Multi-Omic Analysis Reveals the Potential Anti-Disease Mechanism of Disease-Resistant Grass Carp
Abstract
1. Introduction
2. Results
2.1. Intestinal Histology
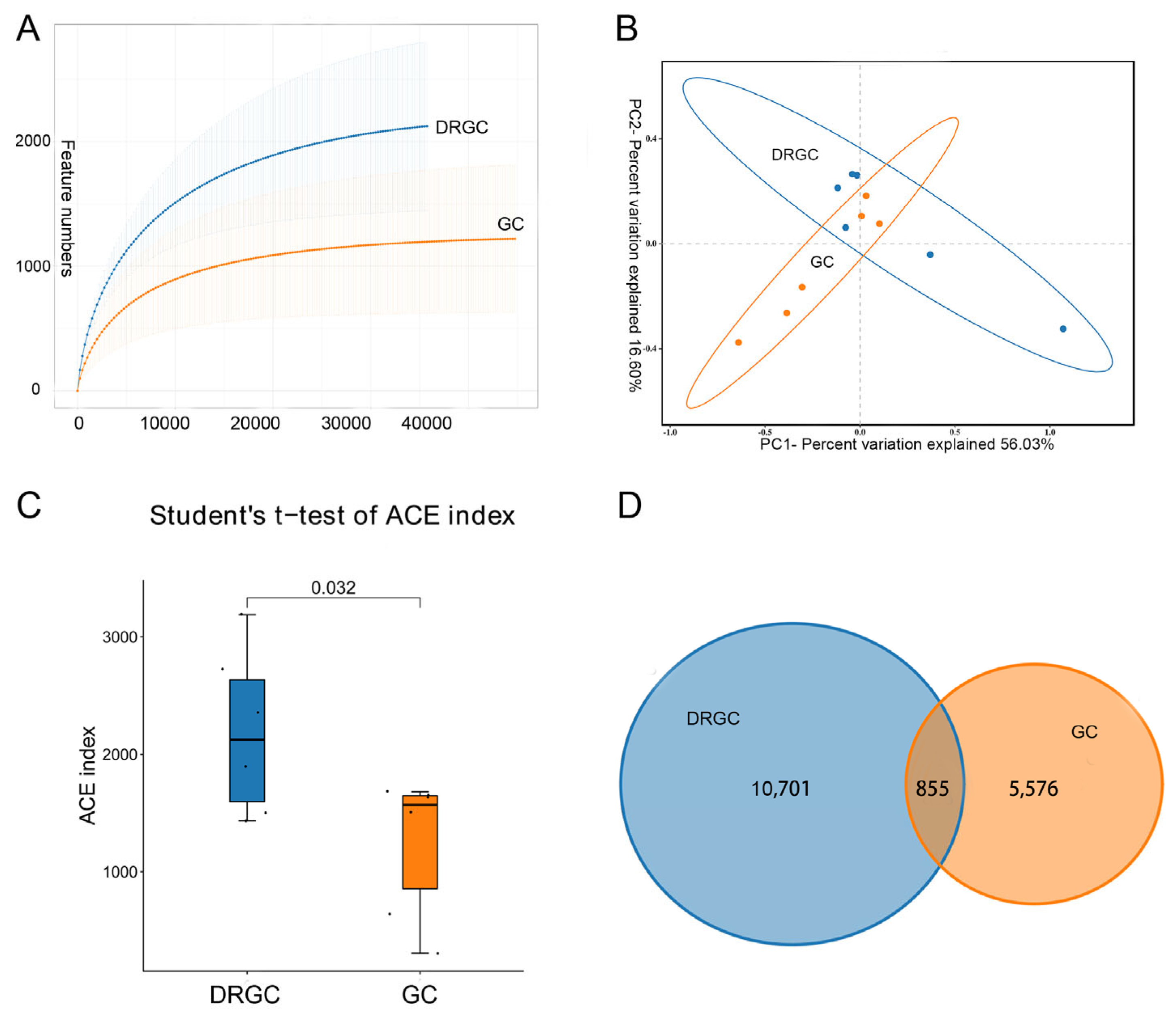
2.2. 16S rRNA Sequencing Analysis and Taxonomic Annotation
2.3. Intestinal Microbial Composition
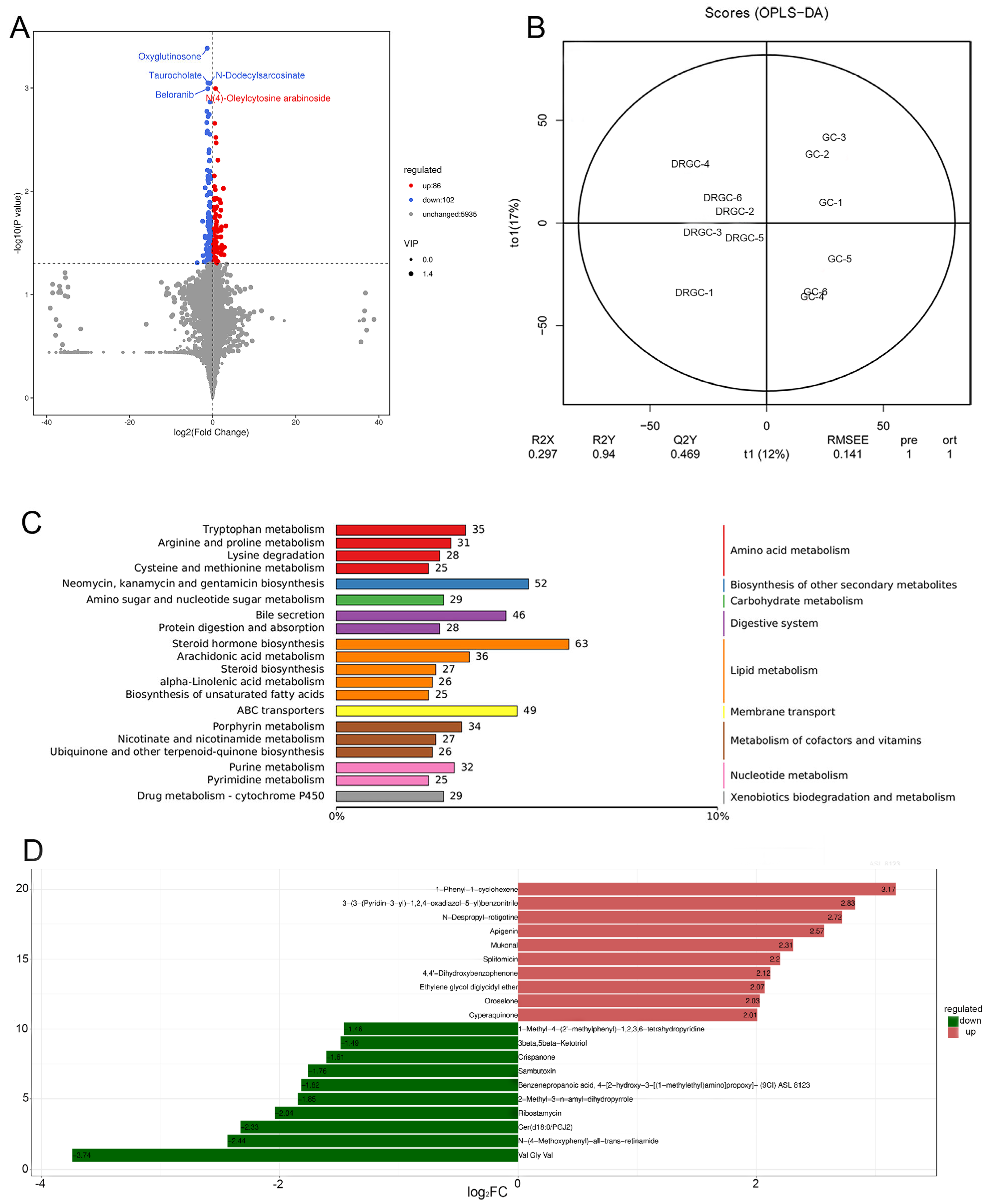
2.4. Metabolome Analysis
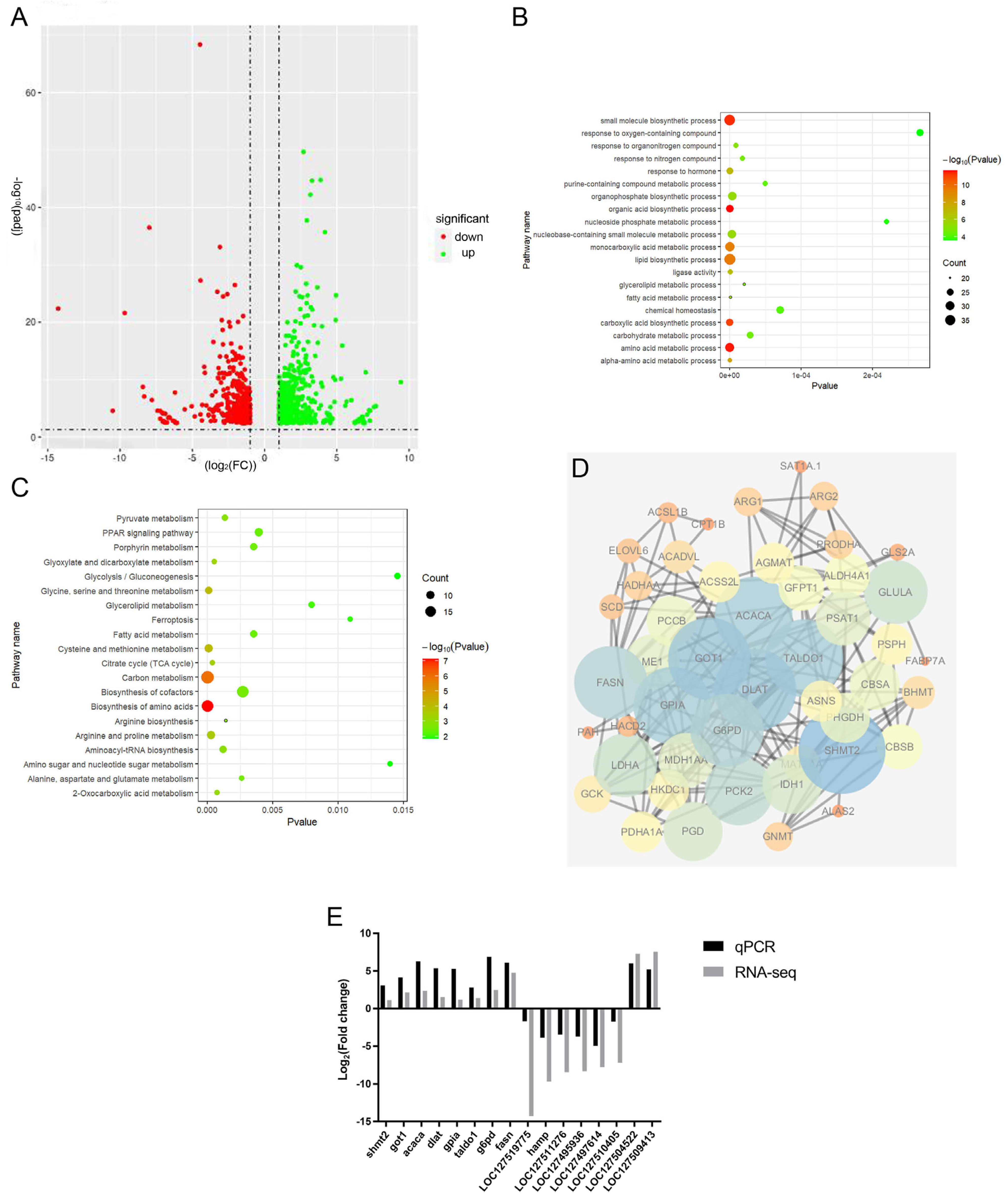
2.5. Transcriptome
2.6. Identification of Differentially Expressed Genes (DEGs)
2.7. Verification
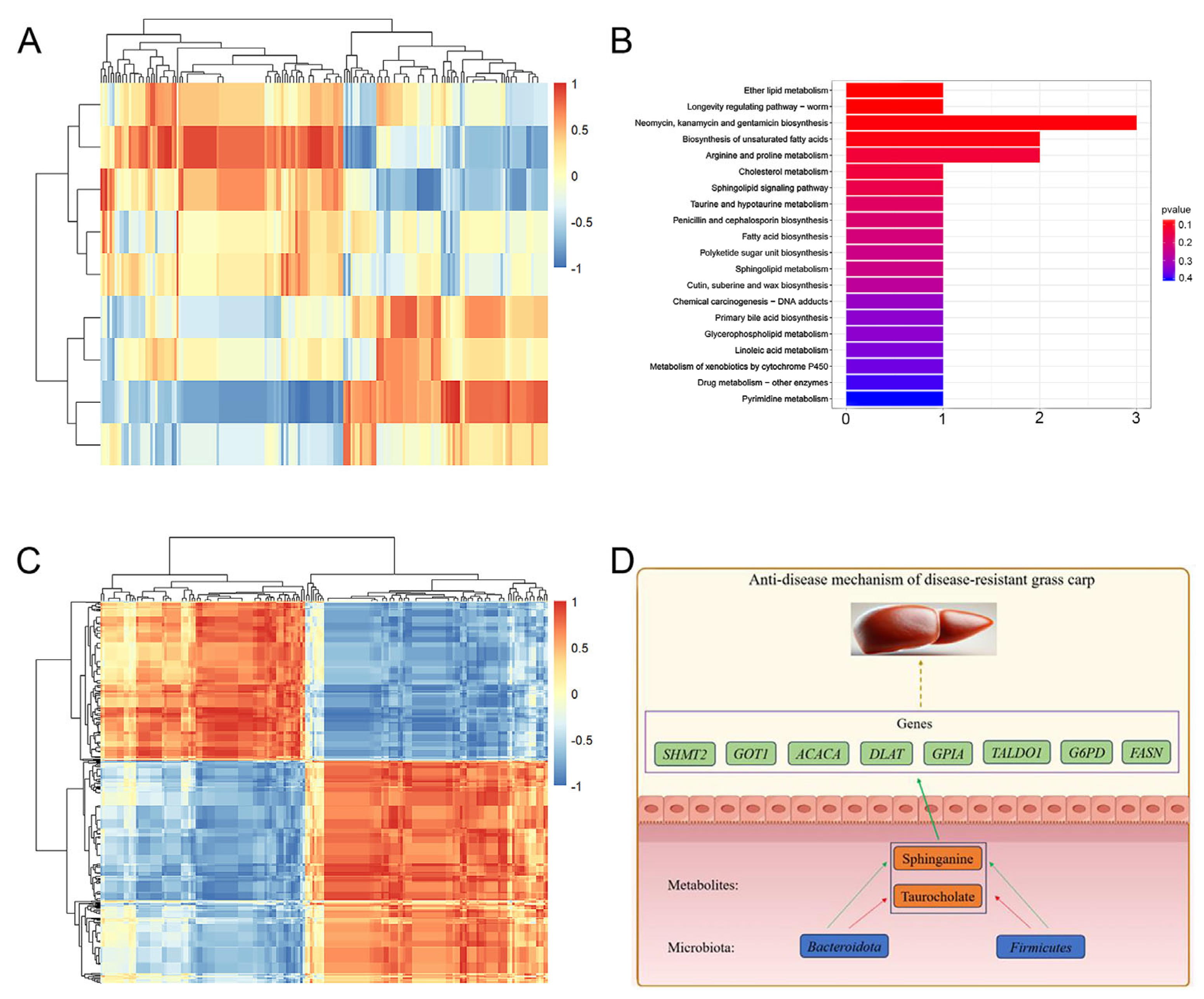
2.8. Multi-Omic Joint Analysis
2.8.1. Correlation Analysis Between Microbiome and Metabolome
2.8.2. Correlation Analysis Between Metabolome and Transcriptome
3. Discussion
4. Materials and Methods
4.1. Animal Materials
4.2. Determination of the Morphological Structure of Intestinal Tissue
4.3. 16S rRNA Gene Sequencing Analysis
4.4. Metabolomic Analysis
4.5. Transcriptome Analysis
4.6. Verification of Transcriptome Data
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- O’Hara, A.M.; Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef] [PubMed]
- Rajeev, R.; Dwivedi, A.P.; Sinha, A.; Agarwaal, V.; Dev, R.R.; Kar, A.; Khosla, S. Epigenetic interaction of microbes with their mammalian hosts. J. Biosci. 2001, 46, 94. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, J.R.; Adams, D.H.; Fava, F.; Hermes, G.D.; Hirschfield, G.M.; Hold, G.; Quraishi, M.N.; Kinross, J.; Smidt, H.; Tuohy, K.M.; et al. The gut microbiota and host health: A new clinical frontier. Gut 2016, 65, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef]
- Postler, T.S.; Ghosh, S. Understanding the holobiont: How microbial metabolites affect human health and shape the immune system. Cell Metab. 2017, 26, 110–130. [Google Scholar] [CrossRef]
- Kim, P.S.; Shin, N.R.; Lee, J.B.; Kim, M.S.; Whon, T.W.; Hyun, D.W.; Yun, J.H.; Jung, M.J.; Kim, J.Y.; Bae, J.W. Host habitat is the major determinant of the gut microbiome of fish. Microbiome 2021, 9, 166. [Google Scholar] [CrossRef]
- Nayak, S.K. Role of gastrointestinal microbiota in fish. Aquac. Res. 2010, 41, 1553–1573. [Google Scholar] [CrossRef]
- Cui, X.; Zhang, Q.; Zhang, Q.; Zhang, Y.; Chen, H.; Liu, G.; Zhu, L. Research progress of the gut microbiome in hybrid fish. Microorganisms 2022, 10, 891. [Google Scholar] [CrossRef]
- Hsu, C.L.; Schnabl, B. The gut-liver axis and gut microbiota in health and liver disease. Nat. Rev. Microbiol. 2023, 21, 719–733. [Google Scholar] [CrossRef]
- Wang, L.; Cao, Z.M.; Zhang, L.L.; Li, J.M.; Lv, W.L. The role of gut microbiota in some liver diseases: From an immunological perspective. Front. Immunol. 2022, 13, 923599. [Google Scholar] [CrossRef]
- Albillos, A.; de Gottardi, A.; Rescigno, M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J. Hepatol. 2020, 72, 558–577. [Google Scholar] [CrossRef] [PubMed]
- Rager, S.L.; Zeng, M.Y. The gut-liver axis in pediatric liver health and disease. Microorganisms 2023, 11, 597. [Google Scholar] [CrossRef] [PubMed]
- Marijani, E. Prevalence and antimicrobial cresistance of bacteria isolated from marine and freshwater fish in Tanzania. Int. J. Microbiol. 2022, 2022, 4652326. [Google Scholar] [CrossRef] [PubMed]
- Fariya, N.; Kaur, H.; Singh, M.; Abidi, R.; El-Matbouli, M.; Kumar, G. Morphological and molecular characterization of a new Myxozoan, Myxobolus grassi sp. nov. (Myxosporea), infecting the grass carp, Ctenopharyngodon idella in the Gomti river, India. Pathogens 2022, 11, 303. [Google Scholar] [CrossRef]
- Niu, H.; Pang, Y.; Xie, L.; Yu, Q.; Shen, Y.; Li, J.; Xu, X. Clustering pattern and evolution characteristic of microRNAs in grass carp (Ctenopharyngodon idella). BMC Genom. 2023, 24, 73. [Google Scholar] [CrossRef]
- Song, X.; Zhao, J.; Bo, Y.; Liu, Z.; Wu, K.; Gong, C. Aeromonas hydrophila induces intestinal inflammation in grass carp (Ctenopharyngodon Idella): An experimental model. Aquaculture 2014, 434, 171–178. [Google Scholar] [CrossRef]
- Rao, Y.; Su, J. Insights into the antiviral immunity against grass carp (Ctenopharyngodon idella) reovirus (GCRV) in grass carp. J. Immunol. Res. 2015, 2015, 670437. [Google Scholar] [CrossRef]
- Liu, S.; Wang, S.; Liu, X.; Wen, L.; Zou, J. Effects of dietary antimicrobial peptides on intestinal morphology, antioxidant status, immune responses, microbiota and pathogen disease resistance in grass carp Ctenopharyngodon idellus. Microb. Pathog. 2022, 165, 105386. [Google Scholar] [CrossRef]
- Chen, Z.; Feng, L.; Wu, P.; Jiang, W.D.; Jiang, J.; Zhou, X.Q.; Liu, Y. From growth promotion to intestinal inflammation alleviation: Unraveling the potential role of Lactobacillus rhamnosus GCC-3 in juvenile grass carp (Ctenopharyngodon idella). Fish Shellfish. Immunol. 2024, 148, 109511. [Google Scholar] [CrossRef]
- Yuan, Z.H.; Feng, L.; Jiang, W.D.; Wu, P.; Liu, Y.; Jiang, J.; Kuang, S.Y.; Tang, L.; Zhou, X.Q. Dietary choline-enhanced skin immune response of juvenile grass carp might be related to the STAT3 and NF-kB signaling pathway (Ctenopharyngodon idella). Front. Nutr. 2021, 8, 652767. [Google Scholar] [CrossRef]
- Ming, J.; Ye, J.; Zhang, Y.; Xu, Q.; Yang, X.; Shao, X.; Qiang, J.; Xu, P. Optimal dietary curcumin improved growth performance, and modulated innate immunity, antioxidant capacity and related genes expression of NF-κB and Nrf2 signaling pathways in grass carp (Ctenopharyngodon idella) after infection with Aeromonas hydrophila. Fish Shellfish Immunol. 2020, 97, 540–553. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Han, R.; Xu, W.; Lu, Z.; Li, Y.; Dan, X.; Mo, Z. The protective effect of inactivated Flavobacterium columnare vaccine in grass carp (Ctenopharyngodon idellus). Front. Immunol. 2023, 14, 1162975. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liao, A.; Tan, H.; Li, M.; Geng, C.; Wang, S.; Zhao, R.; Qin, Q.; Luo, K.; Zhang, C.; et al. The comparative studies on growth rate and disease resistance between improved grass carp and common grass carp. Aquaculture 2022, 560, 738476. [Google Scholar] [CrossRef]
- Yu, C.; Tang, H.; Jiang, Y.; Lu, H.; Chen, Q.; Gui, L.; Qiu, J.; Xu, X.; Li, J.; Shen, Y. Growth performance and selection signatures revealed by whole-genome resequencing in genetically selected grass carp (Ctenopharyngodon idella). Aquaculture 2024, 587, 740885. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, W.; Li, Z.; Qiu, B.; Li, J.; Geng, G.; Hu, B.; Liao, A.; Cai, Y.; Wen, M.; et al. Improvement and application of genetic resources of grass carp (Ctenopharyngodon idella). Reprod. Breed. 2024, 4, 126–133. [Google Scholar] [CrossRef]
- Tan, H.; Hu, B.; Liu, W.; Liao, A.; Wang, Y.; He, W.; Zhang, Y.; Geng, C.; Luo, K.; Tao, M.; et al. Comprehensive analysis of the immunological differences in the intestinal barrier of improved grass carp and their parents. Aquaculture 2023, 577, 739931. [Google Scholar] [CrossRef]
- Elliott, J.P.; Bellwood, D.R. Alimentary tract morphology and diet in three coral reef fish families. J. Fish. Biol. 2003, 63, 1598–1609. [Google Scholar] [CrossRef]
- Horn, M.H.; Gawlicka, A.K.; German, D.P.; Logothetis, E.A.; Cavanagh, J.W.; Boyle, K.S. Structure and function of the stomachless digestive system in three related species of New World silverside fishes (Atherinopsidae) representing herbivory, omnivory, and carnivory. Mar. Biol. 2006, 149, 1237–1245. [Google Scholar] [CrossRef]
- Li, W.; Liu, J.; Tan, H.; Yang, C.; Ren, L.; Liu, Q.; Wang, S.; Hu, F.; Xiao, J.; Zhao, R.; et al. Genetic Effects on the gut microbiota assemblages of hybrid fish from parents with different feeding habits. Front. Microbiol. 2018, 9, 2972. [Google Scholar] [CrossRef]
- Shi, X.; Xu, W.; Che, X.; Cui, J.; Shang, X.; Teng, X.; Jia, Z. Effect of arsenic stress on the intestinal structural integrity and intestinal flora abundance of Cyprinus carpio. Front. Microbiol. 2023, 14, 1179397. [Google Scholar] [CrossRef]
- Fietz, K.; Rye Hintze, C.O.; Skovrind, M.; Kjærgaard Nielsen, T.; Limborg, M.T.; Krag, M.A.; Palsbøll, P.J.; Hestbjerg Hansen, L.; Rask Møller, P.; Gilbert, M.T.P. Mind the gut: Genomic insights to population divergence and gut microbial composition of two marine keystone species. Microbiome 2018, 6, 82. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Ortiz, L.; Amato, K.R. Host genetics influence the gut microbiome. Science 2021, 373, 159–160. [Google Scholar] [CrossRef] [PubMed]
- López Nadal, A.; Ikeda-Ohtsubo, W.; Sipkema, D.; Peggs, D.; McGurk, C.; Forlenza, M.; Wiegertjes, G.F.; Brugman, S. Feed, Microbiota, and gut immunity: Using the zebrafish model to understand fish health. Front. Immunol. 2020, 11, 114. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, J.A.; Villumsen, K.R.; von Gersdorff Jørgensen, L.; Forberg, T.; Zuo, S.; Kania, P.W.; Buchmann, K.; Kristiansen, K.; Bojesen, A.M.; Limborg, M.T. Integrative analyses of probiotics, pathogenic infections and host immune response highlight the importance of gut microbiota in understanding disease recovery in rainbow trout (Oncorhynchus mykiss). J. Appl. Microbiol. 2022, 132, 3201–3216. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, S.; Nie, Q.; He, H.; Tan, H.; Geng, F.; Ji, H.; Hu, J.; Nie, S. Gut firmicutes: Relationship with dietary fiber and role in host homeostasis. Crit. Rev. Food Sci. Nutr. 2023, 63, 12073–12088. [Google Scholar] [CrossRef]
- Dehler, C.E.; Secombes, C.J.; Martin, S.A. Environmental and physiological factors shape the gut microbiota of Atlantic salmon parr (Salmo salar L.). Aquaculture 2017, 467, 149–157. [Google Scholar] [CrossRef]
- Schmidt, V.; Amaral-Zettler, L.; Davidson, J.; Summerfelt, S.; Good, C. Influence of fishmeal-free diets on microbial communities in Atlantic salmon (Salmo salar) recirculation aquaculture systems. Appl. Environ. Microbiol. 2016, 82, 4470–4481. [Google Scholar] [CrossRef]
- Balcázar, J.L.; de Blas, I.; Ruiz-Zarzuela, I.; Cunningham, D.; Vendrell, D.; Múzquiz, J.L. The role of probiotics in aquaculture. Vet. Microbiol. 2006, 114, 173–186. [Google Scholar] [CrossRef]
- Yi, Y.; Zhang, Z.; Zhao, F.; Liu, H.; Yu, L.; Zha, J.; Wang, G. Probiotic potential of Bacillus velezensis JW: Antimicrobial activity against fish pathogenic bacteria and immune enhancement effects on Carassius auratus. Fish Shellfish Immunol. 2018, 78, 322–330. [Google Scholar] [CrossRef]
- Giri, S.S.; Kim, H.J.; Jung, W.J.; Bin Lee, S.; Joo, S.J.; Gupta, S.K.; Park, S.C. Probiotics in addressing heavy metal toxicities in fish farming: Current progress and perspective. Ecotoxicol. Environ. Saf. 2024, 282, 116755. [Google Scholar] [CrossRef]
- Buruiana, C.T.; Georgiana, P.A.; Vizireanu, C. Effects of probiotic bacillus species in aquaculture—An overview. Ann. Univ. Dunarea De Jos Galati 2014, 38, 9–17. [Google Scholar]
- Xue, M.; Wu, Y.; Hong, Y.; Meng, Y.; Xu, C.; Jiang, N.; Li, Y.; Liu, W.; Fan, Y.; Zhou, Y. Effects of dietary Bacillus amyloliquefaciens on the growth, immune responses, intestinal microbiota composition and disease resistance of yellow catfish, Pelteobagrus fulvidraco. Front. Cell Infect. Microbiol. 2022, 12, 1047351. [Google Scholar] [CrossRef] [PubMed]
- Van Doan, H.; Wangkahart, E.; Thaimuangphol, W.; Panase, P.; Sutthi, N. Effects of Bacillus spp. Mixture on growth, immune responses, expression of immune-related genes, and resistance of Nile tilapia against Streptococcus agalactiae infection. Probiotics Antimicrob. Proteins 2023, 15, 363–378. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.T.; Hu, Y.F.; Lee, B.H.; Huang, C.Y.; Lin, Y.R.; Huang, S.N.; Chen, Y.Y.; Chang, J.J.; Nan, F.H. Dietary of Lactobacillus paracasei and Bifidobacterium longum improve nonspecific immune responses, growth performance, and resistance against Vibrio parahaemolyticus in Penaeus vannamei. Fish Shellfish Immunol. 2022, 128, 307–315. [Google Scholar] [CrossRef]
- Yang, Y.; He, J.; Suo, Y.; Lv, L.; Wang, J.; Huo, C.; Zheng, Z.; Wang, Z.; Li, J.; Sun, W.; et al. Anti-inflammatory effect of taurocholate on TNBS-induced ulcerative colitis in mice. Biomed. Pharmacother. 2016, 81, 424–430. [Google Scholar] [CrossRef]
- Rivera, J.; Proia, R.L.; Olivera, A. The alliance of sphingosine-1-phosphate and its receptors in immunity. Nat. Rev. Immunol. 2018, 8, 753–763. [Google Scholar] [CrossRef]
- Ma, S.; Sandhoff, R.; Luo, X.; Shang, F.; Shi, Q.; Li, Z.; Wu, J.; Ming, Y.; Schwarz, F.; Madi, A.; et al. Serine enrichment in tumors promotes regulatory T cell accumulation through sphinganine-mediated regulation of c-Fos. Sci. Immunol. 2024, 9, eadg8817. [Google Scholar] [CrossRef]
- Kong, W.; Wang, Z.; Chen, N.; Mei, Y.; Li, Y.; Yue, Y. SHMT2 regulates serine metabolism to promote the progression and immunosuppression of papillary renal cell carcinoma. Front. Oncol. 2022, 12, 914332. [Google Scholar] [CrossRef]
- Subramanya, S.; Goswami, M.T.; Miller, N.; Weh, E.; Chaudhury, S.; Zhang, L.; Andren, A.; Hager, H.; Weh, K.M.; Lyssiotis, C.A.; et al. Rod photoreceptor-specific deletion of cytosolic aspartate aminotransferase, GOT1, causes retinal degeneration. Front. Ophthalmol. 2023, 3, 1306019. [Google Scholar] [CrossRef]
- Xu, W.; Patel, C.H.; Zhao, L.; Sun, I.H.; Oh, M.H.; Sun, I.M.; Helms, R.S.; Wen, J.; Powell, J.D. GOT1 regulates CD8+ effector and memory T cell generation. Cell Rep. 2023, 42, 111987. [Google Scholar] [CrossRef]
- Wang, L.; Dong, X.; Wu, Y.; Zhou, Q.; Xu, R.; Ren, L.; Zhang, C.; Tao, M.; Luo, K.; Zeng, Y.; et al. Proteomics-based molecular and functional characteristic profiling of muscle tissue in triploid crucian carp. Mol. Omics 2022, 18, 967–976. [Google Scholar] [CrossRef] [PubMed]
- Abu-Elheiga, L.; Almarza-Ortega, D.B.; Baldini, A.; Wakil, S.J. Human acetyl-CoA carboxylase 2. Molecular cloning, characterization, chromosomal mapping, and evidence for two isoforms. J. Biol. Chem. 1997, 272, 10669–10677. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Peng, L.; Zhang, Y.; Liu, Z.; Li, W.; Chen, S.; Li, G. The identification of key genes and pathways in hepatocellular carcinoma by bioinformatics analysis of high-throughput data. Med. Oncol. 2017, 34, 101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhou, Y.; Hu, J.; Yu, X.; Xu, H.; Ba, Z.; Zhang, H.; Sun, Y.; Wang, R.; Du, X.; et al. Comprehensive analysis identifies cuproptosis-related gene DLAT as a potential prognostic and immunological biomarker in pancreatic adenocarcinoma. BMC Cancer 2023, 23, 560. [Google Scholar] [CrossRef]
- Chen, K.; Shi, Y.; Zhu, H. Analysis of the role of glucose metabolism-related genes in dilated cardiomyopathy based on bioinformatics. J. Thorac. Dis. 2023, 15, 3870–3884. [Google Scholar] [CrossRef]
- Kydonopoulou, K.; Rousso, D.; Ilonidis, G.; Mandala, E. Association of GPIa C807T polymorphism with unexplained female infertility and IVF implantation failure. J. Biol. Regul. Homeost. Agents 2019, 33, 883–887. [Google Scholar]
- Adorno-Cruz, V.; Liu, H. Regulation and functions of integrin α2 in cell adhesion and disease. Genes Dis. 2018, 6, 16–24. [Google Scholar] [CrossRef]
- Jin, L.; Duan, Y.; Li, X.; Li, Z.; Hu, J.; Shi, H.; Su, Z.; Li, Z.; Du, B.; Chen, Y.; et al. High expression ITGA2 affects the expression of MET, PD-L1, CD4 and CD8 with the immune microenvironment in pancreatic cancer patients. Front. Immunol. 2023, 14, 1209367. [Google Scholar] [CrossRef]
- Kausar, S.; Abbas, M.N.; Gul, I.; Liu, Y.; Tang, B.P.; Maqsood, I.; Liu, Q.N.; Dai, L.S. Integrins in the immunity of insects: A review. Front. Immunol. 2022, 13, 906294. [Google Scholar] [CrossRef]
- Jiang, P.; Du, W.; Wu, M. Regulation of the pentose phosphate pathway in cancer. Protein Cell 2014, 5, 592–602. [Google Scholar] [CrossRef]
- Cen, K.; Lu, C. Prognostic and immune infiltration analysis of transaldolase 1 (TALDO1) in hepatocellular carcinoma. Int. J. Gen. Med. 2023, 16, 5779–5788. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.S.; Stone, E.F.; Francis, R.O.; Karafin, M.S. The global role of G6PD in infection and immunity. Front. Immunol. 2024, 15, 1393213. [Google Scholar] [CrossRef] [PubMed]
- Du, G.; Xiao, M.; Wei, X.; Zhou, C.; Li, S.; Cai, W. Hepatic transcriptional profiling response to fava bean-induced oxidative stress in glucose-6-phosphate dehydrogenase-deficient mice. Gene 2018, 652, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Bohorquez, D.; Gallego López, I.M.; Jaeger, B.N.; Pfammatter, S.; Bowers, M.; Semenkovich, C.F.; Jessberger, S. FASN-dependent de novo lipogenesis is required for brain development. Proc. Natl. Acad. Sci. USA 2022, 119, e2112040119. [Google Scholar] [CrossRef]
- Xiao, Y.; Yang, Y.; Xiong, H.; Dong, G. The implications of FASN in immune cell biology and related diseases. Cell Death Dis. 2024, 15, 88. [Google Scholar] [CrossRef]
- Deng, Y.; Zhang, Y.; Chen, H.; Xu, L.; Wang, Q.; Feng, J. Gut-liver immune response and gut microbiota profiling reveal the pathogenic mechanisms of Vibrio harveyi in pearl gentian grouper (Epinephelus lanceolatus♂ × E. fuscoguttatus♀). Front. Immunol. 2020, 11, 607754. [Google Scholar] [CrossRef]
- Yan, B.; Sun, Y.; Fu, K.; Zhang, Y.; Lei, L.; Men, J.; Guo, Y.; Wu, S.; Han, J.; Zhou, B. Effects of glyphosate exposure on gut-liver axis: Metabolomic and mechanistic analysis in grass carp (Ctenopharyngodon idellus). Sci. Total Environ. 2023, 902, 166062. [Google Scholar] [CrossRef]
- Del Piano, F.; Mateu, B.; Coretti, L.; Borrelli, L.; Piccolo, G.; Addeo, N.F.; Esposito, S.; Mercogliano, R.; Turco, L.; Meli, R.; et al. Polystyrene microplastic exposure modulates gut microbiota and gut-liver axis in gilthead seabream (Sparus aurata). Sci. Total Environ. 2024, 957, 177857. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, J.; Wang, G.; Chen, H.; Cao, J.; Xie, L.; Luo, Y. Interactive effects of fluoride and seleno-l-methionine at environmental related concentrations on zebrafish (Danio rerio) liver via the gut-liver axis. Fish Shellfish Immunol. 2022, 127, 690–702. [Google Scholar] [CrossRef]
- Duan, Y.; Yang, Y.; Li, H.; Zhang, Z.; Chen, X.; Xiao, M.; Nan, Y. The toxic effects of tetracycline exposure on the physiological homeostasis of the gut-liver axis in grouper. Environ. Res. 2024, 258, 119402. [Google Scholar] [CrossRef]
- Song, Y.; Huang, H.; Jia, K.; Zou, S.; Yang, Y.; Yi, M. Multi-omics analysis reveals toxicity and gut-liver axis disruption induced by polychlorinated biphenyls exposure in yellowfin seabream (Acanthopagrus latus). J. Hazard. Mater. 2025, 487, 137296. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Luo, X.; Zhang, Y.; Zhou, Y.; Xiao, Q.; Huang, X.; Xu, X.; Xu, X.; Qin, Q.; Liu, S. Triploidization modulates intestinal microbiota and promotes growth in Carassius auratus. Aquaculture 2023, 571, 739480. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, C.; Gao, Z.; Pan, Z.; Peng, M.; Ma, J.; Qian, Y.; Guo, J.; Fu, F. Anti-obesity mechanism of Ganpu tea revealed by microbiome, metabolome and transcriptome analyses. Food Chem. 2023, 412, 135048. [Google Scholar] [CrossRef] [PubMed]



| Parameters | DRGC | GC |
|---|---|---|
| Villus length (μm) | 533.35 ± 22.43 a | 376.81 ± 21.23 |
| Thickness of gut wall (μm) | 77.36 ± 4.22 a | 57.81 ± 4.77 |
| Villus width (μm) | 79.60 ± 6.53 a | 109.45 ± 9.30 |
| Sample | Clean Reads | GC Content (%) | Q30 (%) | Uniquely Mapped Reads (Ratio) | Multiple Mapped Reads (Ratio) | Total Mapped Reads (Ratio) |
|---|---|---|---|---|---|---|
| DRGC-1 | 50,043,924 (99.80%) | 46.52 | 96.16 | 43,520,258 (86.96%) | 2,771,962 (5.54%) | 46,292,220 (92.5%) |
| DRGC-2 | 47,104,516 (99.79%) | 46.41 | 96.20 | 40,953,442 (86.94%) | 2,490,242 (5.29%) | 43,443,684 (92.23%) |
| DRGC-3 | 45,800,350 (99.81%) | 46.56 | 96.25 | 39,678,432 (86.63%) | 2,449,190 (5.35%) | 42,127,622 (91.98%) |
| GC-1 | 44,884,616 (99.82%) | 46.57 | 96.19 | 38,596,774 (85.99%) | 2,267,454 (5.05%) | 40,864,228 (91.04%) |
| GC-2 | 47,683,850 (99.80%) | 46.70 | 96.22 | 40,773,060 (85.51%) | 2,640,728 (5.54%) | 43,413,788 (91.05%) |
| GC-3 | 43,187,696 (99.85%) | 46.62 | 96.56 | 37,276,028 (86.31%) | 2,461,866 (5.70%) | 39,737,894 (92.01%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Li, Z.; Huang, X.; Xu, X.; Xu, X.; Zhang, K.; Zhou, Y.; Bai, J.; Liu, Z.; Jiang, Y.; et al. Multi-Omic Analysis Reveals the Potential Anti-Disease Mechanism of Disease-Resistant Grass Carp. Int. J. Mol. Sci. 2025, 26, 3619. https://doi.org/10.3390/ijms26083619
Wang C, Li Z, Huang X, Xu X, Xu X, Zhang K, Zhou Y, Bai J, Liu Z, Jiang Y, et al. Multi-Omic Analysis Reveals the Potential Anti-Disease Mechanism of Disease-Resistant Grass Carp. International Journal of Molecular Sciences. 2025; 26(8):3619. https://doi.org/10.3390/ijms26083619
Chicago/Turabian StyleWang, Chongqing, Zeyang Li, Xu Huang, Xidan Xu, Xiaowei Xu, Kun Zhang, Yue Zhou, Jinhai Bai, Zhengkun Liu, Yuchen Jiang, and et al. 2025. "Multi-Omic Analysis Reveals the Potential Anti-Disease Mechanism of Disease-Resistant Grass Carp" International Journal of Molecular Sciences 26, no. 8: 3619. https://doi.org/10.3390/ijms26083619
APA StyleWang, C., Li, Z., Huang, X., Xu, X., Xu, X., Zhang, K., Zhou, Y., Bai, J., Liu, Z., Jiang, Y., Tang, Y., Deng, X., Li, S., Hu, E., Peng, W., Xiong, L., Xiao, Q., Yang, Y., Qin, Q., & Liu, S. (2025). Multi-Omic Analysis Reveals the Potential Anti-Disease Mechanism of Disease-Resistant Grass Carp. International Journal of Molecular Sciences, 26(8), 3619. https://doi.org/10.3390/ijms26083619





