Evaluation of Thrombomodulin, hsa-miR-18a-5p, and hsa-miR-18b-5p as Potential Prognostic Biomarkers in Uterine Corpus Endometrial Carcinoma
Abstract
1. Introduction
2. Results
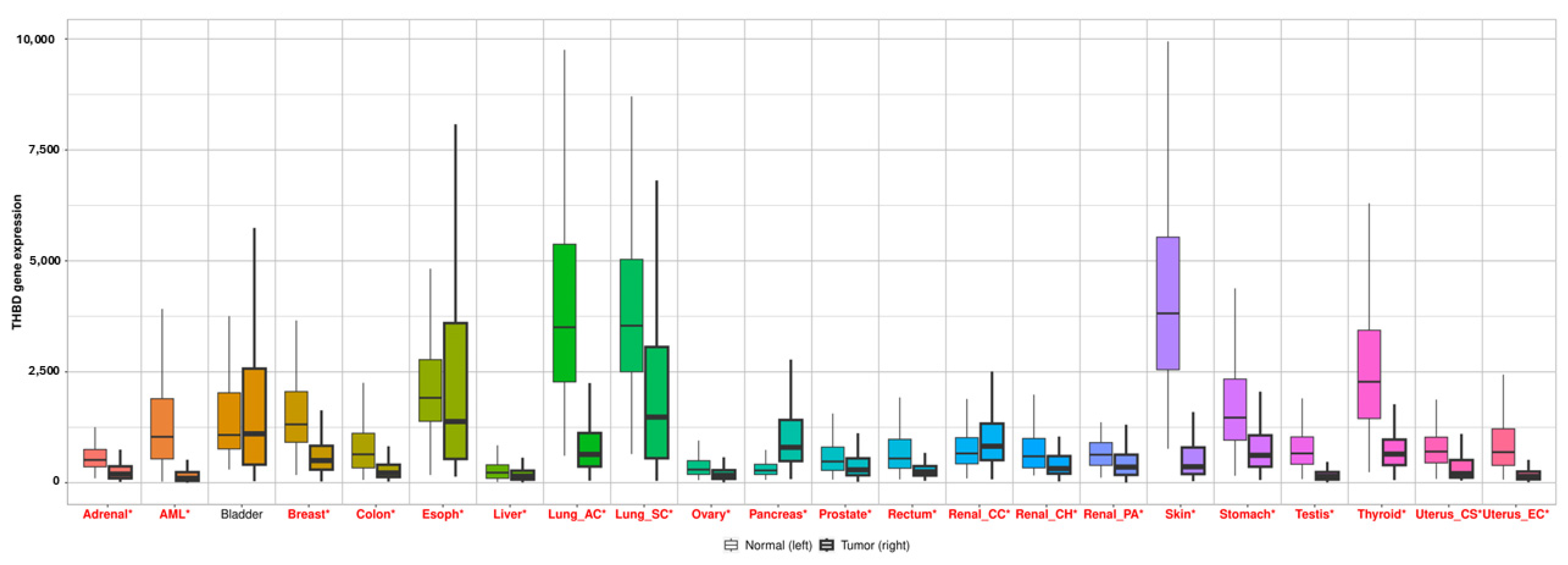
2.1. The Expression Profile of THBD, hsa-miR-18a-5p and hsa-miR-18b-5p
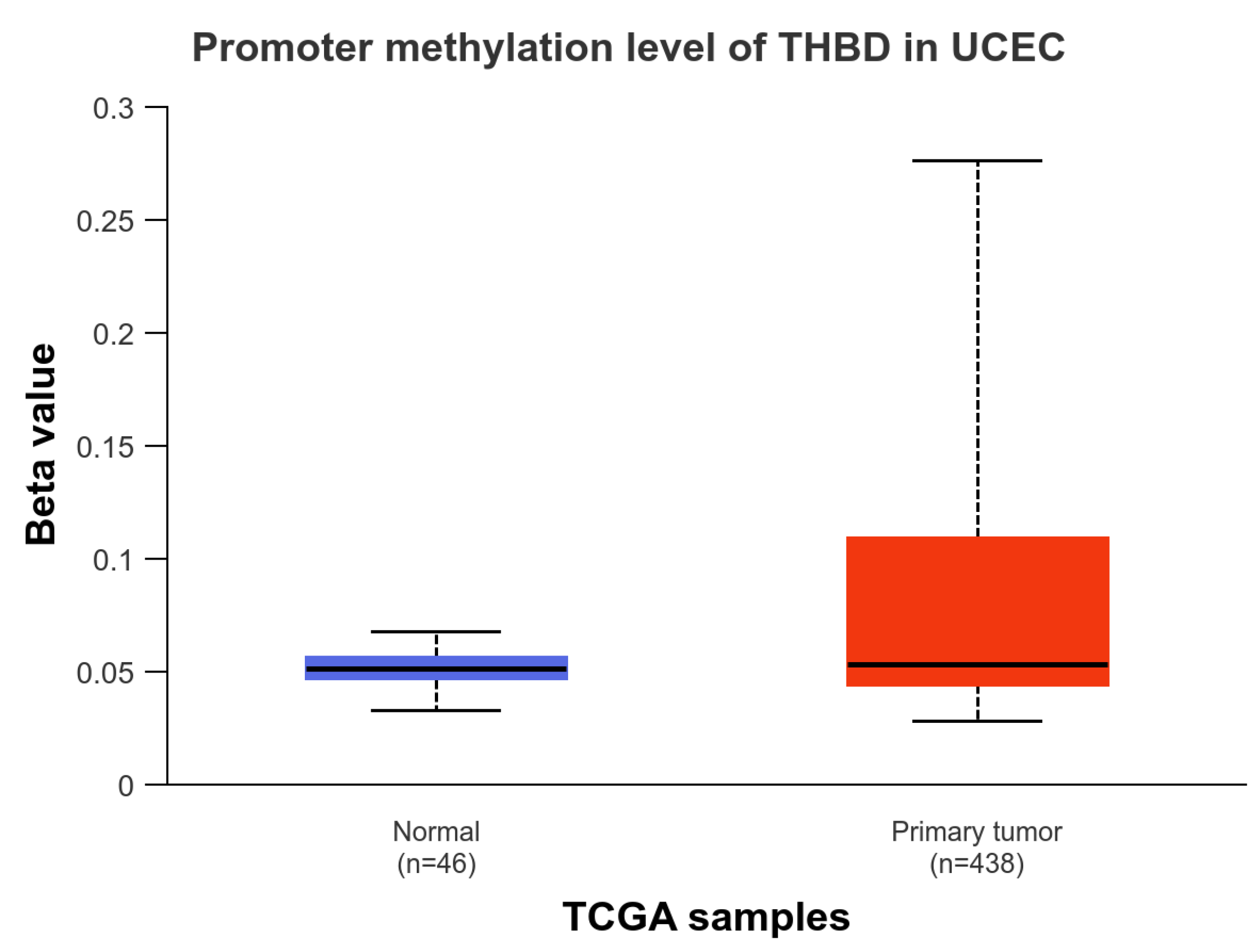
2.2. The Promotor Methylation Status of THBD
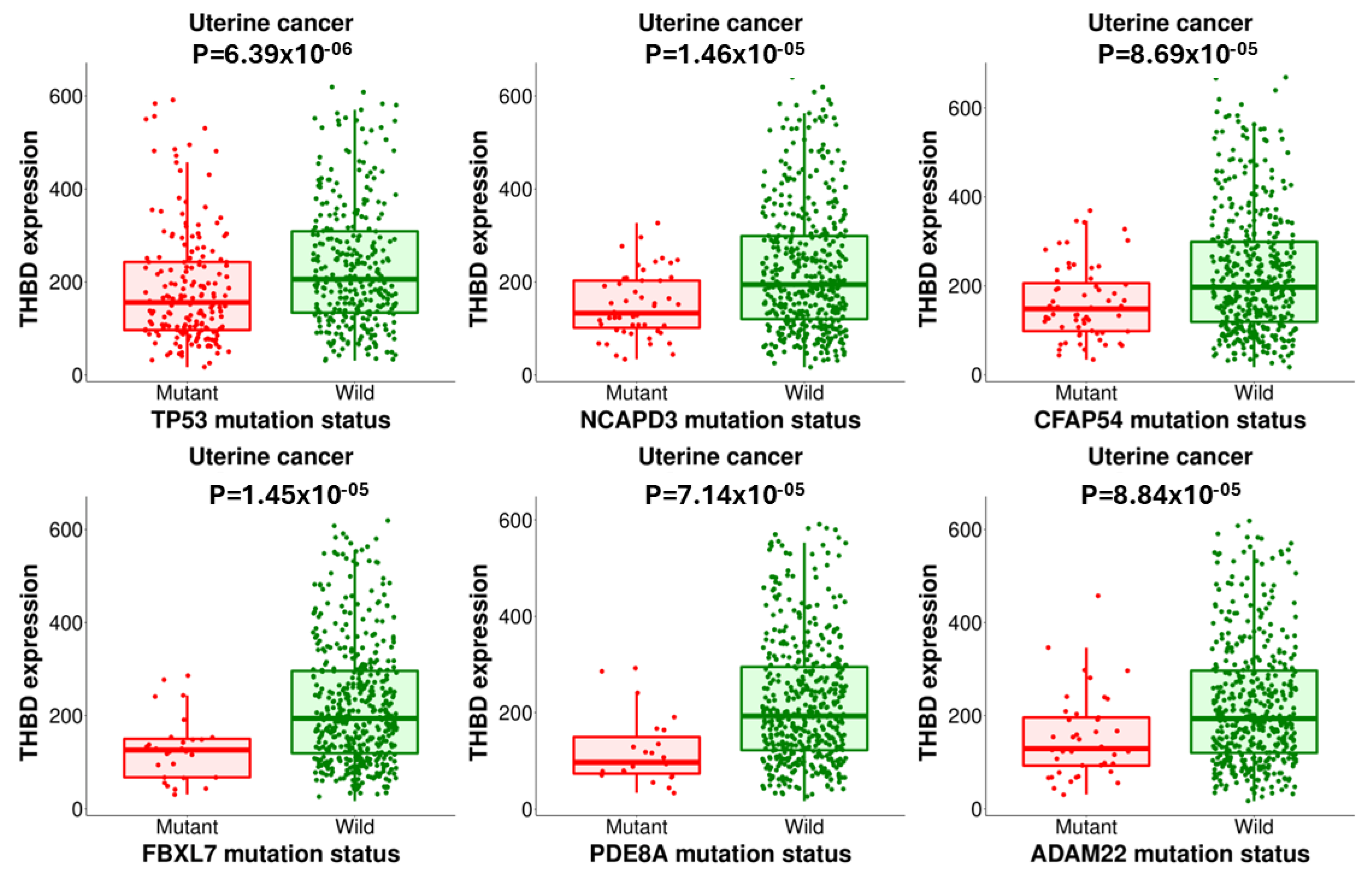
2.3. The TNMplot and muTarget Analysis of THBD
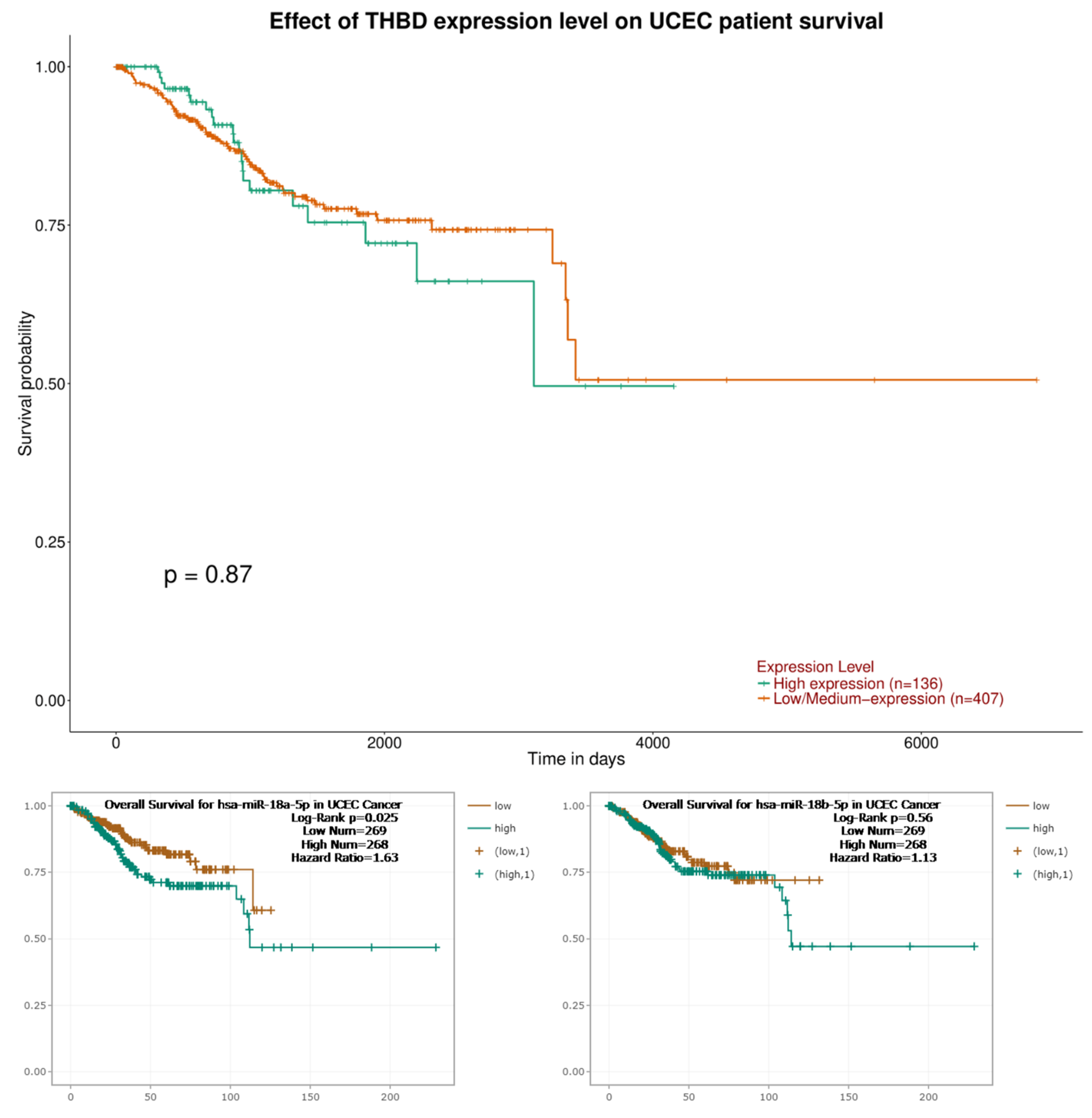
2.4. The Survival Analysis Results of THBD, hsa-miR-18a-5p and hsa-miR-18b-5p
2.5. The Gene–Gene Interaction Results
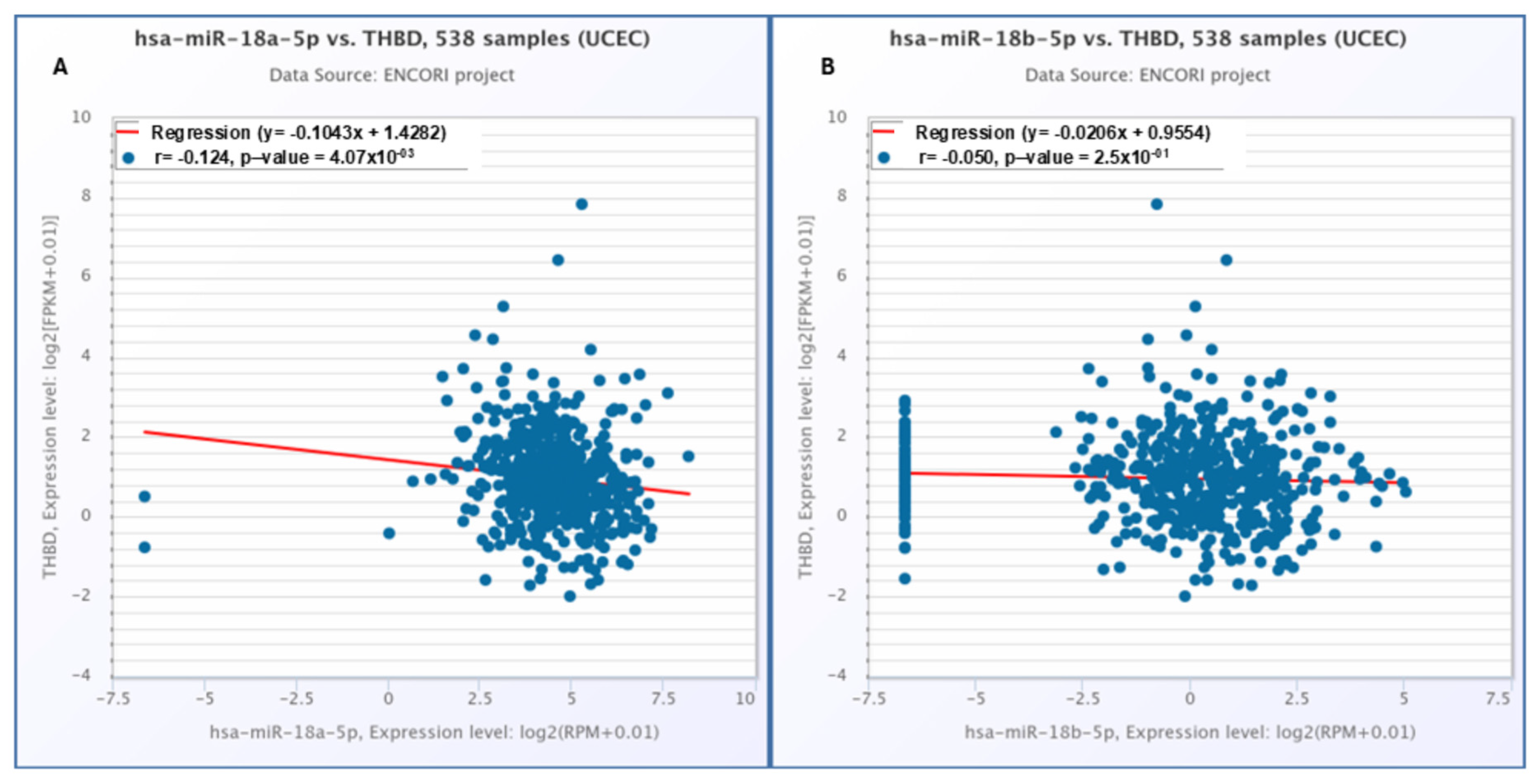
2.6. The Correlation Results Between THBD with hsa-miR-18a-5p and hsa-miR-18b-5p
2.7. DEGs Analysis Result
3. Discussion
4. Limitation
5. Materials and Methods
5.1. The Expression Profile Analysis of THBD
5.2. The Methylation Status and Survival Analysis of THBD
5.3. The Correlation and muTarget Analysis of THBD
5.4. The Gene–Gene Interaction
5.5. MicroRNA Target Analysis
5.6. The Relationship of MicroRNA vs. UCEC and MicroRNA vs. RNA
5.7. Detection of Differentially Expressed Genes (DEGs)
5.8. Statement of Ethics
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| THBD | Thrombomodulin |
| UCEC | Uterine Corpus Endometrial Carcinoma |
| EC | Endometrial Cancer |
| PMB | Postmenopausal Bleeding |
| OS | Overall Survival |
| GEPIA2 | Gene Expression Profiling Interactive Analysis |
| UALCAN | The University of Alabama at Birmingham CANcer data analysis Portal |
| HPA | Human Protein Atlas |
| TNM Plot | Tumor-Normal-Metastatic Plot Database |
| STRING | Search Tool for the Retrieval of Interacting Genes/Proteins |
| TargetScan | MicroRNA Target Prediction Tool |
| ENCORI | The Encyclopedia of RNA Interactomes |
| TCGA | The Cancer Genome Atlas |
| muTarget | Mutation and Target Expression Correlation Analysis Tool |
| RBPs | RNA-Binding Proteins |
| EMT | Epithelial–Mesenchymal Transition |
| PROCR | Endothelial Protein C Recepto |
References
- Zhou, C.; Li, C.; Yan, F.; Zheng, Y. Identification of an immune gene signature for predicting the prognosis of patients with uterine corpus endometrial carcinoma. Cancer Cell Int. 2020, 20, 541. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Yang, Y.; Li, C.; Li, J.; Lv, H.; Jia, X.; Yang, J.; Lei, J.; Li, X.; Guo, H.; et al. Development and validation of matrix metalloproteinase for predicting prognosis and tumour microenvironment immune profiles in uterine corpus endometrial carcinoma. J. Cancer 2024, 15, 4020–4039. [Google Scholar] [CrossRef] [PubMed]
- Makker, V.; MacKay, H.; Ray-Coquard, I.; Levine, D.A.; Westin, S.N.; Aoki, D.; Oaknin, A. Endometrial cancer. Nat. Rev. Dis. Primers 2021, 7, 88. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Le, T.; Hopkins, L.; Fung-Kee-Fung, M.; Jooya, A.; Lupe, K.; Gaudet, M.; Samant, R. Treatment of early-stage high-risk endometrioid-type endometrial cancer and patterns of disease relapse: A retrospective analysis. Adv. Radiat. Oncol. 2020, 5, 910–919. [Google Scholar] [CrossRef]
- Shen, Y.; Yang, W.; Liu, J.; Zhang, Y. Minimally invasive approaches for the early detection of endometrial cancer. Mol. Cancer 2023, 22, 53. [Google Scholar] [CrossRef]
- Li, H.; Zhou, Q.; Wu, Z.; Lu, X. Identification of novel key genes associated with uterine corpus endometrial carcinoma progression and prognosis. Ann. Transl. Med. 2023, 11, 100. [Google Scholar] [CrossRef]
- Manderstedt, E.; Halldén, C.; Lind-Halldén, C.; Elf, J.; Svensson, P.J.; Engström, G.; Melander, O.; Baras, A.; Lotta, L.A.; Zöller, B.; et al. Thrombomodulin (THBD) gene variants and thrombotic risk in a population-based cohort study. J. Thromb. Haemost. 2022, 20, 929–935. [Google Scholar] [CrossRef]
- Dong, H.; Li, Y.; Zhou, J.; Song, J. MiR-18a-5p promotes proliferation, migration, and invasion of endometrial cancer cells by targeting THBD. Crit. Rev. Eukaryot. Gene Expr. 2021, 31, 63–73. [Google Scholar] [CrossRef]
- Giri, H.; Biswas, I.; Rezaie, A.R. Thrombomodulin: A multifunctional receptor modulating the endothelial quiescence. J. Thromb. Haemost. 2024, 22, 905–914. [Google Scholar] [CrossRef]
- Huang, C.-Y.; Wei, P.-L.; Prince, G.M.S.H.; Batzorig, U.; Lee, C.-C.; Chang, Y.-J.; Hung, C.-S. The Role of Thrombomodulin in Estrogen-Receptor-Positive Breast Cancer Progression, Metastasis, and Curcumin Sensitivity. Biomedicines 2023, 11, 1384. [Google Scholar] [CrossRef]
- Hamza, M.S.; Mousa, S.A. Cancer-associated thrombosis: Risk factors, molecular mechanisms, future management. Clin. Appl. Thromb. 2020, 26, 1076029620954282. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.L.; Tsai, J.R.; Chiu, C.C.; Hwang, J.J.; Chou, S.H.; Wang, C.K.; Wu, S.J.; Chen, Y.L.; Chen, W.C.; Chen, Y.H.; et al. Decreased expression of thrombomodulin is correlated with tumor cell invasiveness and poor prognosis in nonsmall cell lung cancer. Mol. Carcinog. 2010, 49, 874–881. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Huo, Z.; Zhang, B.; Meng, M.; Cao, Z.; Wang, Z.; Zhou, Q. Thrombomodulin reduces tumorigenic and metastatic potential of lung cancer cells by up-regulation of E-cadherin and down-regulation of N-cadherin expression. Biochem. Biophys. Res. Commun. 2016, 476, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Ward, M.P.; Ibrahim, E.M.; O’Toole, S.A.; Marchocki, Z.; O’Leary, J.J.; Saadeh, F.A.; Norris, L.A. Chemotherapy alters thrombomodulin and factor VIIIc expression resulting in acquired activated protein C resistance and enhanced thrombin generation in cancer associated thrombosis. Thromb. Res. 2025, 246, 109251. [Google Scholar] [CrossRef]
- Hou, Y.; Li, T.; Gan, W.; Lv, S.; Zeng, Z.; Yan, Z.; Wang, W.; Yang, M. Prognostic significance of mutant-allele tumor heterogeneity in uterine corpus endometrial carcinoma. Ann. Transl. Med. 2020, 8, 339. [Google Scholar] [CrossRef]
- Yan, Q.; Xiaorong, Z.; Zhang, Z.; Bing, W.; Feng, Y.; Hong, B. Prevalence of protein C receptor (PROCR) is associated with inferior clinical outcome in breast invasive ductal carcinoma. Pathol. Res. Pract. 2017, 213, 1173–1179. [Google Scholar] [CrossRef]
- Xue, F.; Xu, Y.H.; Shen, C.C.; Qin, Z.L.; Zhou, H.B. Non-coding RNA LOXL1-AS1 exhibits oncogenic activity in ovarian cancer via regulation of miR-18b-5p/VMA21 axis. Biomed. Pharmacother. 2020, 125, 109568. [Google Scholar] [CrossRef]
- Xue, M.; Tao, W.; Yu, S.; Yan, Z.; Peng, Q.; Jiang, F.; Gao, X. lncRNA ZFPM2-AS1 promotes proliferation via miR-18b-5p/VMA21 axis in lung adenocarcinoma. J. Cell Biochem. 2020, 121, 313–321. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Yan, L.; Yang, S.; Xu, H.N.; Chen, T.T.; Dong, Z.Y.; Chen, S.L.; Wang, W.R.; Yang, Q.L.; Chen, C.J. Long noncoding RNA AC073284.4 suppresses epithelial-mesenchymal transition by sponging miR-18b-5p in paclitaxel-resistant breast cancer cells. J. Cell Physiol. 2019, 234, 23202–23215. [Google Scholar] [CrossRef]
- Mohan Rao, L.V.; Esmon, C.T.; Pendurthi, U.R. Endothelial cell protein C receptor: A multiliganded and multifunctional receptor. Blood 2014, 124, 1553–1562. [Google Scholar] [CrossRef]
- Murali, R.; Soslow, R.A.; Weigelt, B. Classification of endometrial carcinoma: More than two types. Lancet Oncol. 2014, 15, e268–e278. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hu, D.; Lin, Y.; Chen, X.; Yang, R.; Li, L.; Zhan, Y.; Bao, H.; Zang, L.; Zhu, M.; et al. Early detection of uterine corpus endometrial carcinoma utilizing plasma cfDNA fragmentomics. BMC Med. 2024, 22, 310. [Google Scholar] [CrossRef]
- Wise, M.R.; Gill, P.; Lensen, S.; Thompson, J.M.D.; Farquhar, C.M. Body mass index trumps age in decision for endometrial biopsy: Cohort study of symptomatic premenopausal women. Am. J. Obstet. Gynecol. 2016, 215, 598.e1–598.e8. [Google Scholar] [CrossRef] [PubMed]
- Hong, R.; Liu, W.; DeLair, D.; Razavian, N.; Fenyö, D. Predicting endometrial cancer subtypes and molecular features from histopathology images using multi-resolution deep learning models. Cell Rep. Med. 2021, 2, 100400. [Google Scholar] [CrossRef]
- Ejlalidiz, M.; Mehri-Ghahfarrokhi, A.; Saberiyan, M. Identification of hub genes and pathways in Uterine corpus endometrial carcinoma (UCEC): A comprehensive in silico study. Biochem. Biophys. Rep. 2024, 40, 101860. [Google Scholar] [CrossRef]
- Zhao, M.; Li, W. Metabolism-associated molecular classification of uterine corpus endometrial carcinoma. Front. Genet. 2023, 14, 955466. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Bartha, Á.; Győrffy, B. TNMplot.com: A web tool for the comparison of gene expression in normal, tumor, and metastatic tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Nagy, Á.; Győrffy, B. muTarget: A platform linking gene expression changes and mutation status in solid tumors. Int. J. Cancer 2021, 148, 502–511. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA, and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef]
- Hever, A.; Roth, R.B.; Hevezi, P.; Marin, M.E.; Acosta, J.A.; Acosta, H.; Rojas, J.; Herrera, R.; Grigoriadis, D.; White, E.; et al. Human endometriosis is associated with plasma cells and overexpression of B lymphocyte stimulator. Proc. Natl. Acad. Sci. USA 2007, 104, 12451–12456. [Google Scholar] [CrossRef]
- Crispi, S.; Piccolo, M.T.; D’Avino, A.; Donizetti, A.; Viceconte, R.; Spyrou, M.; Calogero, R.A.; Baldi, A.; Signorile, P.G. Transcriptional profiling of endometriosis tissues identifies genes related to organogenesis defects. J. Cell Physiol. 2013, 228, 1927–1934. [Google Scholar] [CrossRef]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef]




| Gene-1 | Gene-2 | Protein Annotation | Combine Scor |
|---|---|---|---|
| PROCR | THBD | Endothelial protein C receptor | 0.999 |
| F2 | THBD | Activation peptide fragment 1 | 0.999 |
| PROC | THBD | Vitamin K-dependent protein C heavy chain | 0.992 |
| F3 | THBD | Tissue factor | 0.982 |
| F2R | THBD | Proteinase-activated receptor 1 | 0.966 |
| TFPI | THBD | Tissue factor pathway inhibitor | 0.963 |
| CPB2 | THBD | Carboxypeptidase B2 | 0.958 |
| F7 | THBD | Coagulation factor VII | 0.942 |
| C3 | THBD | Complement C3c alpha’ chain fragment 1 | 0.933 |
| PLAT | THBD | Tissue-type plasminogen activator chain A | 0.913 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karaman, E.; Bayram, E.; Ayan, D. Evaluation of Thrombomodulin, hsa-miR-18a-5p, and hsa-miR-18b-5p as Potential Prognostic Biomarkers in Uterine Corpus Endometrial Carcinoma. Int. J. Mol. Sci. 2025, 26, 3649. https://doi.org/10.3390/ijms26083649
Karaman E, Bayram E, Ayan D. Evaluation of Thrombomodulin, hsa-miR-18a-5p, and hsa-miR-18b-5p as Potential Prognostic Biomarkers in Uterine Corpus Endometrial Carcinoma. International Journal of Molecular Sciences. 2025; 26(8):3649. https://doi.org/10.3390/ijms26083649
Chicago/Turabian StyleKaraman, Enes, Ergul Bayram, and Durmus Ayan. 2025. "Evaluation of Thrombomodulin, hsa-miR-18a-5p, and hsa-miR-18b-5p as Potential Prognostic Biomarkers in Uterine Corpus Endometrial Carcinoma" International Journal of Molecular Sciences 26, no. 8: 3649. https://doi.org/10.3390/ijms26083649
APA StyleKaraman, E., Bayram, E., & Ayan, D. (2025). Evaluation of Thrombomodulin, hsa-miR-18a-5p, and hsa-miR-18b-5p as Potential Prognostic Biomarkers in Uterine Corpus Endometrial Carcinoma. International Journal of Molecular Sciences, 26(8), 3649. https://doi.org/10.3390/ijms26083649





