1. Introduction
As the auxin signal plays a significant role in the development of plants, auxin early-response genes also play a pivotal role in plant development. They are a unique set of genes that are rapidly modulated and activated when internal auxin concentrations change. Specifically, once the auxin signal is received in the cell nucleus, the auxin early-response genes immediately initiate their expression process. This mechanism ensures that plants can promptly respond to the fluctuations of the auxin signals and adjust their growth strategies accordingly [
1]. These genes function as “information relay stations” throughout the auxin signal transduction process, situated downstream in the auxin signaling pathway, bridging the upstream signals with the downstream responses. Essentially, they serve as the switches, regulating various reactions in plant growth and developmental processes. Due to their pivotal role in connecting auxin signals with actual growth responses, they are also indispensable in the plant’s response to external environmental changes.
Aux/IAAs, GH3s, and SAURs are considered the three primary auxin early-response gene families [
2,
3]. Each family has its specific structural characteristics and functions. Notably, the SAUR family in plants contains more genes than the other families, reflecting its unique role in plant development. In fact, genes belonging to the SAUR family have been widely identified in various plants.
This began with the discovery of the
SAUR genes in soybean hypocotyls by McClure and Guilfoyle [
4]. As the research progressed, it was found that an increasing number of plant species contain gene members belonging to the SAUR family. This not only underscores the ubiquity of these genes but also broadens the scope for subsequent functional research and application. The
SAUR family genes encode a class of plant-specific proteins, and these proteins perform vital functions in the process of plant growth, development, and response to environmental signal changes. Subsequent studies revealed that these gene family members exist in most plant species, such as
Arabidopsis, corn, rice, sorghum, cotton, tomato, apple, lychee, potato, pepper, peach, citrus, and others [
5,
6,
7,
8,
9,
10].
The
SAUR gene members play crucial roles in the various growth and developmental stages of different plants. For example, they are involved in the elongation of the hypocotyl, phototropic growth, the formation of apical hooks for adaptive growth, and leaf growth and senescence. More specifically, based on the existing research, the
SAUR family genes can be classified into two main categories: those that promote growth and those that inhibit growth. For instance, genes like
AAM1,
OsSAUR39, and
OsSAUR45, when overexpressed, can lead to the inhibition of plant growth, while others like
SAUR63 and
SAUR19-24 act as the growth activators, promoting plant growth [
11]. Such a growth regulatory mechanism highlights the complexity and diversity of the
SAUR gene family members in plant development.
Strawberry (
Fragaria spp.), widely cultivated and consumed globally, is not only beloved for its enticing flavor and rich nutritional value but also has become a significant subject in scientific research. During the early stages of strawberry fruit development, the regulatory role of auxin signaling is particularly pronounced. Auxin plays a crucial role in cell division and elongation in fruit, directly influencing the morphological formation and maturation process of the fruit. In non-climacteric fruits like strawberry, variations in auxin concentration have a direct and profound impact on the fruit size, shape, and ultimate quality [
12]. For instance, during the initial maturation stage of the strawberry fruit, auxin facilitates the increase in fruit volume by promoting cell expansion and division. As the fruit matures further, the concentration of auxin gradually decreases, which is closely associated with physiological changes as the fruit transitions from the growth phase to maturation.
Notably, the high-quality genome sequence of the wild-type woodland strawberry (
Fragaria vesca) provides a solid foundation for the in-depth exploration of the gene family members and their functions in strawberry. As previously mentioned, the
SAUR gene family plays a pivotal role in the auxin signal transduction. However, few reports have addressed the potential roles of SAUR genes in woodland strawberry fruit development, and there remains some ambiguity regarding the specific functions and mechanisms of action of these gene family members [
13].
In light of this, our research has focused on the systematic identification and detection of the expression response of the SAUR gene family members in strawberry to the auxin signals. By integrating genomic structure, chromosomal location, and sequence analysis, we have successfully identified and characterized the members of the SAUR gene family in strawberry. The aim of this study is to deepen the understanding of the SAUR genes in strawberry and to provide a basic theoretical foundation for the future exploration of the specific roles of FvSAUR genes in the development of strawberry fruit, especially their roles in the auxin pathway.
2. Results
2.1. Sixty-Four SAUR Members Are Identified in Fragaria vesca
In the present study, we conducted a comprehensive bioinformatics analysis of the
Fragaria vesca genome and identified 64 genes belonging to the
SAUR family. These genes were consecutively designated as
FvSAUR1 to
FvSAUR64, based on their chromosomal order. A detailed list of these genes, along with their relevant genomic information, can be found in
Supplementary Table S1.
The amino acid lengths of the predicted SAUR proteins in Fragaria vesca displayed considerable variation, with their values ranging from 84 residues (in FvSAUR60) to 336 residues (in FvSAUR27). Correspondingly, the molecular weights of these proteins spanned between 9492.18 Da (FvSAUR60) and 40013.41 Da (FvSAUR27). From an isoelectric point (pI) perspective, the range extended from a low of 5.16 (FvSAUR46) to a high of 10.71 (FvSAUR29). These findings underscore the heterogeneity of SAUR proteins in strawberries, with some members exhibiting acidic properties while others lean toward basicity.
To delve deeper into the characteristics of the FvSAUR gene family members in strawberry, we analyzed several bioinformatics parameters, including their instability index, aliphatic index, and GRAVY. The instability index serves as a metric to predict the protein stability within a cellular context. Proteins with values above 40 are usually considered unstable. Notably, the majority of proteins encoded by the FvSAUR genes demonstrated that their indices are greater than 40, indicating their potential in vivo instability. FvSAUR1 and FvSAUR20 stood out with values over 60, suggesting their heightened instability. The aliphatic index, on the other hand, provides insights into the thermal stability of proteins. Proteins with elevated values are considered stable across an expansive temperature spectrum. Among the identified FvSAUR proteins, FvSAUR17 showed the highest aliphatic index, indicating its superb heat stability, while FvSAUR33 exhibited the lowest aliphatic index.
Lastly, the GRAVY index offers a glimpse into the hydrophilic or hydrophobic tendencies of a protein. Our observations revealed the varied GRAVY values across different FvSAUR proteins. Notably, FvSAUR9, FvSAUR12, FvSAUR17, FvSAUR45, FvSAUR46, FvSAUR47, and FvSAUR63 all possessed positive GRAVY values, suggesting that they may be hydrophobic.
2.2. Evolutionary and Structural Intricacies of the FvSAURs
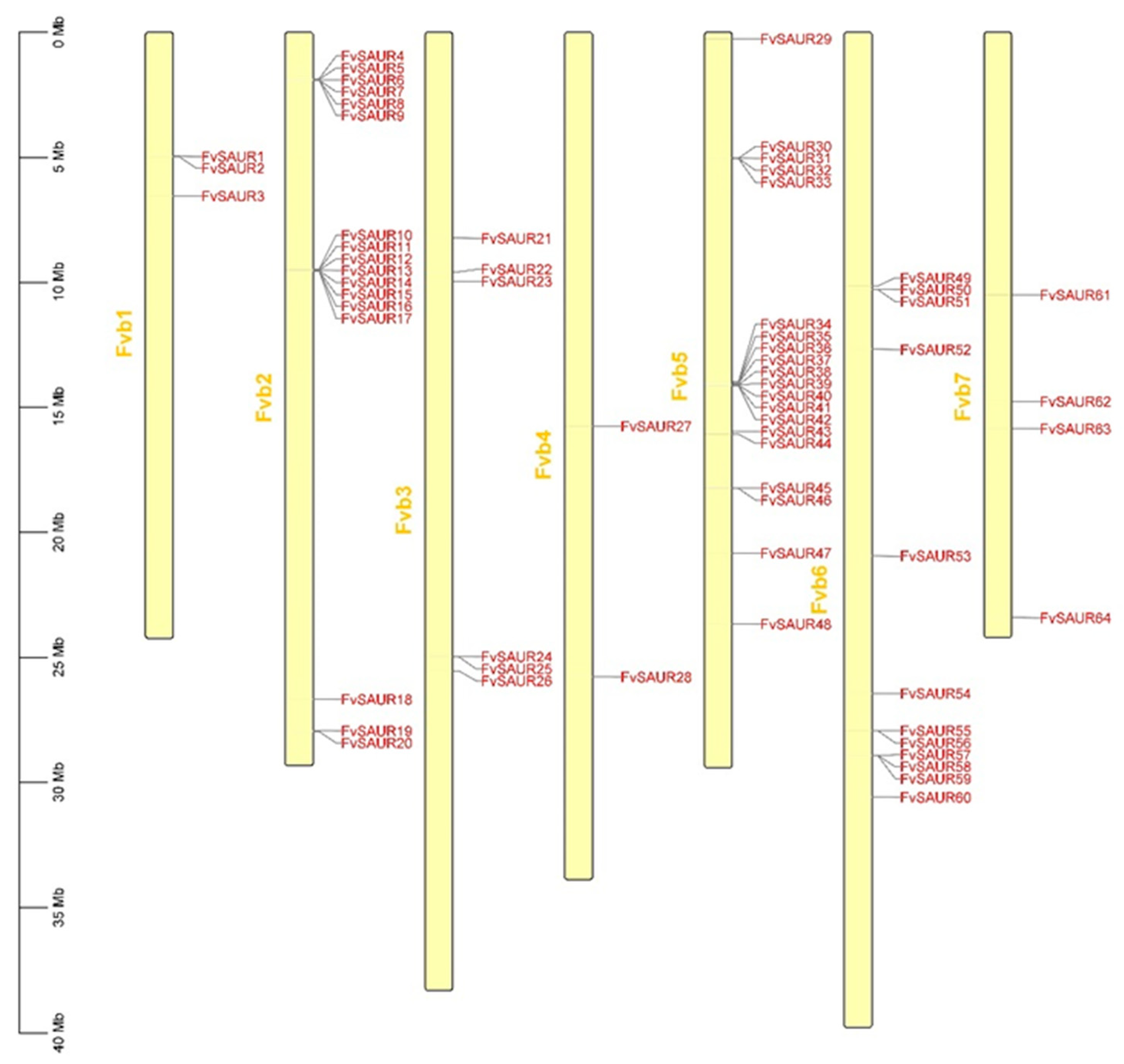
The chromosomal distribution of these genes was initially ascertained. Our results (
Figure 1) revealed that the
SAUR members of
Fragaria vesca were not evenly distributed across its chromosomes. Chromosome Fvb1 contains the genes
FvSAUR1 to
FvSAUR3; chromosome Fvb2 harbors 17 genes ranging from
FvSAUR4 to
FvSAUR20; chromosome Fvb3 consists of 6 genes from
FvSAUR21 to
FvSAUR26; chromosome Fvb4 includes
FvSAUR27 and
FvSAUR28; chromosome Fvb5 comprises 20 genes from
FvSAUR29 to
FvSAUR48; chromosome Fvb6 contains 12 genes from
FvSAUR49 to
FvSAUR60; chromosome Fvb7 encompasses 4 genes from
FvSAUR61 to
FvSAUR64. The chromosome map illustrates the genomic distribution of these genes, which may provide clues to their evolutionary and functional relevance.
Furthermore, the inherent conserved motifs within the FvSAUR members play a pivotal role in the protein–protein interactions, as evidenced by their recurrent presence across various genes. All FvSAUR proteins contain the conserved structural domain of the Auxin Inducible Superfamily (cl23790), suggesting that the characteristic structural domains of the SAUR family of proteins are highly evolutionarily conserved.
The MEME [
14] online tool was used to investigate the structural diversity of FvSAUR proteins in greater detail and to analyze their conserved motifs. A total of 11 conserved motifs were predicted (
Figure 2), and all FvSAUR proteins were found to contain at least one or more of motif 1, motif 2, motif 3, and motif 4.
In addition, it was found that only a small subset of the FvSAURs contain introns, a structural characteristic that may be closely associated with gene expression regulation and functional diversity. Moreover, the length and number of introns vary among different genes, reflecting the structural and functional diversity of specific gene structures.
2.3. Phylogenetic Insights into the FvSAUR Gene Family: Unraveling Evolutionary Affiliations in Fragaria vesca
To elucidate the evolutionary lineage of the FvSAUR members in Fragaria vesca, we referenced the AtSAUR genes and constructed a phylogenetic tree based on the amino acid sequences encoded by these genes. The resulting unrooted phylogenetic tree demarcated distinct clusters, thereby highlighting the evolutionary relationships among these genes.
Our phylogenetic tree analysis has revealed that these genes can be clustered into three main groups (
Figure 3). Group 1 primarily includes the
Arabidopsis SAUR genes
AtSAUR58,
43,
55,
41,
40,
72,
71,
74,
76,
79,
78,
and 77 along with 15 FvSAUR genes:
FvSAUR45,
46,
28,
32,
31,
33,
30,
55,
60,
22,
48,
27,
23,
2, and
56. Group 2 comprises
Arabidopsis SAUR genes
AtSAUR42,
48,
44,
57,
46,
47,
49,
53,
69,
52,
45,
17,
70,
39,
38, and
37, in addition to 16 FvSAUR genes:
FvSAUR25,
24,
50,
51,
63,
49,
58,
10,
20,
52,
57,
47,
1,
18,
59,
54, and
26. Group 3 is formed by the remaining genes in the phylogenetic tree, which include 33
Fragaria vesca SAUR genes.
High bootstrap values further substantiate the divisions within these groups. For example, the ensemble of FvSAUR24, FvSAUR50, and AtSAUR48 indicates a pronounced evolutionary kinship among them, possibly suggesting a shared ancestral origin or similar gene functions. Analogously, additional clusters delineate relationships between the FvSAUR and AtSAUR genes, providing critical insights into the probable evolutionary dynamics of the FvSAUR gene family members when compared to the AtSAUR reference.
Within Group 2, the proximal clustering of genes like FvSAUR25 and FvSAUR54 suggests their shared evolutionary trajectory within the SAUR lineage. A notable observation in Group 3 is the distinct cluster formed by AtSAUR60 and AtSAUR2, indicating a unique evolutionary lineage.
This classification not only reveals the potential evolutionary relationships among members of the SAUR gene family in woodland strawberry but also establishes the foundation for understanding their functional diversification. The genes within each group share a degree of sequence homology, which may suggest either functional similarities or evolutionary relatedness. In summary, this detailed phylogenetic analysis offers a comprehensive perspective of the evolutionary affiliations and tendencies inherent to the FvSAUR members, with AtSAUR genes serving as an insightful benchmark.
2.4. Analysis of the Cis-Acting Elements in FvSAURs Promoters
Cis-acting elements are critical modulators that dictate the magnitude and spatiotemporal framework of gene transcription. To characterize the cis-acting elements distributed in the
FvSAUR promoter region, the 2000 bp sequences upstream of the initiation codon of each
FvSAUR gene were extracted. Subsequent predictions of the cis-acting elements within the promoters were performed using the PlantCARE database. An abundance of light-responsive elements was identified, implicating their potential roles in the light-mediated signaling pathways. Intriguingly, a variety of hormone-responsive elements were also detected. For instance,
FvSAUR61 and 10 other
FvSAUR members exhibited gibberellin-responsive elements (
Figure 4). Similarly,
FvSAUR1 and
FvSAUR7 were found to harbor salicylic acid-responsive elements. Moreover,
FvSAUR30 and 11 additional members contain Jasmonic acid-responsive elements, while
FvSAUR25 and its corresponding genes possess abscisic acid-responsive elements. In addition, the promoter regions of
FvSAUR5,
FvSAUR19, and
FvSAUR54 were also found to contain growth hormone response elements. These data highlight the broad-spectrum involvement of the
SAUR gene members in orchestrating hormone signaling cascades.
Additionally, the prediction pinpointed the cis-acting elements related to growth, development, and environmental stress adaptation. These encompass the endosperm-specific expression elements, meristematic tissue expression elements, elements conferring stress resistance, and elements responsive to cold temperatures.
In essence, the SAUR gene members in Fragaria vesca appear poised to modulate diverse growth, developmental processes, and stress responses, primarily through the intricate regulation of their associated cis-acting elements.
2.5. The Evolutionary Dynamics and Duplication Patterns of FvSAUR Genes
The duplication events within the FvSAUR gene members were discerned utilizing the MCSCAN analysis. Specifically, tandem duplication, segmental duplication, and whole-genome duplication events were all identified, which likely contributed to the expansion of the SAUR gene repertoire in Fragaria vesca.
To decipher the genetic interrelationships among SAUR members in Arabidopsis thaliana, tomato (Solanum lycopersicum), and Fragaria vesca, we performed a collinearity analysis across their respective genomes, focusing on the SAUR genes within Fragaria vesca.
The analysis revealed several notable gene pair associations. For instance,
FvSAUR1 was found to be collinear with
SlSAUR64 (Solyc07g042470), and
FvSAUR3 with
SlSAUR32 (Solyc02g062230) (
Figure 5). Such pairings suggest that the selected
SAUR genes might be evolutionarily conserved across disparate species.
The collinearity analysis further suggested the existence of 15 FvSAUR gene pairs that occupy collinear genomic blocks. Of particular interest, FvSAUR3 displayed significant evolutionary dynamism, showing strong collinearity with two distinct tomato SAUR genes.
Intriguingly, our analysis revealed no evidence of singleton duplications or proximal duplications in the chromosomal regions containing FvSAUR genes. This absence implies that certain duplication mechanisms, such as whole-genome or segmental duplications, may have played a dominant role in the expansion of this gene family, while small-scale (singleton/proximal) duplications were either suppressed or selectively disadvantageous.
In summation, the SAUR gene lineage within Fragaria vesca expanded through distinct duplication events and retained critical functions throughout evolutionary time. This study provides insights into the complex evolutionary and functional dynamics of the SAUR gene members.
2.6. Different Expression Dynamics of FvSAUR Genes Across Developmental Stages and Tissues in Fragaria vesca
To elucidate the roles of the FvSAUR genes in Fragaria vesca development, we conducted a comprehensive investigation into their expression profiles, spanning the various developmental phases and the distinct tissue types of the strawberry, using the expression data, which were also obtained from the YW5AF7 line from the eFP database. Our meticulous analysis encompassed five distinct developmental stages of the cortex and pith of strawberry and delved into specialized tissues, including the microspore, perianth, receptacle, anther, whole flower, leaf, and seedling.
Several prominent expression patterns were observed in their expression (
Figure 6). Notably,
FvSAUR11,
FvSAUR19,
FvSAUR7,
FvSAUR21,
FvSAUR15, and
FvSAUR38 exhibited strong expression levels in the pith and cortex of strawberry, consistently observed across all examined developmental stages. In contrast, genes such as
FvSAUR62,
FvSAUR17,
FvSAUR61,
FvSAUR39, and
FvSAUR56 showed higher expression during development stages 2 to 5 of the pith. It is noteworthy that
FvSAUR3,
FvSAUR64,
FvSAUR28, and
FvSAUR39 were preferentially expressed during the seedling phase. Other members like
FvSAUR16,
FvSAUR35,
FvSAUR43, and others were predominantly expressed in the microspore, perianth, receptacle, and anther but exhibited low expression levels in leaf and seedling tissues. Additionally,
FvSAUR15,
FvSAUR38,
FvSAUR56, and
FvSAUR62 exhibited elevated expression in both the leaves and seedlings. Importantly,
FvSAUR8,
FvSAUR21,
FvSAUR61,
FvSAUR11, and
FvSAUR19 displayed pronounced expression levels across all tissue types investigated.
2.7. Differential Expression Patterns of FvSAUR Genes in Various Tissues: Insights from qRT-PCR Analysis
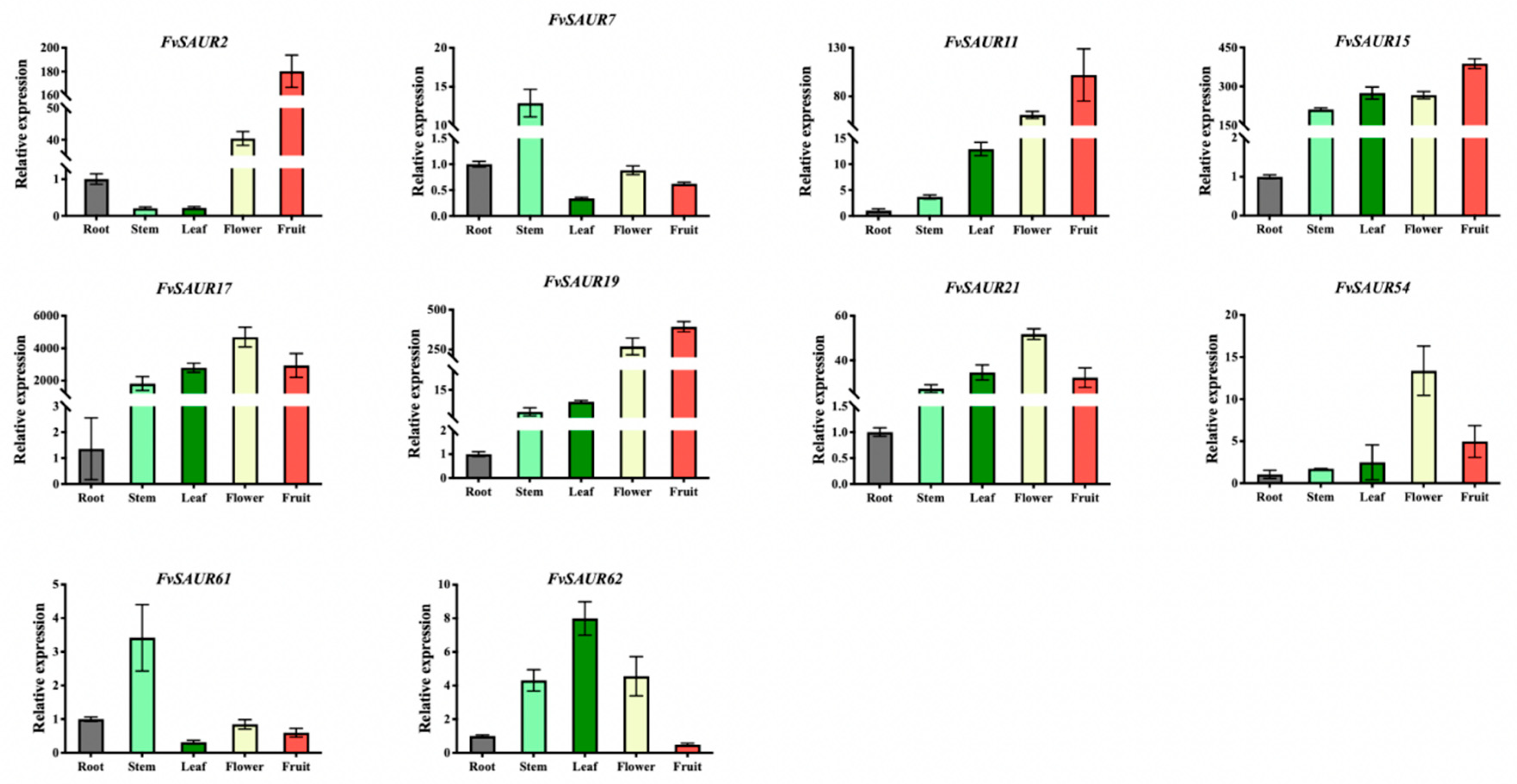
In order to further investigate whether FvSAUR gene members function in the early development of strawberry fruits, 10 gene members with significant expression in the medulla and cortex of strawberry fruits screened in the eFP database were further screened and analyzed by quantitative assays, including FvSAUR2, FvSAUR7, FvSAUR11, FvSAUR15, FvSAUR17, FvSAUR19, FvSAUR21, FvSAUR54, FvSAUR61, and FvSAUR62, based on the results of the heatmap analysis of pre-transcriptomic data. Notably, the expression trends revealed by quantitative PCR were consistent with the expression level changes shown in the RNA-seq heatmap.
The expression patterns of these 10 selected genes in roots, stems, leaves, flowers, and fruits of the ‘YW5AF7’ strawberry cultivar at the same developmental period were first examined using real-time quantitative PCR (qRT-PCR). The quantitative PCR results (
Figure 7) showed that
FvSAUR2 and
FvSAUR61 exhibited higher expression levels in leaves, and
FvSAUR62 also showed an elevated relative expression in leaves. In contrast,
FvSAUR17 and
FvSAUR54 displayed the highest relative expression in flowers. Meanwhile,
FvSAUR2,
FvSAUR11,
FvSAUR15, and
FvSAUR19 all showed the most significant expression levels in fruits.
Considering the important roles of the auxin signal in the fruit development of strawberry, we further investigated the role of FvSAUR genes in early fruit development, and we also examined the expression changes in these genes at 0, 4, 8, and 12 days after pollination was detected. The results are shown in
Figure 8; six genes (
FvSAUR2,
FvSAUR11,
FvSAUR15,
FvSAUR17,
FvSAUR19, and
FvSAUR21) exhibited their highest expression levels on the day of pollination (day 0). The expression levels of
FvSAUR11,
FvSAUR17,
FvSAUR19, and
FvSAUR21 showed a decreasing trend as fruit development progressed. In contrast, the expression of the other nine genes, except
FvSAUR62, was significantly higher on day 8 and before pollination compared to day 12.
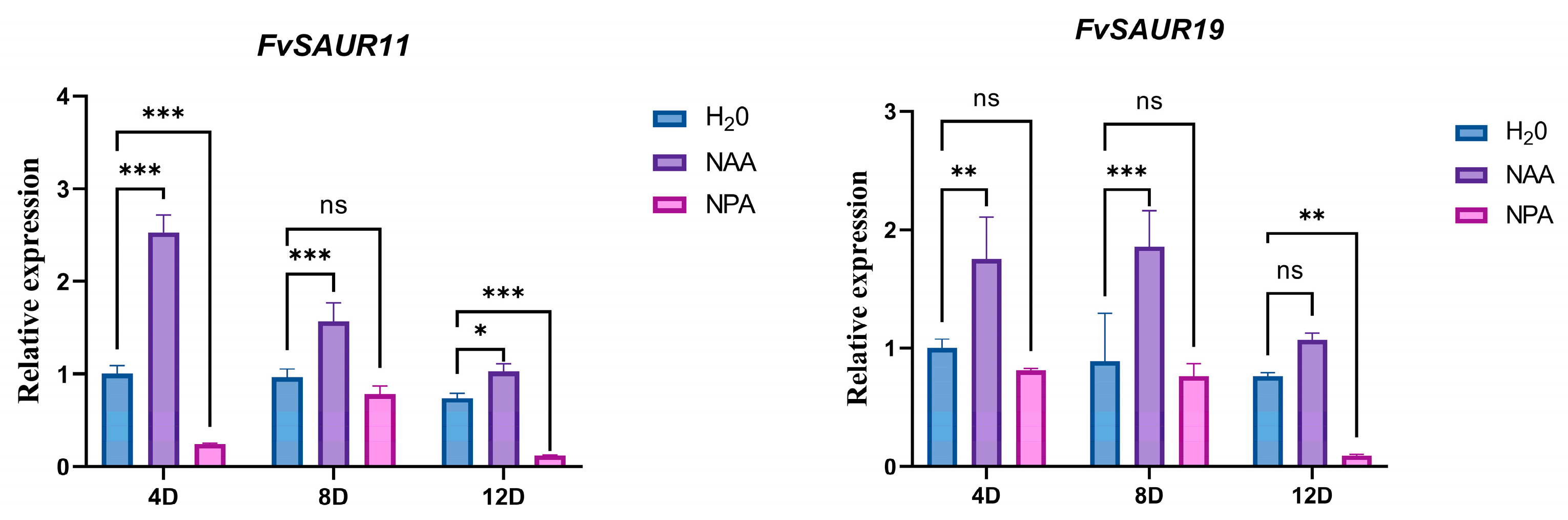
2.8. Auxin Treatment Upregulates the Gene Expression of FvSAUR11 and FvSAUR19
Based on the expression patterns of
FvSAUR genes in strawberry fruits at different developmental stages and the pooled expression data of these genes in the pith and cortex from the eFP database (
Figure S1), we selected
FvSAUR11 and
FvSAUR19, which showed a significant decreasing trend in early fruit development, for the exogenous auxin signal (NAA) and its transport inhibitor (NPA) treatment experiment to detect their expressional responses.
As shown in
Figure 9, the expression levels of both
FvSAUR11 and
FvSAUR19 were significantly increased at 4 and 8 days after NAA treatment, indicating that these two genes are clearly responsive to the auxin signaling. Conversely, the expression levels of these two genes were down-regulated after 12 days of NPA treatment.
2.9. FvSAUR11 and FvSAUR19 Are Subcellularly Located in the Nucleus
As noted, FvSAUR11 and FvSAUR19 show peak expression during the early strawberry fruit development stage and are both NAA-responsive, suggesting that they may function in the regulation of early fruit development. Most SAUR members function in plant development by regulating the transcription levels of target genes, indicating that SAURs should be transcriptional factors (TFs) localized in the nucleus. To further investigate this, we employed the GFP fusion protein technique to determine the precise cellular localization of the proteins encoded by these two genes.
As shown in the images (
Figure 10), the first row serves as the control group, displaying widespread GFP expression with uniform green fluorescence throughout the entire cell, thereby establishing a baseline for a fluorescence background. Following this, the second and third rows illustrate the localization of the SAUR11 and SAUR19 GFP fusion proteins, respectively. The green fluorescence of these proteins reveals a localization pattern that is distinctly different from that of the control group, indicating a concentration of these SAUR proteins within the cell nucleus, which is consistent with their role as TFs.