Abstract
In China, soil contamination by heavy metals is a widespread issue, with substantial increases in lead(Pb), cadmium(Cd), copper(Cu), and zinc(Zn) levels observed across various regions. Particularly, the concentrations of Pb and Cd significantly exceed their natural background levels. P-ATPases, a group of proteins, utilize energy from ATP hydrolysis to support the transmembrane movement of metal ions. This group encompasses several Heavy Metal Associated Transporter (HMA) ATPases. Studies on hyperaccumulators have shown the critical role of HMAs in the movement and reduction in Zn and Cd toxicity in plant systems. This research identifies a protein encoded by the SpHMA gene from Sedum plumbizincicola, a species noted for aiding Zn/Cd hyperaccumulators, which enhances tolerance to Cd and Zn. We detail a protein encoded by SpH/A within the HMA family that enhances Cd tolerance. Real-time fluorescence quantification (RT-PCR) indicates that SpHMA3 expression in Arabidopsis thaliana and Zea mays KN5585 correlates with high Cd tolerance, linked to Cd accumulation in Zea mays. In addition, homozygous Arabidopsis thaliana AtHMA3 mutants exhibited increased Cd sensitivity compared to the wild type (WT). Notably, plants of Arabidopsis thaliana and maize overexpressing SpHMA3 showed enhanced Cd stress tolerance compared to WT. Enhanced Cd accumulation in tissues was observed when SpHMA3 was overexpressed, as revealed by subcellular distribution analysis. We propose that SpHMA3 augments maize tolerance to Cd and Zn stresses through enhanced cellular uptake and translocation of Cd ions. This investigation clarifies the gene function of SpHMA3 in Cd and Zn stress response, offering insights for enhancing heavy metal absorption traits in maize varieties and phytoremediation methods for soils contaminated with heavy metals.
1. Introduction
Heavy metals refer to metallic elements with densities greater than 5 g/cm3, such as lead, mercury, cadmium, chromium, etc. They are widely found in nature, but heavy metal pollution is a growing problem due to human practices such as industrial activities, mining, and agriculture. The importance of studying heavy metals lies in their potential harm to the environment and health. Heavy metals are not easy to degrade, can accumulate in soil and water, and enter the human body through the food chain, resulting in chronic poisoning, damage to the nervous system, kidney, and immune system. In addition, heavy metal pollution can damage ecosystems and affect biodiversity. Therefore, it is very important for environmental protection and human health to study the sources, migration rules, and treatment methods of heavy metals. The swift pace of industrialization has led to significant environmental degradation, notably soil contamination with heavy metals in China, now a pressing issue [1]. Lead (Pb) and cadmium (Cd) concentrations are notably high in soils, with approximately ten percent of soil in China affected by Cd contamination, impacting farmlands across various regions to varying extents. Such contamination poses severe threats to ecological health, endangers public health, and impacts human activities and well-being [2,3]. Cd, recognized as a non-essential and harmful element, disrupts the growth of diverse biological entities [4,5]. Its inherent toxicity, persistence, and resistance to biodegradation mean even minimal exposure can culminate in significant Cd accumulation in humans, adversely affecting vital organs such as the kidneys, liver, and lungs [6]. Cd toxicity can hinder plant growth and development; high levels can cause symptoms including stunted growth, leaf and stem yellowing, reduced biomass [7], and in extreme cases, cessation of growth or plant death due to dehydration [8]. Maize is a crucial cereal crop globally. Since 1998, its total production has exceeded that of rice and wheat, making it the most widely produced cereal. By 2023, the global maize cultivation area will reach 2.04 × 104 million hectares, with a production output of 1236 million tonnes [9,10,11,12,13]. Due to its high yield and adaptability compared to crops like wheat and rice, maize is frequently used as a model crop for research on soil heavy metal remediation. Some previous studies have indicated that lower concentrations of Cd stress did not adversely affect maize plants [14,15]. When exposed to higher concentrations of Cd, maize exhibited a marked decline in chlorophyll content, weakened photosynthetic activity, and reduced root and stem growth, ultimately leading to decreased yields [16,17].
PIB-type ATPases, a subfamily of P-type ATPases (HMAs), utilize the energy from ATP hydrolysis to facilitate the transmembrane transport of ions [18,19,20]. These transporters primarily mediate the transport of Zn2+ but are also involved in the movement of heavy metal ions harmful to plants, such as Cd2+ and Pb2+. Based on the substrate specificity, HMA transporters are classified into the Zn/Co/Cd/Pb subgroup and the Cu/Ag subgroup [21]. Studies have shown that the genomes of Arabidopsis thaliana and the hyperaccumulator Sedum plumbizincicola contain eight HMA genes, while Oryza sativa Japonica Group possesses nine HMA genes. Notably, AtHMA1-AtHMA4, OsHMA1-OsHMA3, and SpHMA4-SpHMA8 belong to the Cu/Ag subgroup, whereas AtHMA5-AtHMA8, OsHMA4-OsHMA9, and SpHMA1-SpHMA3 are part of the Zn/Co/Cd/Pb subgroup [21]. In Arabidopsis thaliana, AtHMA1 resides within the chloroplast periplasm, facilitating the Zn expulsion from chloroplasts; the upregulation of AtHMA3 in these plants heightens their resilience to and accumulation of Cd, Zn, Pb, and Co [22]. AtHMA4-deficient mutants exhibit heightened sensitivity to Cd and Zn compared to their wild-type counterparts. In the Japonica Group of Oryza sativa, OsHMA1 plays a role in Zn transportation [23], while OsHMA3 is exclusive to Cd transport and serves as a Cd barrier within root cell vesicles [24]. The mRNA of the AtHMA3 protein is extensively expressed across various tissues of Arabidopsis thaliana and is located in the vesicular membrane, where it facilitates the transport of Cd into vesicles, thereby contributing to Cd compartmentalization. Overexpression of this gene in other plant species led to a significant increase in Cd accumulation in transgenic plants compared to wild-type plants, indicating that AtHMA3 plays a role in Cd storage within plants [25].
Phytoremediation offers an environmentally friendly and safe approach to restoring contaminated land. Sedum plumbizincicola, a perennial succulent herb, thrives in areas abundant in Pb and Zn minerals [26]. However, its narrow range of adaptation, low economic value, and other limitations have significantly restricted its application and promotion. In contrast, advancements in genetic engineering to enhance plant uptake of heavy metals provide new opportunities for the phytoremediation of contaminated soils. SpHMA3, a heavy metal transporter found in hyperaccumulator plants, exhibits greater efficiency in transporting cadmium (Cd) and zinc (Zn) than other HMA3 genes, and is able to more effectively sequester heavy metal ions into vacuoles, thereby enhancing plant tolerance and accumulation. The transport mechanism of SpHMA3 may have higher substrate specificity or stronger affinity, which makes it better in the recognition and transport of heavy metal ions. In terms of practical application, SpHMA3 shows significant potential in the field of phytoremediation. Through genetic engineering, plants can enhance the repair ability of heavy metal contaminated soil. Compared with other HMA3 genes, SpHMA3 has more advantages in efficiency and application prospects. This plant species is capable of extracting and hyperaccumulating Zn and Cd from soils contaminated with heavy metals, making it a viable candidate for phytoremediation of Zn and Cd contamination [27,28]. It is also important to note that traditional maize cultivation not only suffers yield losses due to Cd stress but also experiences a decline in kernel quality as a result of Cd presence. Therefore, enhancing Cd tolerance in maize and diminishing Cd accumulation in kernels significantly enhance both the yield and quality of maize. The HMA family, notable for its role as a heavy metal transport protein, participates in Cd transport across various plant species; however, its association with maize remains underexplored. This research identifies the SpHMA3 gene as a close counterpart to the AtHMA3 gene from Arabidopsis. Moreover, Arabidopsis mutants lacking the AtHMA3 gene exhibited heightened Cd sensitivity. By introducing the SpHMA3 gene into Arabidopsis and maize, plants overexpressing this transgene were developed, affirming the influence of the gene on Cd absorption and transportation. Thus, the findings from this investigation are poised to furnish a theoretical framework for employing biotechnological strategies to bolster Cd resistance in maize and genetically enhance the capacity of maize to manage Cd accumulation. At the same time, this study also provides a new bioremediation path for improving crops to solve the problem of heavy metal pollution in agricultural systems.
2. Results
2.1. Phylogenetic Tree and Conserved Structural Domain Analysis of HMA Family Genes in Several Major Crops
To obtain the CDS of the SpHMA3 gene from Sedum plumbizincicola (KY312109.1), we referred to the sequence available on the NCBI database (NCBI: https://www.ncbi.nlm.nih.gov/search/all/?term=KY312109.1, accessed on 10 September 2019) and downloaded the complete sequence. To explore the evolutionary relationships between the SpHMA3 gene of Sedum plumbizincicola and the HMA gene family members of Zea mays, Sorghum bicolor, Oryza sativa Japonica Group, Cicer arietinum, Nicotiana tomentosiformis, Triticum aestivum, and Arabidopsis thaliana, we retrieved the gene sequences of these species through NCBI (NCBI: https://www.ncbi.nlm.nih.gov/, accessed on 20 September 2019). An evolutionary tree was constructed using MEGA based on amino acid multiple sequence alignments, comparing Sedum plumbizincicola (SpHMA3) with Zea mays (ZmHMA1-ZmHMA5), Sorghum bicolor (SbHMA1, SbHMA3, SbHMA5), Oryza sativa Japonica Group (OsHMA1-OsHMA5), Arabidopsis thaliana (AtHMA1-AtHMA5), Cicer arietinum (CaHMA1-CaHMA5), Nicotiana tomentosiformis (NtHMA1-NtHMA5), and Triticum aestivum (TaHMA1-TaHMA5) (Figure 1A). Investigations revealed that the SpHMA3 gene exhibits the closest relation to AtHMA3 from Arabidopsis thaliana, and shares similarities with AtHMA2 and AtHMA4 genes as well. The DNAMAN 9 software was utilized to assess sequence similarities among SpHMA3, AtHMA2, AtHMA3 and AtHMA4, revealing a 52.3% homology between SpHMA3 and AtHMA3. The SpHMA3 gene was identified to possess the E1-E2 ATPase and the haloacid-like dehalogenase hydrolase structural domains, as determined by Pfam (Pfam: http://pfam.xfam.org/, accessed on 10 October 2019) and independently confirmed by SOSUI (SOSUI: https://harrier.nagahama-i-bio.ac.jp/sosui/, accessed on 23 October 2019), which also indicated eight transmembrane structural domains within SpHMA3 (Figure 1B).
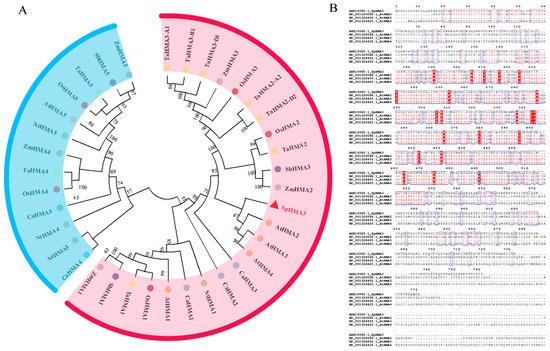
Figure 1.
Phylogenetic construction and sequence conservation analysis of the HMA gene family (A) phylogenetic analysis of the HMA family in Sedum plumbizincicola, Zea mays, Sorghum bicolor, Oryza sativa Japonica Group, Cicer arietinum, Nicotiana tomentosiformis, Triticum aestivum, and Arabidopsis thaliana; (B) conservative sequence analysis of SpHMA3 and AtHMA2, 3, and 4 in Arabidopsis thaliana.
2.2. Identification of Arabidopsis thaliana athma3 Pure Mutants and Gene Transcription Levels
Protein sequence comparison revealed that the homologous gene to SpHMA3 in Arabidopsis thaliana is AtHMA3. Therefore, AtHMA3 mutant seeds were obtained from the American Arabidopsis thaliana Seed Resource Center (ABRC). Based on the information available from the TAIR database (https://www.arabidopsis.org/, accessed on 5 December 2022) and sequencing results, the T-DNA insertion site in the mutant was identified to be located in the promoter region of the AtHMA3 gene, specifically within the first 155 bp upstream of the start codon ATG (Supplementary Figure S1A). The athma3 mutant was propagated in a soil-based culture system, and its purity was verified using the three-primer method. Primers LP, RP, and LB3.1 were designed for detecting the homozygous mutant strain, and the design is available online at http://signal.salk.edu/. The purity of the Arabidopsis thaliana athma3 mutants was confirmed to be high (Supplementary Figure S1B).
Variations in the T-DNA insertion sites may influence gene transcript levels. To explore how the homozygous loss of SpHMA3 function impacts Arabidopsis under Cd stress, the AtHMA3 gene expression in the mutant was analyzed. qT-PCR results (Supplementary Figure S1C) revealed that AtHMA3 expression in the mutant was significantly lower, at 23.4% compared to WT. This suggests that the AtHMA3 gene was only partially knocked out in the mutant.
2.3. Mutant of AtHMA3 Decreased Resistance to Cd Stress in Arabidopsis
To investigate the impact of varying Cd stress concentrations on the growth of Arabidopsis thaliana mutant seedlings, three CdCl2 concentrations were used: 0 μmol/L, 40 μmol/L, and 80 μmol/L, with parallel experiments conducted for each. Germination rates were recorded on the 7th day (Figure 2B). Under normal conditions, the germination rate of athma3 mutants was comparable to WT at 100%. Under 40 μmol/L CdCl2 stress, the germination rate of the mutant decreased to 71.7%, whereas that of WT was 91.7%. When the stress concentration was increased to 80 μmol/L, the germination rate of the mutant further decreased to 53.9%, while that of WT was 70.6% (Figure 2A). Under Cd stress, root length of athma3 mutants was significantly shorter than WT. Root lengths were measured and showed notable differences between the mutant and WT (Figure 2C). Under non-stress conditions, primary root length of the athma3 mutant was similar to WT. Under 40 μmol/L CdCl2 stress, the primary and total root lengths of WT were 1.83 and 1.94 times that of the athma3 mutant. At 80 μmol/L CdCl2, these values increased to 3.97 and 3.28 times, respectively (Figure 2D,E). The inhibition of root length became more pronounced with higher Cd concentrations, indicating that the AtHMA3 gene plays a crucial role in the response of Arabidopsis thaliana to Cd stress.
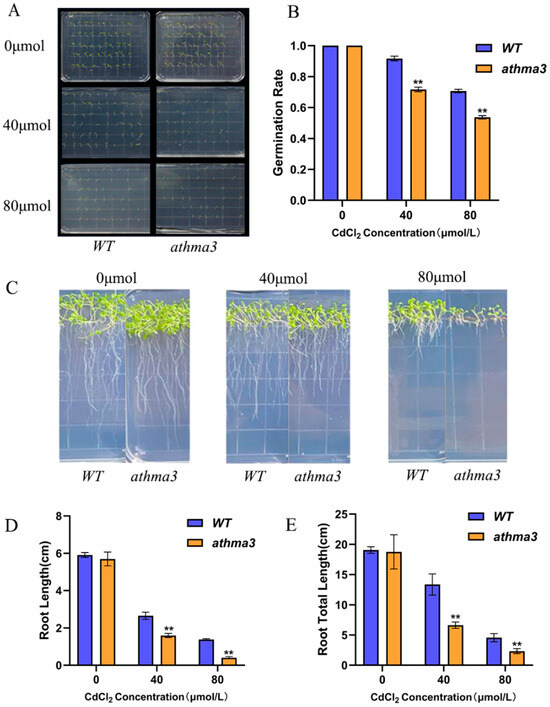
Figure 2.
Analysis of phenotypic and root characteristics in Arabidopsis thaliana AtHMA3 homozygous mutants: (A) Comparison of phenotype between Arabidopsis thaliana AtHMA3 homozygous mutants and WT during germination exposed to varying Cd concentrations; (B) Germination rates for both Arabidopsis thaliana AtHMA3 homozygous mutants and WT seeds assessed on day 7 across different Cd stress levels; (C) Graphical representation depicting variations in root length and overall root system development in Arabidopsis thaliana AtHMA3 homozygous mutants and WT under varied Cd concentrations; (D,E) Charts displaying primary root length and total root system extent under Cd stress conditions; ** denotes p < 0.01; analyzed using Student’s t-test.
2.4. Decreased Cd Accumulation in the Arabidopsis thaliana athma3 Mutant
To examine the capability of Arabidopsis thaliana athma3 mutant plants to accumulate Cd under Cd stress conditions, experiments were performed using Cd stress at concentrations of 40 μmol/L and 80 μmol/L. Both WT and athma3 mutant seeds were sterilized and cultivated on 1/2MS medium supplemented with varying gradients of CdCl2 for 14 days. Arabidopsis thaliana seedlings subjected to different Cd treatments were collected, dried, and weighed for nitrification, and their Cd content was measured using an inductively coupled plasma mass spectrometer. The results showed that the athma3 mutant had lower Cd content compared to WT under both stress concentrations. Under the 40 μmol/L treatment, the Cd content in the athma3 mutant was 87.6% of that in the WT, while under the 80 μmol/L treatment, it was 74.8% of that in the WT (Figure 3). These findings suggest that a significant reduction or loss of AtHMA3 gene expression resulted in decreased Cd accumulation in the plants.
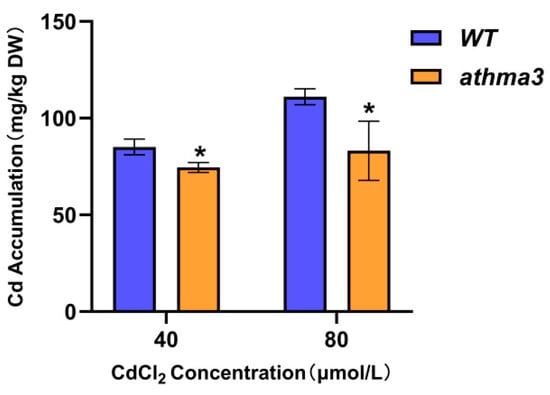
Figure 3.
Variation in Cd levels in Arabidopsis thaliana athma3 mutant and WT under varying Cd stress conditions; * denotes p ≤ 0.05; analyzed using Student’s t-test.
2.5. Overexpression of ZmHMA3 Increases Tolerance to Heavy Metal Stress in Arabidopsis thaliana
To investigate the role of SpHMA3 in response to Cd stress, the SpHMA3 gene was introduced into Arabidopsis thaliana, resulting in nine transgenic lines, designated SpHMA3OE1-SpHMA3OE9, later renamed AtOE1-AtOE9. During subsequent cultivation, due to sterility issues, only three lines, AtOE2, AtOE4, and AtOE6, were retained for further study. The germination rates and agronomic characteristics of these transgenic Arabidopsis seedlings were assessed under various heavy metal stress conditions (Figure 4A–E).
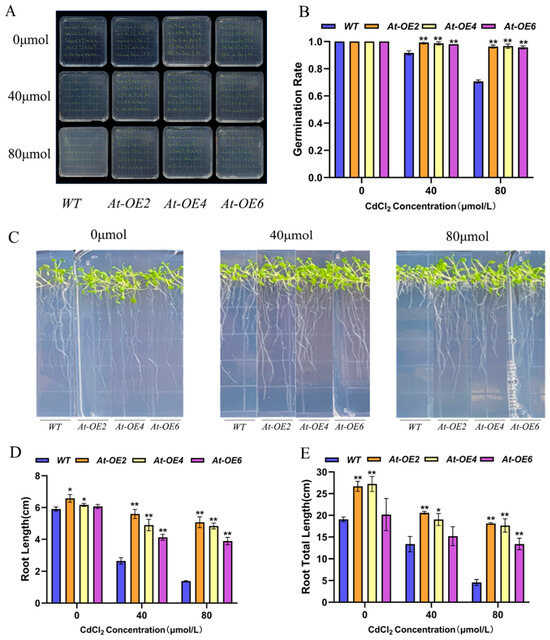
Figure 4.
Phenotypic characterization and root trait analysis of Arabidopsis thaliana overexpressing SpHMA3 gene transformation events: (A) Seventh-day germination phenotypes of Arabidopsis thaliana SpHMA3 overexpression materials and WT seeds under varying Cd stress concentrations; (B) Seventh-day germination rates of Arabidopsis thaliana SpHMA3 overexpression materials and WT seeds under varying Cd stress concentrations. (C) Identification of root growth of Arabidopsis thaliana SpHMA3 overexpression and WT plants at 14 d under varying Cd stress concentrations; (D) Determination of primary root length of Arabidopsis thaliana SpHMA3 overexpression material and WT seeds at 14 d under varying Cd stress concentrations; (E) Determination of total root length of Arabidopsis thaliana SpHMA3 overexpression material and WT seeds at 14 d under varying Cd stress concentrations; * denotes p ≤ 0.05; ** denotes p ≤ 0.01; analyzed using Student’s t-test.
Seeds of both Arabidopsis thaliana overexpression lines (OE) and WT were germinated on 1/2MS medium containing CdCl2 concentrations of 0 μmol/L, 40 μmol/L, and 80 μmol/L in three sets of parallel experiments, with germination rates recorded on day 7. Under no-stress conditions, both the WT and OE plants grew normally. Cd stress reduced germination in both plant types, though the OE lines exhibited higher germination rates compared to WT under stress (Figure 4A). Under 40 μmol CdCl2 stress, the germination rates of At-OE2, At-OE4, and At-OE6 were 99.4%, 98.9%, and 98.3%, respectively, while the WT showed a rate of 91.7%. Under 80 μmol CdCl2 stress, At-OE2, At-OE4, and At-OE6 had germination rates of 96.1%, 96.7%, and 95.6%, respectively, with the WT at 70.6% (Figure 4B). Meanwhile, after 14 days of growth under the same conditions, the root lengths of transgenic and WT plants exposed to varying Cd concentrations were measured. Overexpression of the SpHMA3 gene enhanced seedling growth, particularly under Cd stress, showing more pronounced growth compared to the WT (Figure 4C). Under 40 μmol CdCl2 stress, the primary roots of At-OE2, At-OE4, and At-OE6 were 2.11, 1.85, and 1.56 times longer than those of the WT, while total root lengths were 1.54, 1.43, and 1.14 times longer. At 80 μmol CdCl2 stress, the primary roots of At-OE2, At-OE4, and At-OE6 were 3.67, 3.5, and 2.72 times longer, with total root lengths 3.98, 3.87, and 2.93 times longer than the WT (Figure 4D,E). This indicates that when the Arabidopsis thaliana SpHMA3 gene was overexpressed, the overexpression plants showed better tolerance and environmental adaptation under Cd stress compared with the WT.
2.6. Overexpression of SpHMA3 Improves Tolerance to Heavy Metal Stress in Maize
To determine the function of SpHMA3 in maize in response to Cd stress, we transformed the SpHMA3 gene into Zea mays KN5585 (WT) and obtained a total of two transgenic maize materials, SpHMA3-OE1 and SpHMA3-OE2, named Zm-OE1 and Zm-OE2. We compared the differences in germination rates between overexpressed maize and the WT under different heavy metal stresses (Figure 5A,B). The KN5585 (WT) and OE plants were exposed to varying concentrations of CdCl2·2.5H2O solutions, specifically 0.3 mg/L, 3 mg/L, 10 mg/L, and 20 mg/L, for a set period. A comparison of growth phenotypes between KN5585 and OE plants is illustrated in Figure 5B. It is observed that the seed germination rates of WT, Zm-OE1, and Zm-OE2 initially increased and then decreased as the Cd stress concentration rose. The highest germination rates, 83.33%, 93.33%, and 95.00%, respectively, were recorded at 0.3 mg/L Cd, while the lowest rate, 68.33%, was seen at 20 mg/L. As the Cd concentration increased, OE plants consistently showed significantly higher germination rates than the WT, indicating better adaptation to Cd stress. These results suggest that Cd stress considerably inhibits WT seed germination, while the OE material may possess a protective mechanism that reduces Cd toxicity, thereby enhancing plant survival.
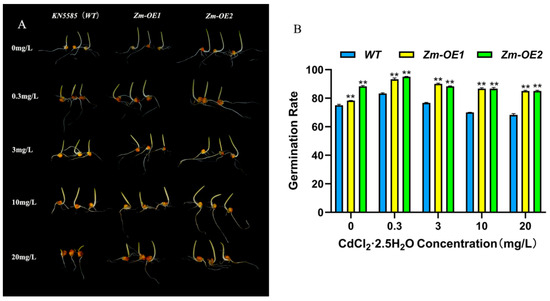
Figure 5.
Germination of WT and OE seeds under Cd stress conditions: (A) Seed germination comparison between WT and OE under varying Cd stress concentrations; (B) Statistical analysis of seed germination rates for WT and OE under varying Cd stress concentrations; ** denotes p ≤ 0.01; analyzed using Student’s t-test.
2.7. Functional Expression of SpHMA3 Under Cd Stress
To investigate the function of SpHMA3 under Cd stress, we examined the relative expression levels in transgenic maize subjected to Cd stress. When exposed to 80 μmol/L CdCl2, RNA samples from Arabidopsis thaliana transformation events At-OE2, At-OE4, and At-OE6 were collected at various time points for analysis of SpHMA3 expression using qT-PCR (Figure 6A). Under 800 μmol/L Cd stress, RNA was extracted from the roots and leaves of transgenic Zea mays lines Zm-OE1 and Zm-OE2, and the SpHMA3 expression patterns were determined via qRT-PCR, with untreated plants serving as the control (Figure 6B). Subcellular localization predictions for SpHMA3, performed using Softberry (http://www.softberry.com/, accessed on 10 September 2020), indicated a strong likelihood of localization in the cytoplasmic membrane based on homology to known localization genes in LocDB, suggesting that SpHMA3 might reside in the cell membrane. To verify this conclusion, the recombinant plasmid was introduced into maize protoplasts alongside the cytoplasmic marker vector PM-RFP. After 12 h of expression, fluorescence confocal microscopy revealed green fluorescence on the cytoplasmic membrane, with yellow fluorescence observed after co-localization with the marker vector (Figure 6C), confirming that SpHMA3 is localized to the maize cell membrane.
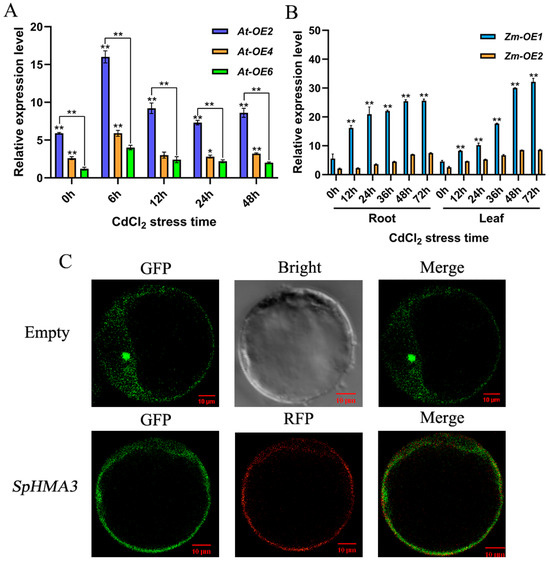
Figure 6.
Tissue-specific expression and subcellular localization of SpHMA3 in maize and Arabidopsis thaliana: (A) Analysis of SpHMA3 expression in converted material of Arabidopsis thaliana; (B) Analysis of SpHMA3 expression in various tissues of maize; (C) Observation of the localization of SpHMA3 in maize protoplasts; * denotes p ≤ 0.05; ** denotes p ≤ 0.01; analyzed using Student’s t-test. Note: (A), Graphs present the relative expression levels of the SpHMA3 gene in the entire Arabidopsis thaliana plant at 6, 12, 24, and 48 h after exposure to 80 μmol/L Cd stress; (B), Graphs depict the relative expression of the SpHMA3 gene in maize root and leaf tissues subjected to 12, 24, 36, 48, and 72 h of Cd stress at a concentration of 800 μmol/L; (C), GFP server represents green fluorescent protein, the RFP server refers to red fluorescent protein, the bright field image shows cell morphology, and the merged image integrates dark fluorescence with cell structure. The scale bar represents 10 μm.
2.8. Overexpression of the SpHMA3 Gene Enhances the Accumulation Enrichment Capacity of Plants to Heavy Metal Stresses
The SpHMA3 gene plays a crucial role in Cd accumulation in the hyperaccumulator Sedum plumbizincicola. To evaluate the Cd enrichment potential of transgenic materials developed by introducing this gene into maize and Arabidopsis thaliana, Cd content was measured and compared between the transgenic lines and wild types under Cd stress conditions. WT, At-OE2, At-OE4, and At-OE6 were grown in media containing 40 μmol/L and 80 μmol/L CdCl2, and whole-plant Cd content was assessed after 14 days (Figure 7A). Meanwhile, maize WT and OE plants were cultivated in soil containing Cd at concentrations of 0 mg/kg, 0.3 mg/kg, 3 mg/kg, 10 mg/kg, and 20 mg/kg. Samples were taken for Cd content analysis during the nodulation stage (Maize-V6), stamen withdrawal (Maize-VT), and maturity (Maize-R6) (Figure 7B–D). The data demonstrated that Cd content in Arabidopsis thaliana overexpression plants was consistently higher than in WT under both concentration treatments (Figure 8A). The Cd levels in At-OE2, At-OE4, and At-OE6 at 40 μmol/L were 1.35, 1.34, and 1.15 times higher, respectively, than in WT, while at 80 μmol/L, the increases were 1.16, 1.11, and 1.06 times, respectively. In maize, Cd levels in overexpression plants surpassed those in WT at all tested concentrations (Figure 7B–D). During the Maize-V6 period, Cd levels in OE plants remained low, but a significant increase was observed in the Maize-VT period, suggesting enhanced Cd uptake and translocation in OE plants. Across all Cd treatments, OE plants consistently showed higher Cd accumulation than the WT, indicating that the SpHMA3 gene markedly enhances Cd enrichment in maize.
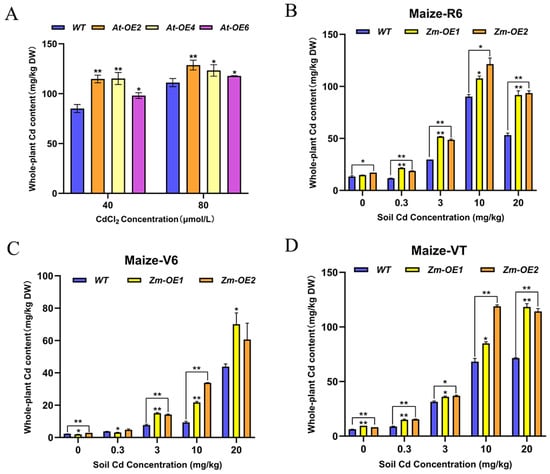
Figure 7.
Comparison of the differences in the ability of WT and SpHMA3 overexpression on Cd accumulation: (A) Cd content of Arabidopsis thaliana SpHMA3 overexpression plants and WT; (B) Cd content of maize SpHMA3 overexpression plants and WT in whole plant of Maize-R6; (C) Cd content of maize SpHMA3 overexpression plants and WT in whole plant of Maize-V6; (D) maize SpHMA3 overexpressing plants and WT in Maize-VT whole plant Cd content; * denotes p ≤ 0.05; ** denotes p ≤ 0.01; analyzed using Student’s t-test.
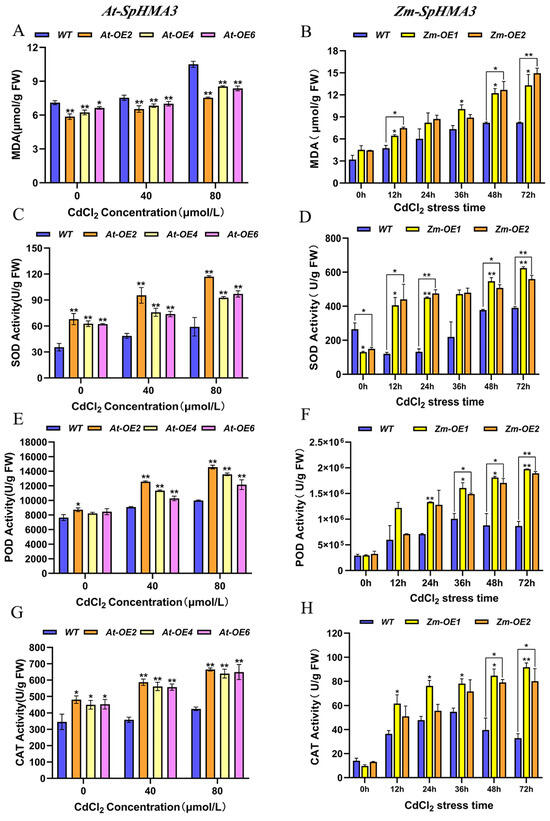
Figure 8.
Physiological and biochemical expression level analysis of SpHMA3 transgenic maize and Arabidopsis thaliana under Cd stress conditions: (A) Influence of Cd stress on MDA levels in Arabidopsis thaliana MDA; (B) Influence of Cd stress on MDA levels in maize; (C) Influence of Cd stress on SOD activity in Arabidopsis thaliana; (D) Influence of Cd stress on SOD activity in maize; (E) Influence of Cd stress on POD activity in Arabidopsis thaliana; (F) Influence of Cd stress on POD activity in maize; (G) Influence of Cd stress on CAT activity in Arabidopsis thaliana; (H) Influence of Cd stress on CAT activity in maize; * denotes p ≤ 0.05; ** denotes p ≤ 0.01; analyzed using Student’s t-test.
2.9. Physiological Response of SpHMA3 Transgenic Maize and Arabidopsis Under Cd Stress
To examine the physiological and biochemical responses of transgenic Arabidopsis thaliana plants under Cd stress, both WT and OE Arabidopsis thaliana were cultivated in media containing 40 μmol/L and 80 μmol/L CdCl2 for 14 days. The MDA content and activities of the enzymes CAT, POD, and SOD were measured (Figure 8A,C,E,G). Meanwhile, transgenic maize and WT plants were evaluated, with maize WT and OE grown to the three-leaf stage and then subjected to 80 μmol/LCdCl2 solution for 0, 12, 24, 36, 48, and 72 h. Enzymatic activities and MDA content were assessed in the root systems of maize (Figure 8B,D,F,H). As seen in Figure 8A,B, under Cd stress, the MDA level in OE Arabidopsis thaliana was lower than in WT, while the transgenic maize showed higher MDA levels than WT at certain times. This variation might be due to differences in stress concentration and gene expression. Both Arabidopsis thaliana and maize demonstrated an upward trend in MDA content as Cd concentration and stress duration increased, consistent with the known effect of heavy metal stress on MDA accumulation. Figure 8C–H illustrate that CAT, POD, and SOD enzyme activities increased under Cd stress in both transgenic and WT plants. In particular, the enzymatic activities in transgenic Arabidopsis thaliana were more pronounced under 80 μmol/L CdCl2 than under 40 μmol/L CdCl2 (Figure 8C,E,G). A similar pattern was observed in maize, where CAT, POD, and SOD enzyme activities increased over time in transgenic maize, reaching significantly higher levels compared to WT under certain stress conditions (Figure 8D,F,H). These findings suggest that the expression of the SpHMA3 gene was enhanced as stress levels intensified, leading to increased antioxidant enzyme activity, which likely played a role in protecting the cells from stress.
2.10. Remediation of Cd-Contaminated Soil by Overexpressing SpHMA3 Maize
Maize, as the most widely cultivated crop, exhibits significantly enhanced adaptability to Cd environments when SpHMA3 is overexpressed. Prior research has indicated that maize can absorb Cd from contaminated environments, contributing to the reduction in soil Cd levels. To explore whether SpHMA3-overexpressing maize could be utilized for soil Cd remediation, a study was conducted. In Cd-contaminated farmland (pH 6.87, Cd concentration 14.6 mg/kg), WT and OE KN5585 maize plants were grown under standard field conditions. During the maturation stage, data on plant height, and fresh/dry weights of roots, stems, leaves, male ears, and thousand kernel weights (TKW) were collected (Figure 9A–D). Results showed that Zm-OE1 and Zm-OE2 plants exhibited greater plant height and higher fresh/dry weights of leaves, stems, and male ears compared to WT under Cd stress (Figure 9A–C). Although Cd treatment reduced TKW for both WT and OE materials, the OE consistently maintained higher TKW than the WT, indicating superior bioaccumulation (Figure 9D). Zm-OE2 outperformed the WT in all measured traits, while Zm-OE1 surpassed the WT in all traits except female spikes under Cd stress. Further analysis of Cd content revealed that Zm-OE1 and Zm-OE2 accumulated more Cd in leaves, roots, stems, male spikes, female spikes, and seeds than WT. Among them, in the root system, the Cd contents of SpHMA3-OE1 and SpHMA3-OE2 were highly significant higher than that of WT, elevated by 55.12% and 127.37%, respectively. The Cd content in the stalk, leaves, female and male ears also showed significantly higher overexpression material than the WT (Figure 9E). Determining the Cd content of the whole maize plant, we found that the Cd content of Zm-OE1 and Zm-OE2 was extremely significantly higher than that of WT by 89.14% and 166.87%, respectively (Figure 9F), suggesting that SpHMA3 overexpression materials are more suitable for growth in the Cd environment than the WT.
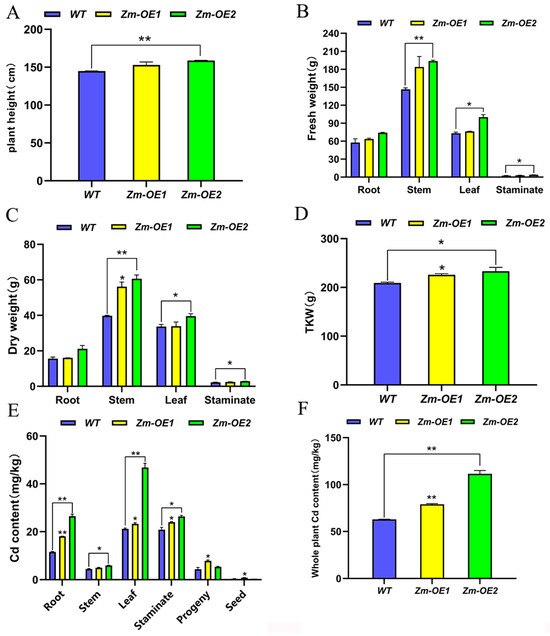
Figure 9.
Changes in agronomic traits and Cd content of maize on Cd-contaminated soil: (A) Changes in maize plant height; (B) Fresh weight distribution across different maize parts; (C) Dry weight accumulation in various parts of maize; (D) Thousand-kernel weight comparison; (E) Examination of Cd distribution across maize parts; and (F) Comparison of the accumulation of Cd by the material as a whole; * denotes p ≤ 0.05; ** denotes p ≤ 0.01; analyzed using Student’s t-test.
2.11. Characterization of Cd Enrichment Transfer in SpHMA3 Overexpression Material
Based on the above studies, we analyzed the enrichment coefficients (BCF) and transfer coefficients (BTF) of WT and overexpressed maize materials for soil Cd in various tissues, organs, and the whole of the maize plant under Cd farmland growing conditions. Figure 10A shows that the overexpression of the SpHMA3 gene in maize led to higher BCF in various tissues compared to WT, particularly in the root system, where Zm-OE1 and Zm-OE2 exhibited Cd BCF that were significantly higher than WT by 55.12% and 127.37%, respectively. This demonstrates that maize roots have a stronger capacity for Cd enrichment compared to other tissues, and the absorbed Cd was primarily concentrated in the roots. SpHMA3-overexpressing maize monocotyledons showed a significantly greater Cd enrichment ability than the WT, with an increase of 1.26 and 1.77 times, respectively (Figure 10B). The analysis of Cd translocation in maize monocots revealed that the WT exhibited a stronger ability to transfer Cd than OE lines (Figure 10C). According to Figure 10D, most of the Cd absorbed by the roots was transported to the leaves, and it was notable that WT had a higher capacity to move Cd from the roots to the leaves, stems, and male ears compared to the OE lines. This may result in increased Cd levels in the aerial parts of the plant, potentially impacting photosynthesis. For a hyperaccumulator, both the BCF and BTF should exceed 1.0. In this study, both BCF and BTF in SpHMA3-overexpressing maize exceeded 1.0, indicating that while this transgenic maize had a higher Cd enrichment capacity than the WT, its translocation ability was lower. This suggests that the overexpressed SpHMA3 gene enhanced the absorption of Cd from the soil and its retention in the root system, thereby limiting its movement to the aerial parts and reducing toxicity in those tissues. Thus, this transgenic material holds potential as a hyperaccumulator for remediating Cd-contaminated soils.
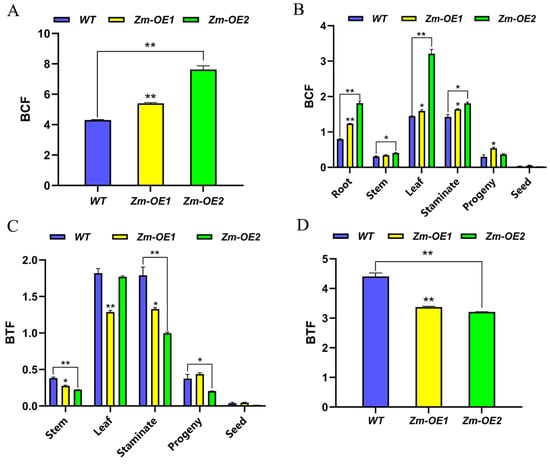
Figure 10.
Enrichment and translocation capacity of maize for soil Cd determined by: (A) enrichment capacity of different parts of maize for Cd; (B) enrichment capacity of maize monocultures for Cd; (C) translocation capacity of different parts of maize for Cd; and (D) translocation capacity of maize monocultures for Cd; * denotes p ≤ 0.05; ** denotes p ≤ 0.01; analyzed using Student’s t-test.
3. Discussion
SpHMA3 was obtained from Zn- and Cd-hyperaccumulating plants located in southeastern China. It is widely acknowledged that most HMA proteins are involved in metal transportation, including Cd transport in vivo. Previous research indicated that SpHMA3 functions as a P-type ATPase heavy metal transporter, enabling Sedum species to accumulate high levels of Cd in their shoots without exhibiting toxicity [29,30]. Cadmium is a highly toxic non-essential element for all organic life forms, including plants and humans. Under Cd stress, plants have evolved multiple protective mechanisms, such as Cd2+ chelation, vesicle sequestration, regulation of Cd2+ uptake, and enhanced antioxidant defense [31]. By isolating the SpHMA3 homolog from the Cd hyperaccumulating S. plumbizincicola, a genetic transformation system was established between this plant and the non-hyperaccumulating ecotype S. alfedii, allowing for the quantification of its expression in various tissues under Cd stress [32]. Although the role of SpHMA3 has been confirmed in organisms such as yeast, southeastern Sedum, and Chinese cabbage, its function in maize under Cd stress remains unexplored. Structural analysis revealed that SpHMA3 shares significant homology with other HMA3 proteins and shows greater similarity to AtHMA2, AtHMA3, and AtHMA4 in Arabidopsis thaliana (Figure 1A). SpHMA3 is part of the Zn/Cd subclass and contains eight transmembrane regions, including two conserved domains, the E1-E2 ATPase and, haloacid dehalogenase hydrolase domains (Figure 1B).
Subcellular localization analysis of maize protoplasts revealed that the SpHMA3 gene was positioned on the cytoplasmic membrane (Figure 6C), which contrasts with its localization on the vesicular membrane in Sedum species [32]. This difference is likely attributed to species-specific variations, resulting in the same gene performing its function in distinct subcellular structures across different species. The results of this study are similar to those of a previous study on the localization of ZmHMA3 genes in maize [33], which are located on cell membranes and are associated with functions such as the transfer and transport of heavy metals in and out of the cell.
Research on the AtHMA3 gene has demonstrated that overexpression of this gene enhances plant tolerance to Cd, whereas the deletion of AtHMA3 reduces tolerance to Cd [34]. AtHMA3 is primarily expressed in the vacuole membrane, and its mechanism of action involves sequestering heavy metals into vacuoles, thereby reducing toxicity to other organelles. The seed germination stage is particularly sensitive to external environmental factors, and heavy metal stress during this phase can negatively impact plant growth and biomass accumulation in later stages. Under Cd stress, the AtHMA3-deficient mutant exhibited a notably lower germination rate compared to the WT (Figure 2A). Cd toxicity primarily affects plant roots by damaging root cell membranes, which impairs normal root development, as evidenced by inhibited root elongation. In this study, the root length of the AtHMA3-deletion mutant was significantly more inhibited under Cd stress (Figure 2C), consistent with previous findings [24] that show increased sensitivity to Cd in the athma3 mutant. By measuring the Cd content in the mutant and WT, we found that the Cd content in the athma3 mutant is significantly lower than that in WT, which indicates that the HMA gene affects the uptake and accumulation of Cd in the plant.
In recent years, substantial research has been conducted on the physiological functions of HMA3 in non-accumulating species, such as AtHMA3 in Arabidopsis thaliana and OsHMA3 in Oryza sativa Japonica Group. Overexpression of AtHMA3 or OsHMA3 in these species significantly enhanced tolerance to toxic Cd [24,34]. In this study, plants expressing SpHMA3 in Arabidopsis thaliana and maize exhibited higher Cd accumulation under Cd treatment (Figure 7), and the SpHMA3 gene showed increased expression in roots and leaves during Cd stress (Figure 6A). Although overall seed germination rates decreased with rising Cd concentrations, SpHMA3-expressing plants demonstrated a significantly higher germination rate, indicating improved Cd stress resistance in these plants. Root growth analysis further revealed that under Cd stress, SpHMA3-expressing plants experienced less inhibition compared to the WT, with better root development and more fully extended lateral roots. These results suggest that SpHMA3 expression helps mitigate the adverse effects of Cd on root systems, thereby enhancing Cd tolerance [35].
Plant cells possess a variety of antioxidant enzyme systems. Under stress conditions, the levels of reactive oxygen species in the cell increase, leading to higher H2O2 concentrations and cellular imbalance. Antioxidant enzymes, such as SOD, POD, CAT, along with some small molecule metabolites, help eliminate free radicals and reduce H2O2 levels [36]. Once Cd enters the cell, it binds to Cys residues, which can deactivate enzymes [34] and disrupt redox balance, causing oxidative stress. The content of MDA, a biomarker for oxidative stress, reflects the degree of lipid peroxidation in the cell and indicates the ability of the plant to adapt under stress conditions. In this study, examining the activities of antioxidant enzymes in SpHMA3-overexpressing Arabidopsis and maize under Cd stress revealed that MDA content increased in both WT and OE plants. However, WT plants showed a larger increase in Arabidopsis, whereas the opposite was observed in maize (Figure 8A,B). This disparity could be attributed to higher Cd accumulation in OE maize plants compared to the WT, which results in greater MDA levels in OE plants. This higher Cd accumulation in OE plants could further stimulate the activities of related enzymes. The activities of SOD, POD, and CAT were also measured in both Arabidopsis and maize, showing that SpHMA3 overexpression significantly enhanced antioxidant enzyme activities compared to WT plants (Figure 8C–H). This suggests that SpHMA3 expression strengthens antioxidant defenses, thereby mitigating Cd-induced free radical damage. Considering the higher Cd accumulation in OE plants compared to the WT, as seen in Figure 7, it can be concluded that overexpression of SpHMA3 enhances Cd uptake, translocation, and accumulation in plants, which in turn promotes gene expression and increases antioxidant enzyme activity, thereby improving the tolerance of plants to Cd. These results are consistent with previous findings [37,38]. Studies have demonstrated that under Cd2+ stress, NH4+ alleviates cadmium toxicity in rice seedlings by activating the bZIP20-APX2/CATA transcription module in an ABA-dependent manner, indicating that different species have evolved a rich array of mechanisms to resist environmental stress during their evolutionary process [39]. With the global spread of maize, significant potential exists for its further development. This study demonstrates that transgenic maize, modified with the SpHMA3 gen, exhibits superior Cd tolerance and transfer properties compared to conventional varieties (Figure 9 and Figure 10). In recent years, numerous successful cases of soil bioremediation have been reported, and the transgenic maize in this research could serve as a reference for future remediation of Cd-contaminated soils [40,41]. Meanwhile, the remediated maize can be utilized as a biofeedstock for ethanol fermentation, promoting a sustainable, low-carbon, and environmentally friendly development model [42]. These results deepen our understanding of the function of SpHMA3 in enhancing maize growth and resilience to Cd stress. Of course, it is worth noting that the premise of using corn as a soil cadmium pollution remediation plant for bioremediation is to enhance people’s recognition of this remediation model. The key lies in how to make corn complete soil restoration and realize sustainable utilization of corn. This is also the starting point of the current research work. We firmly believe that in the future, there will be many successful cases like genetically modified corn to repair cadmium pollution, and this model will also provide a new green way for soil remediation. This research provides valuable insight into improving the metal accumulation traits of current maize varieties and advancing phytoremediation technology for heavy metal-polluted soils.
4. Materials and Methods
4.1. Plant Materials and Growing Conditions
The maize inbred line KN5585 was supplied by Sichuan Agricultural University, while the Colombian wild-type Arabidopsis thaliana and AtHMA mutant were sourced from ABRC. Transient expression receptor maize and maize healing tissues were provided by the group of Haijian Lin at the Maize Research Institute, Sichuan Agricultural University. The E. coli receptor Trans1-T1, Agrobacterium receptor GV3101, Arabidopsis expression vector pRI101-AN, tobacco vector pCAMBIA2300-35S-eGFP for transient expression, and maize expression vector pCAMBIA3300 were also obtained from the group of Haijian Lin. Arabidopsis plants were grown in a short-day incubator for two months before the imbibed seeds were collected and cultured on 1/2 MS solid medium containing CdCl2. Smut was grown in pots and exposed to light for 3 to 4 weeks and used for subcellular localization studies. The soil utilized in this study was sourced from an agricultural site in Chengdu, Sichuan Province, China. The physical and chemical properties of the soil were characterized as follows: the pH value was 7.32, the organic matter content was 46.10 g/kg, the alkali-hydrolyzed nitrogen content was 80.0 mg/kg, the available phosphorus content was 62.8 mg/kg, and the available potassium content was 182.6 mg/kg.
4.2. SpHMA3 and AtHMA3 Expression Analysis
Expression profiles were analyzed using an ABI 7500 real-time PCR system (Torrance, CA, USA). KN5585 maize seedlings were exposed to 800 μmol/L CdCl2, while Arabidopsis seedlings were treated with 80 μmol/L CdCl2. The CdC12·2.5H2O used in the study was provided by Shanghai Test Reagent. Total RNA from KN5585 maize seedling roots, leaves, and Arabidopsis thaliana was isolated using TRIzol reagent (Invitrogen, Gaithersburg, MD, USA). RNA integrity was assessed through 1% agarose gel electrophoresis, and RNA concentration was measured by a nucleic acid protein assay. The cDNA sequence of the SpHMA3 gene was retrieved from NCBI, and after designing primers via Gramene, the sequence was amplified using Novizime high-fidelity enzyme. The primers used are listed in Table S1, while the PCR mix and reaction conditions are shown in Tables S2 and S3. The ABclonal Biologicals ABScript III RT Master Mix for qPCR, coupled with the gDNA Remover kit, and the Nearshore Protein NovoScript®Plus All-in-one 1st Strand cDNA Synthesis SuperMix (gDNA Purge) kit were utilized to reverse transcribed RNA into cDNA at a concentration of 1000 ng/μL−1. The CDS of the SpHMA3 gene (gene ID: KY312109) was obtained from NCBI and used for further analysis. The sequence was analyzed for homologous comparison, quantitative primer design, and primer specificity verification using the website (https://Plants.ensembl.org/Zea_mays/Info/Index?db=core, accessed on 20 March 2024). Primer specificity was assessed via 1% agarose gel electrophoresis, using GAPDH as the internal reference. The primer sequences are listed in Table S4, the quantitative PCR setup is provided in Table S5, and the reaction protocols are detailed in Table S6. Each experiment included three replicates, and relative gene expression levels of SpHMA3 and AtHMA3 were determined by the 2−ΔΔCT method.
4.3. Subcellular Localization Assay
The subcellular localization of target proteins was predicted using the online software Softberry (https://www.softberry.com/, accessed on 22 February 2023). SpHMA3 from Companion Mine Sedum was cloned into pCAMBIA2300-35S-eGFP, and the positive expression vector was transformed into E. coli Trans1-T1 receptor cells. The transformation was carried out using Agrobacterium tumefaciens GV3101 receptor cells. The fusion expression vector plasmid was introduced into maize protoplasts for subcellular localization, and the fluorescent signals were detected using a laser scanning microscope. The gene expression sites were identified through marker prediction and co-localization of the recombinant vectors.
4.4. Genetic Transformation of Arabidopsis thaliana
Arabidopsis homozygous mutant seeds containing T-DNA insertions were obtained from ABRC. These seeds underwent stress verification, and the subsequent generation was screened under varying Cd stress conditions. A basal medium of 1/2 MS solid medium, supplemented with Cd at concentrations of 0, 40, and 80 μmol/L, was used. Sixty seeds were inoculated per Petri dish, with three replicates per treatment. Root length and root hair development were then analyzed.
4.5. Validation of SpHMA3 Gene Overexpression in Arabidopsis thaliana
The Arabidopsis expression vector used was pRI101-AN, containing the 35S promoter, NOS terminator, KanR resistance marker gene, and BamHI digestion site. Following successful amplification, the SpHMA3 gene cDNA was inserted into the overexpression vector through homologous recombination. The recombinant plasmid was first introduced into E. coli for plasmid extraction and then transferred to Agrobacterium. Wild-type Arabidopsis thaliana Columbia (Col) served as the material for transformation. The Agrobacterium containing the target gene was prepared into a transformation solution, in which the flowering Arabidopsis thaliana plants were submerged. The solution was covered with plastic film for 24 h before the film was removed for further cultivation. The seeds of the overexpressed materials were identified, and the T0 generation transgenic Arabidopsis seeds were germinated. DNA extracted from the leaves of Arabidopsis was further confirmed until no phenotypic segregation was observed. At this point, the transgenic Arabidopsis was confirmed to be purely positive and used in the subsequent experimental steps.
4.6. Arabidopsis Germination Phenotype Under Cd Stress
Arabidopsis thaliana T3 generation seeds, both positive and wild-type, were sown on 1/2 MS medium without KanR resistance. A total of 60 seeds were placed in each medium, with three replicates prepared. Following a 48 h vernalization period, the plates were incubated for 7 days. On the 7th day, the germination rate of the overexpressed lines and WT seeds was recorded. Additionally, the primary and total root lengths of Arabidopsis thaliana were measured on day 14.
4.7. Determination of Cd Content in Arabidopsis thaliana
Arabidopsis OE and WT were exposed to Cd stress at concentrations of 0, 40, and 80 µmol/L for 14 days. After treatment, the samples were fully dried to a constant weight, ground, and processed using a microwave ablation system (MARS 6) to prepare an ablation solution. The Cd content in the samples was then measured by an inductively coupled plasma mass spectrometer (NeXLON2000). Set the instrument to its optimum condition. The reference conditions of atomic absorption spectrophotometer are as follows: wavelength 228.8 nm, slit 0.2~1.0 nm, lamp current 2–10 mA, drying temperature 105 °C, drying time 20 s; Ashing temperature: 400~700 °C, ashing time: 20~40 s; The atomization temperature is 1300~2300 °C, and the atomization time is 3~5 s. Background correction was in the form of a deuterium lamp or the Zeeman effect. The LOD and LOQ for Cd were 0.001 mg/kg and 0.003 mg/kg, respectively.
4.8. Measurement of Physiological and Biochemical Indexes in Arabidopsis and Maize Seedlings
Arabidopsis OE and WT plants were exposed to 0, 40, and 80 µmol/LCd stress treatments. After 14 days of stress exposure, samples were collected to measure the levels of MDA, SOD, POD, and CAT in the plants, with the respective reaction systems detailed in Tables S6–S8. Additionally, roots of KN5585 and SpHMA3 overexpression plants were harvested at 0, 12, 24, 36, 48, and 72 h under 800 μmol/L Cd treatment. Each time point was replicated three times, and MDA, SOD, POD, and CAT levels were assessed, with the corresponding systems shown in Tables S6–S8.
4.9. Overexpression of SpHMA3 Gene in Maize
The expression vector used was pCAMBIA3300, with primers incorporating enzyme restriction sites, and downstream primers containing terminators were designed. The method for vector construction followed the same procedure as that used for subcellular localization recombinant vectors, and the primer sequences are listed in Supplementary Table S9. Transgenic T0 plants were generated by infiltrating young maize embryos with Agrobacterium tumefaciens. DNA was extracted from T0 maize leaves, and positive identification was confirmed. The primers used for identification are provided in Table S10.
4.10. Determination of Cd Content in Various Parts of Maize
Overexpressed and WT maize seedlings were exposed to stress treatment using an 800 μmol/L CdCl2 solution for varying time points (0 h, 12 h, 24 h, 36 h, 48 h, and 72 h), and samples of their leaves and roots were collected to assess Cd accumulation in different plant tissues. Soil Cd concentrations were set at 0 mg/kg, 0.3 mg/kg, 3 mg/kg, 10 mg/kg, and 20 mg/kg in pots where both overexpression and WT maize were planted. Samples were collected at three growth stages: Maize-V6, Maize-VT, and Maize-R6, to measure the total Cd accumulation in the plants. Cd content was quantified by atomic absorption spectrophotometry, which involved preparing a Cd standard solution gradient, plotting a standard curve, and constructing a univariate linear regression equation to relate absorbance values to Cd concentration.
The Cd concentration in the sample was determined using the following equation:
where plant enrichment factor (BCF) = Cplant/Csoil; plant transport factor (BTF) = Covergroundpart/Csubterraneanpart; x—Cd content in the specimen, in mg/kg−1; C1—Cd content in the specimen solution in ng/mL−1; C0—Cd content in the blank solution in ng/mL−1; V—the total volume of the specimen solution after calibration in mL. unit is mL; M—a mass of specimen in g; 1000—conversion factor; Cplant—heavy metal content in a tissue of a single plant (e.g., root, stem, leaf, male spike, female spike, seed); Csoil—concentration of the corresponding heavy metal elements in the soil (mg/kg−1); Covergroundpart—concentration of heavy metal elements in the above-ground part of the plant (mg/kg−1); Csubterraneanpart—concentration of a heavy metal element in the underground part of the plant (mg/kg−1).
4.11. Statistical Analysis
The data analysis was conducted with BM SPSS Statistics 26, GraphPad Prism 8, and Excel 2016, and the significant differences between the samples were evaluated by employing Student’s t-test.
5. Conclusions
This research explored SpHMA3, a key regulator of Zn and Cd uptake and translocation in the hyperaccumulator Sedum, which encodes a P-type ATPase family protein. Subcellular localization analysis demonstrated that SpHMA3 protein resides in the maize cell membrane. The overexpression of SpHMA3 significantly improved Cd tolerance in both Arabidopsis thaliana and maize, leading to an increased capacity for Cd uptake and translocation. These transgenic plants exhibited superior growth, enhanced antioxidant enzyme system activity, and greater accumulation of substances under Cd stress. The AtHMA3 homozygous mutant displayed greater Cd sensitivity compared to WT. Analysis of Cd accumulation in overexpressed maize showed that more Cd was retained in the root system compared to the aerial parts, reducing the movement of Cd from soil to maize kernels and thereby mitigating the effects of Cd contamination. This work advances our understanding of the genetic and functional roles of SpHMA3 and proposes a new, green, and efficient model for soil bioremediation of heavy metal Cd pollution while offering a valuable reference for maize in soil Cd pollution management.
6. Glossary
- Heavy Metal ATPase (HMA): the HMA heavy metal transporter protein is a transmembrane transporter protein belonging to the P-type ATPase family, which mainly provides energy for metal transport by hydrolyzing ATP. This protein generally contains 6–8 transmembrane fragments and can selectively transport heavy metal cations, which plays an important role in phytoremediation of heavy metal contaminated soil.
- Coding sequence (CDS): refers to a sequence that encodes a segment of a protein product. CDS is a structural genomics term that denotes the portion of a DNA sequence that is capable of being transcribed into mRNA and further translated into a protein.
- Complementary DNA(cDNA): complementary (sometimes called copy) DNA, specifically a strand of DNA that is complementary to RNA after reverse transcription in vitro. Unlike what we normally call genomic DNA, cDNA does not have introns but only exon sequences.
- Quantitative Real-time PCR(qRT-PCR): it is a method to measure the total amount of product after each polymerase chain reaction (PCR) cycle with fluorescent chemicals in a DNA amplification reaction. It is a method for quantitatively analyzing specific DNA sequences in the sample to be tested by internal or external reference methods.
- Superoxide Dismutase(SOD): superoxide dismutase is also known as superoxide dismutase. It is a class of enzymes that catalyze the disproportionation of superoxide anion radicals (O2−) to H2O2 and O2. This enzyme is extremely widely distributed and has so far been isolated from a variety of organisms including bacteria, fungi, algae, plants, protozoa, insects, fish and mammals.
- Peroxidase(POD): peroxidases are the hallmark enzymes of the peroxisome, its class of oxidoreductases, and they catalyze many reactions. Peroxidases are enzymes that catalyze the oxidation of substrates using hydrogen peroxide as an electron acceptor.
- Catalase(CAT): it is an enzyme that catalyzes the breakdown of hydrogen peroxide into oxygen and water and is found in the peroxisomes of cells.
- Malondialdehyde(MDA): it is an organic compound with the molecular formula C3H4O2, which belongs to the list of group 3 carcinogens.
- Bioconcentration Factor (BCF): BCF is the ratio of the concentration of a compound in biological tissue (dry weight) to the concentration dissolved in water, or it can be considered as the ratio of the rate of uptake of the compound by the organism to the rate of purification of the compound from the organism, and it is used to indicate the magnitude of bioconcentration of organic compounds in the organism. The bioconcentration coefficient is an important indicator to describe the accumulation trend of chemical substances in organisms.
- Plant Transport Coefficient(BTF): it is the ratio of the metal content in the above-ground part of the plant to the metal content in the below-ground part of the plant, and is used as an indicator to evaluate the ability of the plant to transport and enrich heavy metals from the below-ground part of the plant to the above-ground part of the plant.
- Wild Type(WT): it is an individual or gene that has not been artificially mutated in nature. In genetics, a wild type is a phenotype that is present in more than 1% of a population and is often used as a standard control gene.
- Thousand-grain Weight(TKW): it is the weight of 1000 grains of rice in grams. It is an indicator of the size and fullness of the seed, which is used to test the quality of the seed and the content of the crop test, and it is also an important basis for predicting yield in the field.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms26083487/s1.
Author Contributions
H.L. and G.L. conceived and designed the experiments and supervised the research works. R.P., G.H., and Q.J. performed most of the experiments and prepared the data and the draft of the manuscript. B.Z., W.Z., C.X., and J.H. (Jianfeng Hu) performed a part of the experiments and prepared part of the data. W.X., M.L., H.D., S.Z., and J.H. (Jialong Han) participated in material development. H.L. and R.P. analyzed the data and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Zhejiang Province Science and Technology Project of Zhejiang Province of China (2022C04024), the Sichuan Tianfu New Area Rural Revitalization Research Institute “unveils the list of marshal” projects (the first batch NO. XZY1-01).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original data presented in the study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zhuang, P.; McBride, M.B.; Xia, H.; Li, N.; Li, Z. Health risk from heavy metals via consumption of food crops in the vicinity of Dabaoshan mine, South China. Sci. Total Environ. 2009, 407, 1551–1561. [Google Scholar]
- Zhang, X.; Chen, D.; Zhong, T.; Zhang, X.; Cheng, M.; Li, X. Assessment of cadmium (Cd) concentration in arable soil in China. Environ. Sci. Pollut. Res. 2015, 22, 4932–4941. [Google Scholar]
- Zhou, C. Research progress of in situ remediation technology for cadmium contaminated soil. Nonferrous Metall. Des. Res. 2018, 39, 24–28+32, (In Chinese with English Abstract). [Google Scholar]
- Ramakrishnan, S.; Sulochana, K.N.; Selvaraj, T.; Rahim, A.A.; Lakshmi, M.; Arunagiri, K. Smoking of beedies and cataract: Cadmium and vitamin C in the lens and blood. Br. J. Ophthalmol. 1995, 79, 6–202. [Google Scholar] [CrossRef][Green Version]
- Chen, Z.; Mo, D.; Chou, R. Hazards of Cadmium Pollution on Biological Organisms and Countermeasures for Prevention and Control. Environ. Prot. Sci. 2001, 37–39, (In Chinese with English Abstract). [Google Scholar]
- Godt, J.; Scheidig, F.; Grosse-Siestrup, C.; Esche, V.; Brandenburg, P.; Reich, A.; Groneberg, D.A. The toxicity of cadmium and resulting hazards for human health. J. Occup. Med. Toxicol. 2006, 1, 22. [Google Scholar] [CrossRef]
- Xie, H.; Chen, A.; Zhang, F.; Xiong, Y.; Chen, Y.L.; Liu, J.; Zhou, L.Y.; Lin, R.Y. Physiological response of Perilla frutescens to different concentrations of cadmium stress. Chin. J. Ecol. Agric. 2011, 19, 672–675, (In Chinese with English Abstract). [Google Scholar]
- Li, J.; Shi, G.; Wei, Y. Heavy metal cadmium to each other on the grass growth and physiological characteristics influence. J. Anhui Agric. Sci. 2011, 33, 2174–2176+2182, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Dragomir, V.; Ioan Sebastian, B.; Alina, B.; Victor, P.; Tanasă, L.; Horhocea, D. An overview of global maize market compared to Romanian production. Rom. Agric. Res. 2022, 39, 535–544. [Google Scholar]
- Adiaha, M.S.; Agba, O.A.; Attoe, E.E.; Ojikpong, T.O.; Kekong, M.A.; Obio, A.; Undie, U.L. Effect of maize (Zea mays L.) on human development and the future of man-maize survival: A review. World Sci. News 2016, 59, 52–62. [Google Scholar]
- Ghosh, P.K.; Das, A.; Saxena, R.; Banerjee, K.; Kar, G.; Vijay, D. (Eds.) Trajectory of 75 Years of Indian Agriculture After Independence; Springer: Singapore, 2023. [Google Scholar]
- Kaul, J.; Jain, K.; Olakh, D. An overview on role of yellow maize in food, feed and nutrition security. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 3037–3048. [Google Scholar] [CrossRef]
- Sangwan, S.; Kumar, S.; Goyal, S. Maize utilisation in food bioprocessing: An overview. In Maize: Nutrition Dynamics and Novel Uses; Springer: New Delhi, India, 2014; pp. 119–134. [Google Scholar]
- Rascio, N.; Vecchia, F.D.; Ferretti, M.; Merlo, L.; Ghisi, R. Some effects of cadmium on maize plants. Arch. Environ. Contam. Toxicol. 1993, 25, 244–249. [Google Scholar] [CrossRef]
- Florijn, P.J.; Van Beusichem, M.L. Uptake and distribution of cadmium in maize inbred lines. Plant Soil 1993, 150, 25–32. [Google Scholar] [CrossRef]
- Anjum, S.A.; Tanveer, M.; Hussain, S.; Bao, M.; Wang, L.; Khan, I.; Ullah, E.; Tung, S.A.; Samad, R.A.; Shahzad, B. Cadmium toxicity in Maize (Zea mays L.): Consequences on antioxidative systems, reactive oxygen species and cadmium accumulation. Environ. Sci. Pollut. Res. 2015, 22, 17022–17030. [Google Scholar] [CrossRef] [PubMed]
- Pál, M.; Horváth, E.; Janda, T.; Páldi, E.; Szalai, G. Physiological changes and defense mechanisms induced by cadmium stress in maize. J. Plant Nutr. Soil Sci. 2006, 169, 239–246. [Google Scholar] [CrossRef]
- Axelsen, K.B.; Palmgren, M.G. Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 1998, 46, 84–101. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Bertini, I.; Ciofi-Baffoni, S.; Su, X.-C.; Miras, R.; Bal, N.; Mintz, E.; Catty, P.; Shokes, J.E.; Scott, R.A. Structural Basis for Metal Binding Specificity: The N-terminal Cadmium Binding Domain of the P1-type ATPase CadA. J. Mol. Biol. 2006, 356, 638–650. [Google Scholar] [CrossRef][Green Version]
- Toyoshima, C. How Ca2+-ATPase pumps ions across the sarcoplasmic reticulum membrane. Biochim. Biophys. Acta 2009, 1793, 941–946. [Google Scholar] [CrossRef]
- Smith, A.T.; Smith, K.P.; Rosenzweig, A.C. Diversity of the metal-transporting P1B-type ATPases. J. Biol. Inorg. Chem. 2014, 19, 947–960. [Google Scholar] [CrossRef]
- Axelsen, K.B.; Palmgren, M.G. Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiol. 2001, 126, 696–706. [Google Scholar] [CrossRef]
- Kim, Y.; Choi, H.; Segami, S.; Cho, H.; Martinoia, E.; Maeshima, M.; Lee, Y. AtHMA1 contributes to the detoxification of excess Zn(II) in Arabidopsis. Plant J. Cell Mol. Biol. 2009, 58, 737–753. [Google Scholar]
- Morel, M.; Crouzet, J.; Gravot, A.; Auroy, P.; Leonhardt, N.; Vavasseur, A.; Richaud, P. AtHMA3, a P1B-ATPase allowing Cd/Zn/co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 2009, 149, 894–904. [Google Scholar] [PubMed]
- Morin, I.; Gudin, S.; Mintz, E.; Cuillel, M. Dissecting the role of the N-terminal metal-binding domains in activating the yeast copper ATPase in vivo. FEBS J. 2009, 276, 4483–4495. [Google Scholar] [PubMed]
- Lu, S.; Du, Y.; Zhong, D.; Zhao, B.; Li, X.; Xu, M.; Li, Z.; Luo, Y.; Yan, J.; Wu, L. Comparison of Trace Element Emissions from Thermal Treatments of Heavy Metal Hyperaccumulators. Environ. Sci. Technol. 2012, 46, 5025. [Google Scholar]
- Zhong, D.; Zhong, Z.; Wu, L.; Ding, K.; Luo, Y.; Christie, P. Pyrolysis of Sedum plumbizincicola, a zinc and cadmium hyperaccumulator: Pyrolysis kinetics, heavy metal behaviour and bio-oil production. Clean Technol. Environ. Policy 2016, 18, 2315–2323. [Google Scholar]
- Cao, D.; Zhang, H.; Wang, Y.; Zheng, L. Accumulation and Distribution Characteristics of Zinc and Cadmium in the Hyperaccumulator Plant Sedum plumbizincicola. Bull. Environ. Contam. Toxicol. 2014, 93, 171–176. [Google Scholar]
- Wu, L.H.; Liu, Y.J.; Zhou, S.B.; Guo, F.G.; Bi, D.; Guo, X.H.; Baker, A.J.M.; Smith, J.A.C.; Luo, Y.M. Sedum plumbizincicola XH Guo et SB Zhou ex LH Wu (Crassulaceae): A new species from Zhejiang Province, China. Plant Syst. Evol. 2013, 299, 487–498. [Google Scholar]
- Wu, L.H.; Zhou, S.B.; Bi, D.; Guo, X.H.; Qin, W.H.; Wang, H.; Wang, G.J.; Luo, Y.M. Sedum plumbizincicola, a new species of the Crassulaceae from Zhejiang, China. Soils 2006, 38, 632–633. [Google Scholar]
- Luo, P.; Wu, J.; Li, T.T.; Shi, P.; Ma, Q.; Di, D.W. An Overview of the Mechanisms through Which Plants Regulate ROS Homeostasis under Cadmium Stress. Antioxidants 2024, 13, 1174. [Google Scholar] [CrossRef]
- Liu, H.; Zhao, H.; Wu, L.; Liu, A.; Zhao, F.; Xu, W. Heavy metal ATPase 3 (HMA3) confers cadmium hypertolerance on the cadmium/zinc hyperaccumulator Sedum plumbizincicola. New Phytol. 2017, 215, 687–698. [Google Scholar]
- Zhao, X.W. Analysis of the Genetic Basis for the Control of Heavy Metal Cadmium Cd Accumulation in Maize. Ph.D. Thesis, Sichuan Agricultural University, Ya’an, China, 2018. [Google Scholar]
- Sasaki, A.; Yamaji, N.; Ma, J.F. Overexpression of OsHMA3 enhances Cd tolerance and expression of Zn transporter genes in rice. J. Exp. Bot. 2014, 65, 6013–6021. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.Z. Effects of High pH Stress on PCD in Primary Root Genesis of Arabidopsis Seedlings. Master’s Thesis, Shandong Agricultural University, Tai’an, China, 2007. [Google Scholar]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Yao, Y. Effect of zinc on the growth of maize under cadmium stress. J. Green Sci. Technol. 2013, 09, 166–167. (In Chinese) [Google Scholar]
- Chen, Y.; Chao, Z.F.; Jin, M.; Wang, Y.L.; Li, Y.; Wu, J.C.; Xiao, Y.; Peng, Y.; Lv, Q.Y.; Gui, S.; et al. A heavy metal transporter gene ZmHMA3a promises safe agricultural production on cadmium-polluted arable land. J. Genet. Genom. 2023, 50, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Di, D.W.; Li, T.T.; Yu, Z.L.; Cheng, J.; Wang, M.; Liu, C.F.; Wang, Y.; Kronzucker, H.J.; Yu, M.; Shi, W. Ammonium mitigates cadmium toxicity by activating the bZIP20-APX2/CATA transcriptional module in rice seedlings in an ABA-dependent manner. J. Hazard. Mater. 2024, 480, 135874. [Google Scholar] [CrossRef]
- Zhang, T.; Zhou, J.; Zhao, W.; Li, X.; Wang, X.; Li, L.; Xin, Z.; Sun, X.; Jin, W. Research on the safe growth and planting of annual economic plants in cadmium-polluted farmland in red soil area. Jiangxi Sci. 2024, 4, 820–825, (In Chinese with English Abstract). [Google Scholar]
- Tan, C.; Yang, Y. Study on the bioremediation ability of lily on soil heavy metal pollution. Environ. Sci. Manag. 2024, 49, 154–157, (In Chinese with English Abstract). [Google Scholar]
- Shu, Y.; Liu, Y.; Liu, M.; Bi, J. Study on the Energy Potential of Collectable Straw and Bioethanol Production Potential of Major Crops in China. J. Beijing Inst. Technol. (Soc. Sci. Ed.) 2023, 25, 64–72, (In Chinese with English Abstract). [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
