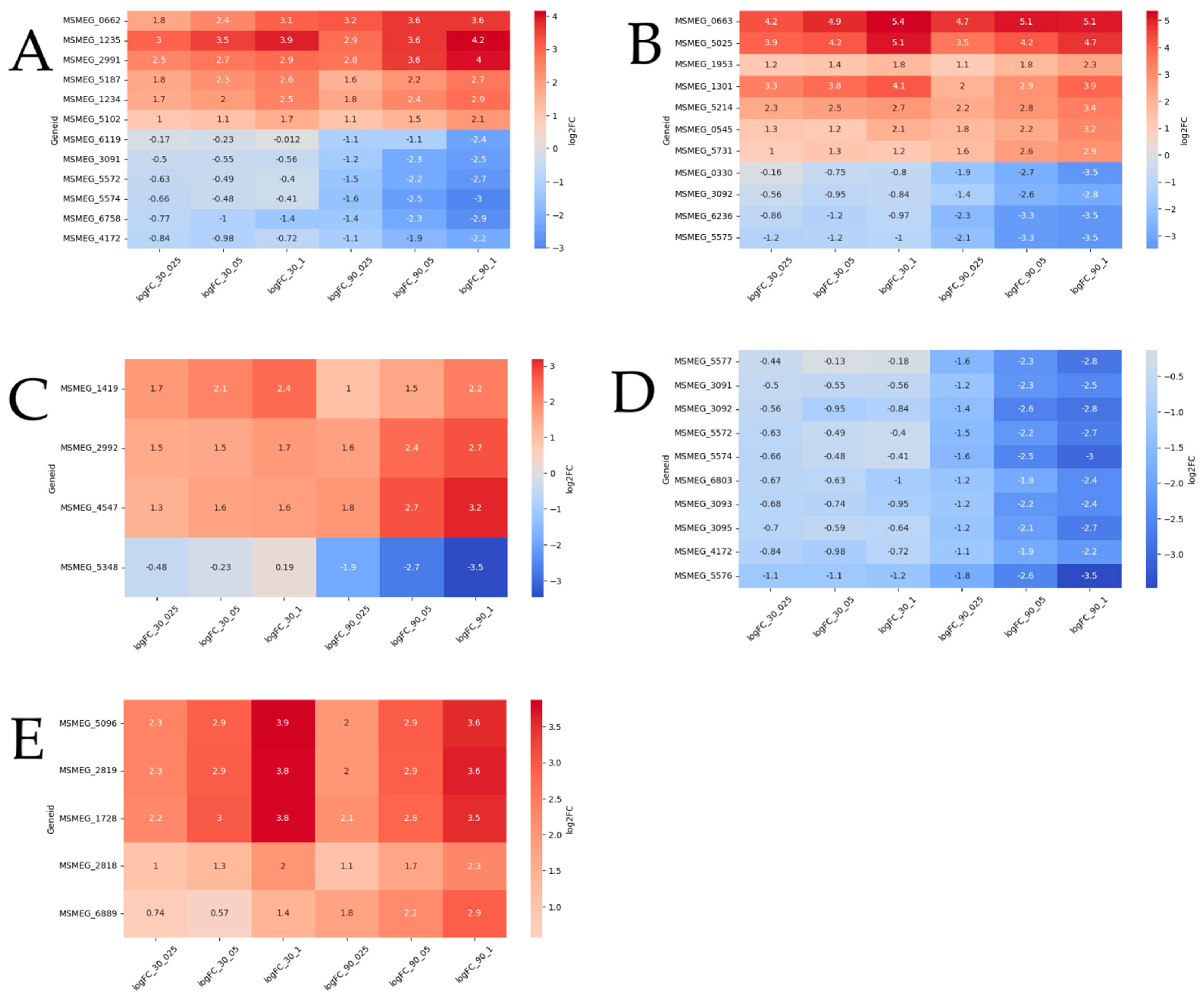
2.2. Functional Groups of Differentially Expressed Genes
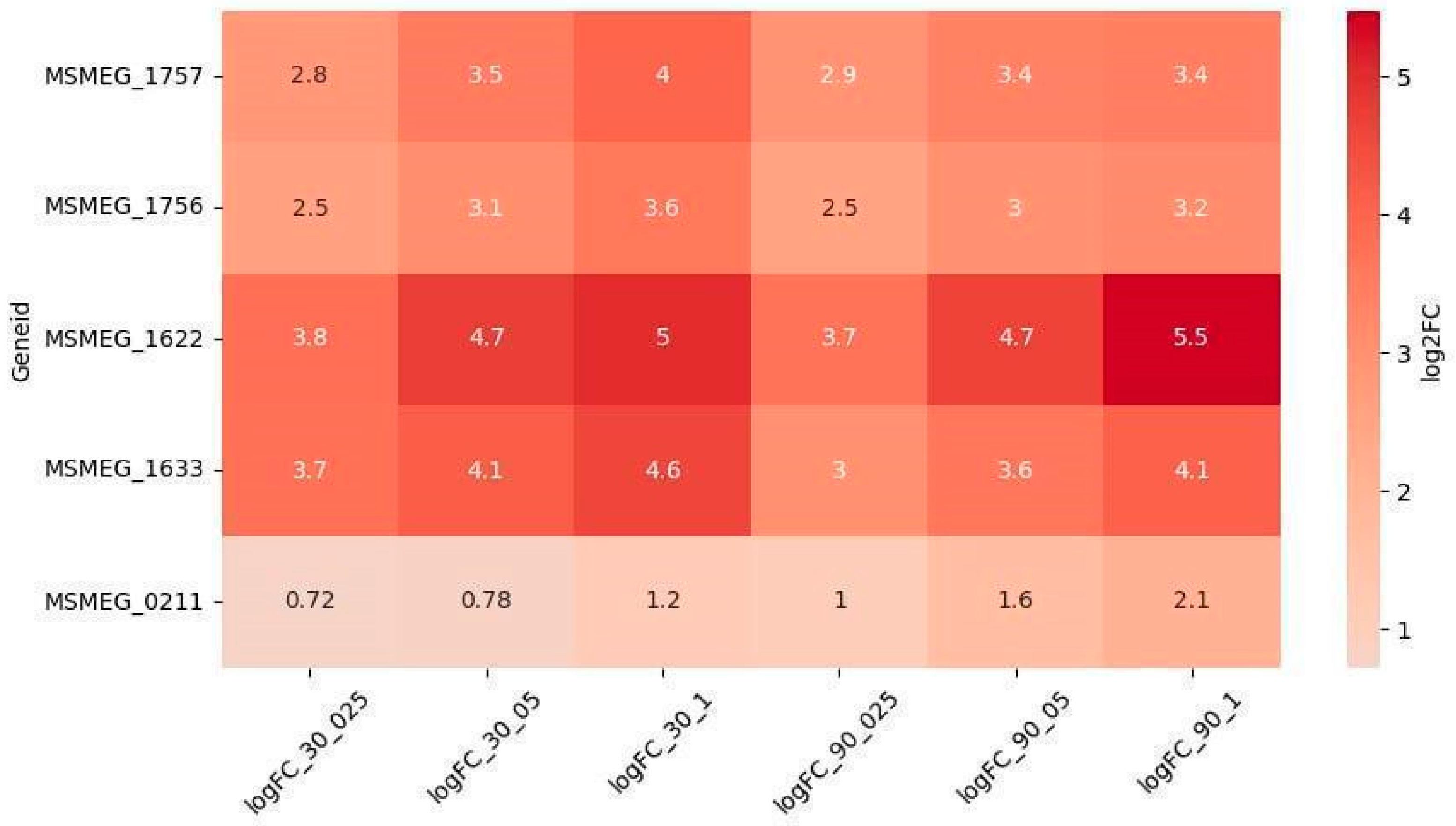
2.2.1. Reparation Genes
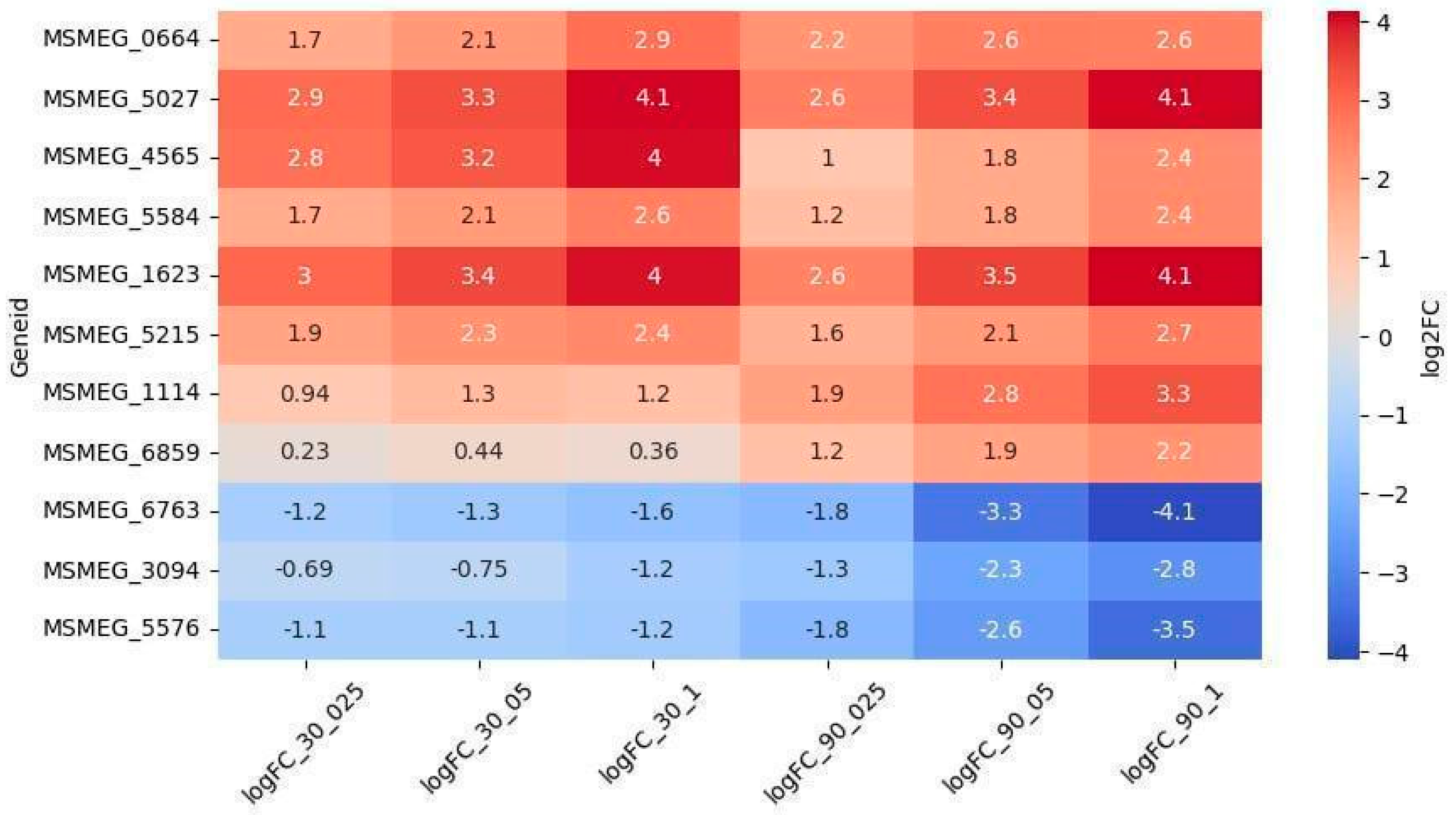
2.2.2. Oxidoreductases
2.2.3. Other Genes
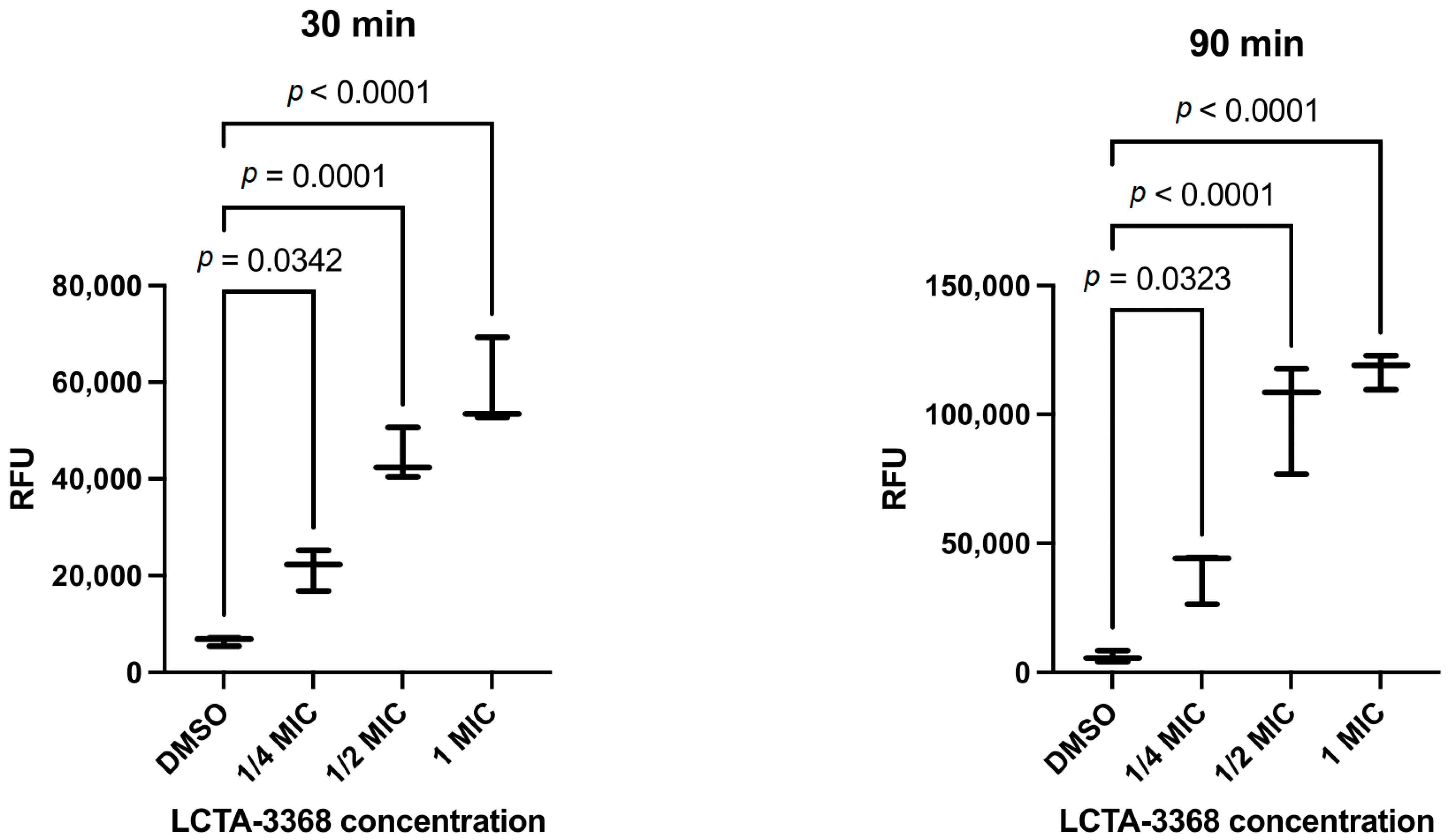
2.2.4. LCTA-3368 Induces Reactive Oxygen Species Formation in M. smegmatis
3. Discussion
4. Materials and Methods
4.1. Microbial Cultures and Growth Conditions
4.2. Dichloro-Dihydro-Fluorescein Diacetate (H2DCF-DA) Assay
4.3. Total RNA Extraction
4.4. Library Preparation and RNA Sequencing
4.5. Bioinformatics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Global Tuberculosis Report 2023. Available online: https://www.who.int/teams/global-programme-on-tuberculosis-and-lung-health/tb-reports/global-tuberculosis-report-2023 (accessed on 8 April 2025).
- Cheng, G.; Sa, W.; Cao, C.; Guo, L.; Hao, H.; Liu, Z.; Wang, X.; Yuan, Z. Quinoxaline 1,4-Di-N-Oxides: Biological Activities and Mechanisms of Actions. Front. Pharmacol. 2016, 7, 64. [Google Scholar] [CrossRef] [PubMed]
- Villar, R.; Vicente, E.; Solano, B.; Pérez-Silanes, S.; Aldana, I.; Maddry, J.A.; Lenaerts, A.J.; Franzblau, S.G.; Cho, S.-H.; Monge, A.; et al. In Vitro and in Vivo Antimycobacterial Activities of Ketone and Amide Derivatives of Quinoxaline 1,4-Di-N-Oxide. J. Antimicrob. Chemother. 2008, 62, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, S.; Sechi, L.A.; Molicotti, P.; Cannas, S.; Bua, A.; Deriu, A.; Carta, A.; Paglietti, G. In Vitro Activity of New Quinoxalin 1,4-Dioxide Derivatives against Strains of Mycobacterium Tuberculosis and Other Mycobacteria. Int. J. Antimicrob. Agents 2005, 25, 179–181. [Google Scholar] [CrossRef]
- Ortega, M.A.; Montoya, M.E.; Jaso, A.; Zarranz, B.; Tirapu, I.; Aldana, I.; Monge, A. Antimycobacterial Activity of New Quinoxaline-2-Carbonitrile and Quinoxaline-2-Carbonitrile 1,4-Di-N-Oxide Derivatives. Pharmazie 2001, 56, 205–207. [Google Scholar] [CrossRef] [PubMed]
- Crawford, P.W.; Scamehorn, R.G.; Hollstein, U.; Ryan, M.D.; Kovacic, P. Cyclic Voltammetry of Phenazines and Quinoxalines Including Mono- and Di-N-Oxides. Relation to Structure and Antimicrobial Activity. Chem. Biol. Interact. 1986, 60, 67–84. [Google Scholar] [CrossRef]
- Chowdhury, G.; Kotandeniya, D.; Daniels, J.S.; Barnes, C.L.; Gates, K.S. Enzyme-Activated, Hypoxia-Selective DNA Damage by 3-Amino-2-Quinoxalinecarbonitrile 1,4-Di-N-Oxide. Chem. Res. Toxicol. 2004, 17, 1399–1405. [Google Scholar] [CrossRef]
- Frolova, S.G.; Vatlin, A.A.; Maslov, D.A.; Yusuf, B.; Buravchenko, G.I.; Bekker, O.B.; Klimina, K.M.; Smirnova, S.V.; Shnakhova, L.M.; Malyants, I.K.; et al. Novel Derivatives of Quinoxaline-2-Carboxylic Acid 1,4-Dioxides as Antimycobacterial Agents: Mechanistic Studies and Therapeutic Potential. Pharmaceuticals 2023, 16, 1565. [Google Scholar] [CrossRef]
- Grzelak, E.M.; Choules, M.P.; Gao, W.; Cai, G.; Wan, B.; Wang, Y.; McAlpine, J.B.; Cheng, J.; Jin, Y.; Lee, H.; et al. Strategies in Anti-Mycobacterium Tuberculosis Drug Discovery Based on Phenotypic Screening. J. Antibiot. 2019, 72, 719–728. [Google Scholar] [CrossRef]
- Cooper, C.B. Development of Mycobacterium Tuberculosis Whole Cell Screening Hits as Potential Antituberculosis Agents. J. Med. Chem. 2013, 56, 7755–7760. [Google Scholar] [CrossRef]
- Junnotula, V.; Sarkar, U.; Sinha, S.; Gates, K.S. Initiation of DNA Strand Cleavage by 1,2,4-Benzotriazine 1,4-Dioxide Antitumor Agents: Mechanistic Insight from Studies of 3-Methyl-1,2,4-Benzotriazine 1,4-Dioxide. J. Am. Chem. Soc. 2009, 131, 1015–1024. [Google Scholar] [CrossRef]
- Briffotaux, J.; Liu, S.; Gicquel, B. Genome-Wide Transcriptional Responses of Mycobacterium to Antibiotics. Front. Microbiol. 2019, 10, 249. [Google Scholar] [CrossRef]
- Vatlin, A.A.; Shitikov, E.A.; Shahbaaz, M.; Bespiatykh, D.A.; Klimina, K.M.; Christoffels, A.; Danilenko, V.N.; Maslov, D.A. Transcriptomic Profile of Mycobacterium Smegmatis in Response to an Imidazo[1,2-b][1,2,4,5]Tetrazine Reveals Its Possible Impact on Iron Metabolism. Front. Microbiol. 2021, 12, 724042. [Google Scholar] [CrossRef]
- Xu, F.; Cheng, G.; Hao, H.; Wang, Y.; Wang, X.; Chen, D.; Peng, D.; Liu, Z.; Yuan, Z.; Dai, M. Mechanisms of Antibacterial Action of Quinoxaline 1,4-Di-N-Oxides against Clostridium Perfringens and Brachyspira Hyodysenteriae. Front. Microbiol. 2016, 7, 1948. [Google Scholar] [CrossRef]
- Suter, W.; Rosselet, A.; Knüsel, F. Mode of Action of Quindoxin and Substituted Quinoxaline-Di-N-Oxides on Escherichia Coli. Antimicrob. Agents Chemother. 1978, 13, 770–783. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.L.; Liu, R.H. Cellular Antioxidant Activity (CAA) Assay for Assessing Antioxidants, Foods, and Dietary Supplements. J. Agric. Food Chem. 2007, 55, 8896–8907. [Google Scholar] [CrossRef] [PubMed]
- Full Article: Bactericidal Efficacy of UV Activated TiO2 Nanoparticles Against Gram-Positive and Gram-Negative Bacteria on Suspension. Available online: https://www.tandfonline.com/doi/full/10.1080/19476337.2019.1590461 (accessed on 8 April 2025).
- Wang, X.; Zhang, H.; Huang, L.; Pan, Y.; Li, J.; Chen, D.; Cheng, G.; Hao, H.; Tao, Y.; Liu, Z.; et al. Deoxidation Rates Play a Critical Role in DNA Damage Mediated by Important Synthetic Drugs, Quinoxaline 1,4-Dioxides. Chem. Res. Toxicol. 2015, 28, 470–481. [Google Scholar] [CrossRef] [PubMed]
- An, H.; Li, Y.; Li, Y.; Gong, S.; Zhu, Y.; Li, X.; Zhou, S.; Wu, Y. Advances in Metabolism and Metabolic Toxicology of Quinoxaline 1,4-Di-N-Oxides. Chem. Res. Toxicol. 2024, 37, 528–539. [Google Scholar] [CrossRef]
- Gupta, R.; Barkan, D.; Redelman-Sidi, G.; Shuman, S.; Glickman, M.S. Mycobacteria Exploit Three Genetically Distinct DNA Double-Strand Break Repair Pathways. Mol. Microbiol. 2011, 79, 316–330. [Google Scholar] [CrossRef]
- Sharma, A.; Nair, D.T. Cloning, Expression, Purification, Crystallization and Preliminary Crystallographic Analysis of MsDpo4: A Y-Family DNA Polymerase from Mycobacterium Smegmatis. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011, 67, 812–816. [Google Scholar] [CrossRef]
- Miggiano, R.; Morrone, C.; Rossi, F.; Rizzi, M. Targeting Genome Integrity in Mycobacterium Tuberculosis: From Nucleotide Synthesis to DNA Replication and Repair. Molecules 2020, 25, 1205. [Google Scholar] [CrossRef]
- Dos Vultos, T.; Mestre, O.; Tonjum, T.; Gicquel, B. DNA Repair in Mycobacterium Tuberculosis Revisited. FEMS Microbiol. Rev. 2009, 33, 471–487. [Google Scholar] [CrossRef] [PubMed]
- Khanam, T.; Rai, N.; Ramachandran, R. Mycobacterium Tuberculosis Class II Apurinic/Apyrimidinic-Endonuclease/3’-5’ Exonuclease III Exhibits DNA Regulated Modes of Interaction with the Sliding DNA β-Clamp. Mol. Microbiol. 2015, 98, 46–68. [Google Scholar] [CrossRef]
- Chen, X.; Bradley, N.P.; Lu, W.; Wahl, K.L.; Zhang, M.; Yuan, H.; Hou, X.-F.; Eichman, B.F.; Tang, G.-L. Base Excision Repair System Targeting DNA Adducts of Trioxacarcin/LL-D49194 Antibiotics for Self-Resistance. Nucleic Acids Res. 2022, 50, 2417–2430. [Google Scholar] [CrossRef]
- Nätt, D.; Thorsell, A. Stress-Induced Transposon Reactivation: A Mediator or an Estimator of Allostatic Load? Environ. Epigenetics 2016, 2, dvw015. [Google Scholar] [CrossRef] [PubMed]
- Frolova, S.G.; Klimina, K.M.; Kumar, R.; Vatlin, A.A.; Salunke, D.B.; Kendrekar, P.; Danilenko, V.N.; Maslov, D.A. Identification of Mutations Conferring Tryptanthrin Resistance to Mycobacterium Smegmatis. Antibiotics 2020, 10, 6. [Google Scholar] [CrossRef] [PubMed]
- Ganley, B.; Chowdhury, G.; Bhansali, J.; Daniels, J.S.; Gates, K.S. Redox-Activated, Hypoxia-Selective DNA Cleavage by Quinoxaline 1,4-Di-N-Oxide. Bioorganic Med. Chem. 2001, 9, 2395–2401. [Google Scholar] [CrossRef]
- Kumar, A.; Farhana, A.; Guidry, L.; Saini, V.; Hondalus, M.; Steyn, A.J.C. Redox Homeostasis in Mycobacteria: The Key to Tuberculosis Control? Expert. Rev. Mol. Med. 2011, 13, e39. [Google Scholar] [CrossRef]
- Greening, C.; Jirapanjawat, T.; Afroze, S.; Ney, B.; Scott, C.; Pandey, G.; Lee, B.M.; Russell, R.J.; Jackson, C.J.; Oakeshott, J.G.; et al. Mycobacterial F420H2-Dependent Reductases Promiscuously Reduce Diverse Compounds through a Common Mechanism. Front. Microbiol. 2017, 8, 1000. [Google Scholar] [CrossRef]
- Gurumurthy, M.; Rao, M.; Mukherjee, T.; Rao, S.P.S.; Boshoff, H.I.; Dick, T.; Barry, C.E.; Manjunatha, U.H. A Novel F(420) -Dependent Anti-Oxidant Mechanism Protects Mycobacterium Tuberculosis against Oxidative Stress and Bactericidal Agents. Mol. Microbiol. 2013, 87, 744–755. [Google Scholar] [CrossRef]
- Vatlin, A.A.; Frolova, S.G.; Bekker, O.B.; Danilenko, V.N. Studying the mechanism of action of new derivatives of quinoxalin-1,4-dioxide on the model organism Mycobacterium smegmatis. RUDN J. Ecol. Life Saf. 2024, 32, 41–50. [Google Scholar] [CrossRef]
- Logsdon, M.M.; Aldridge, B.B. Stable Regulation of Cell Cycle Events in Mycobacteria: Insights From Inherently Heterogeneous Bacterial Populations. Front. Microbiol. 2018, 9, 514. [Google Scholar] [CrossRef] [PubMed]
- Babraham Bioinformatics—FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 8 April 2025).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Conesa, A.; García-Alcalde, F. Qualimap 2: Advanced Multi-Sample Quality Control for High-Throughput Sequencing Data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/index.html (accessed on 8 April 2025).
- Python 3.13 Documentation—DevDocs. Available online: https://devdocs.io/python/ (accessed on 8 April 2025).
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array Programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef]
- GOpiscator 0.1.5. Available online: https://pypi.org/project/gopiscator/ (accessed on 8 April 2025).
- GraphPad. Available online: https://www.graphpad.com/ (accessed on 8 April 2025).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bekker, O.B.; Galanova, O.O.; Vatlin, A.A.; Frolova, S.G.; Shitikov, E.A.; Bespiatykh, D.A.; Klimina, K.M.; Veselovsky, V.A.; Ilyasov, R.A.; Smirnova, S.V.; et al. A Quinoxaline 1,4-Dioxide Activates DNA Repair Systems in Mycobacterium smegmatis: A Transcriptomic Study. Int. J. Mol. Sci. 2025, 26, 3689. https://doi.org/10.3390/ijms26083689
Bekker OB, Galanova OO, Vatlin AA, Frolova SG, Shitikov EA, Bespiatykh DA, Klimina KM, Veselovsky VA, Ilyasov RA, Smirnova SV, et al. A Quinoxaline 1,4-Dioxide Activates DNA Repair Systems in Mycobacterium smegmatis: A Transcriptomic Study. International Journal of Molecular Sciences. 2025; 26(8):3689. https://doi.org/10.3390/ijms26083689
Chicago/Turabian StyleBekker, Olga B., Olesya O. Galanova, Aleksey A. Vatlin, Svetlana G. Frolova, Egor A. Shitikov, Dmitry A. Bespiatykh, Ksenia M. Klimina, Vladimir A. Veselovsky, Rustem A. Ilyasov, Svetlana V. Smirnova, and et al. 2025. "A Quinoxaline 1,4-Dioxide Activates DNA Repair Systems in Mycobacterium smegmatis: A Transcriptomic Study" International Journal of Molecular Sciences 26, no. 8: 3689. https://doi.org/10.3390/ijms26083689
APA StyleBekker, O. B., Galanova, O. O., Vatlin, A. A., Frolova, S. G., Shitikov, E. A., Bespiatykh, D. A., Klimina, K. M., Veselovsky, V. A., Ilyasov, R. A., Smirnova, S. V., Reznikova, D. A., Kochetkov, N. I., Maslov, D. A., & Danilenko, V. N. (2025). A Quinoxaline 1,4-Dioxide Activates DNA Repair Systems in Mycobacterium smegmatis: A Transcriptomic Study. International Journal of Molecular Sciences, 26(8), 3689. https://doi.org/10.3390/ijms26083689






