Transcriptome Screening of Hormone-Regulated Genes Related to Fruit Development in Zizyphus jujuba Mill. cv. Goutou Fruits at Different Ripening Stages
Abstract
1. Introduction
2. Results
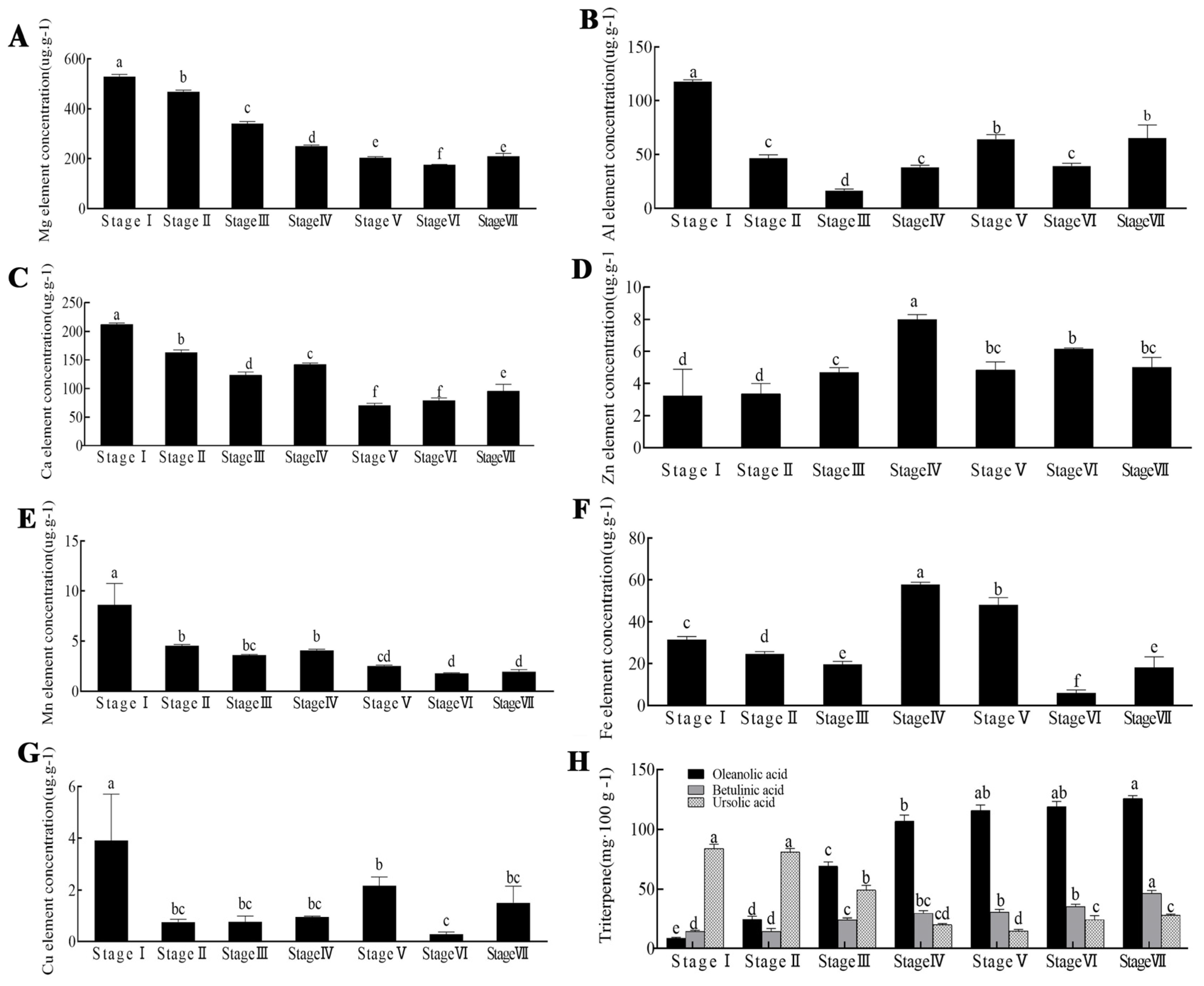
2.1. Detection of the Metal Element, Oleanolic Acid, Betulinic Acid, and Ursolic Acid Content of the Z. jujuba Mill. cv. Goutou Fruits at Different Ripening Stages
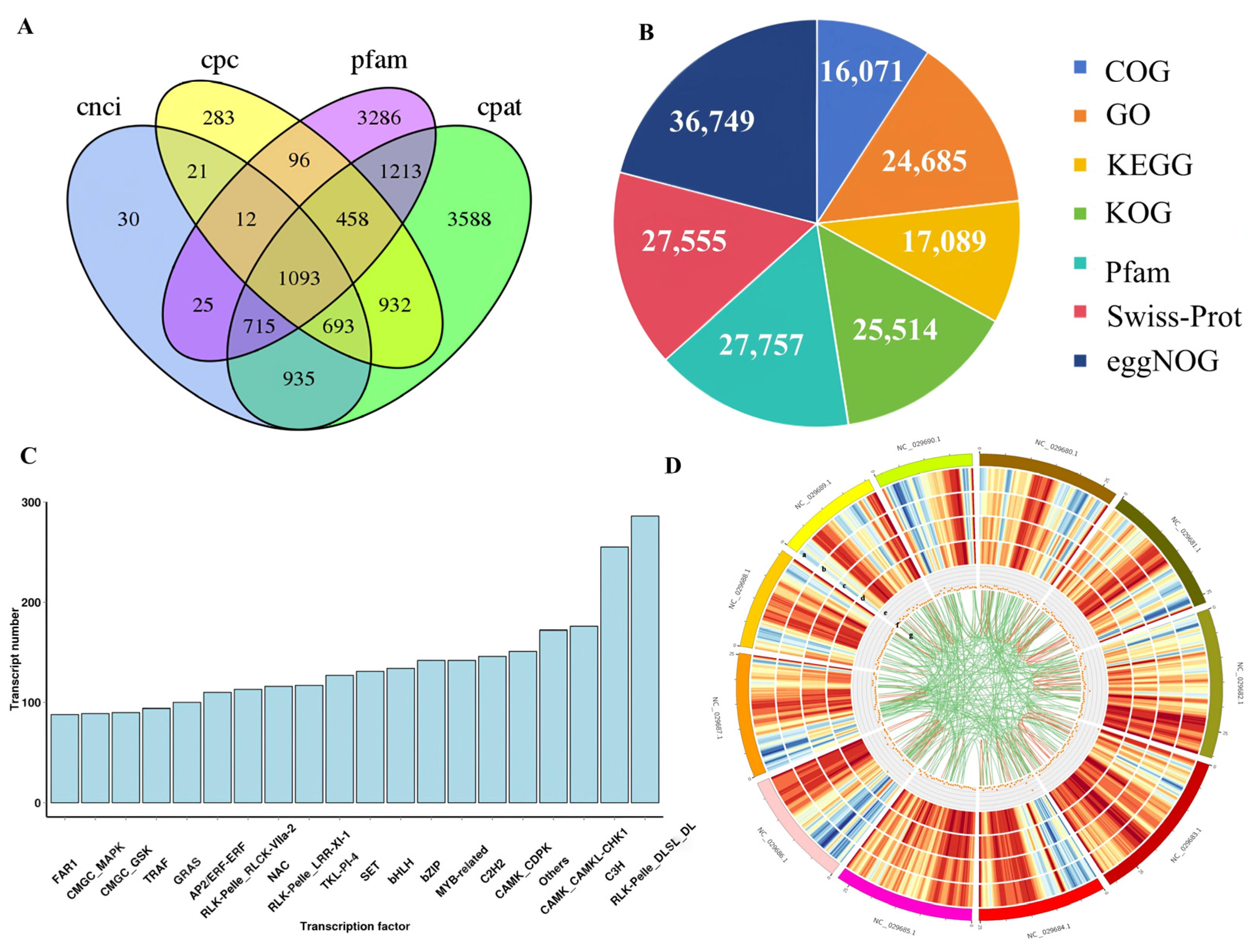
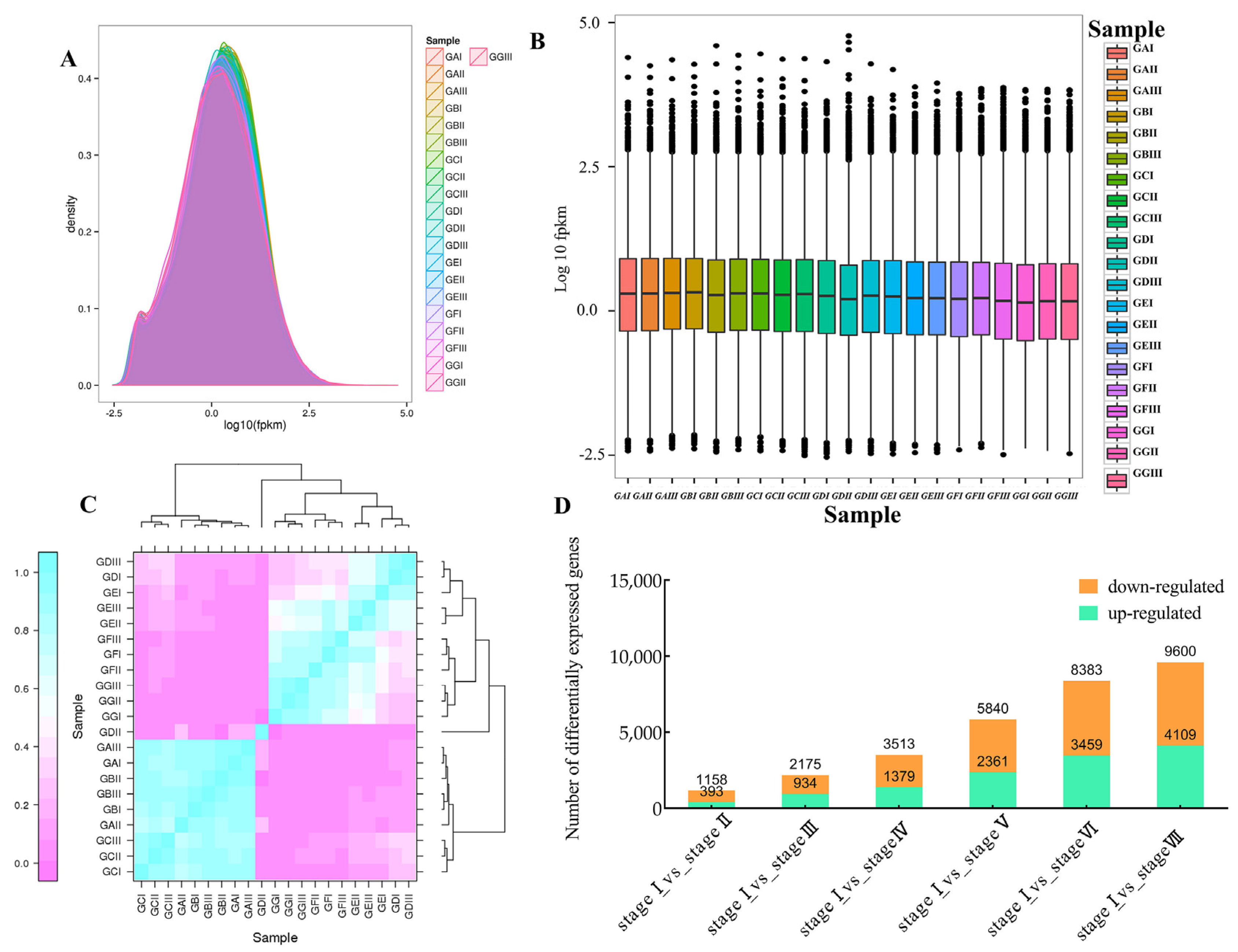
2.2. Transcriptome Sequencing Analysis of Z. jujuba Mill. cv. Goutou Fruits at Different Ripening Stages
2.3. Screening and Validation of Differentially Expressed Genes
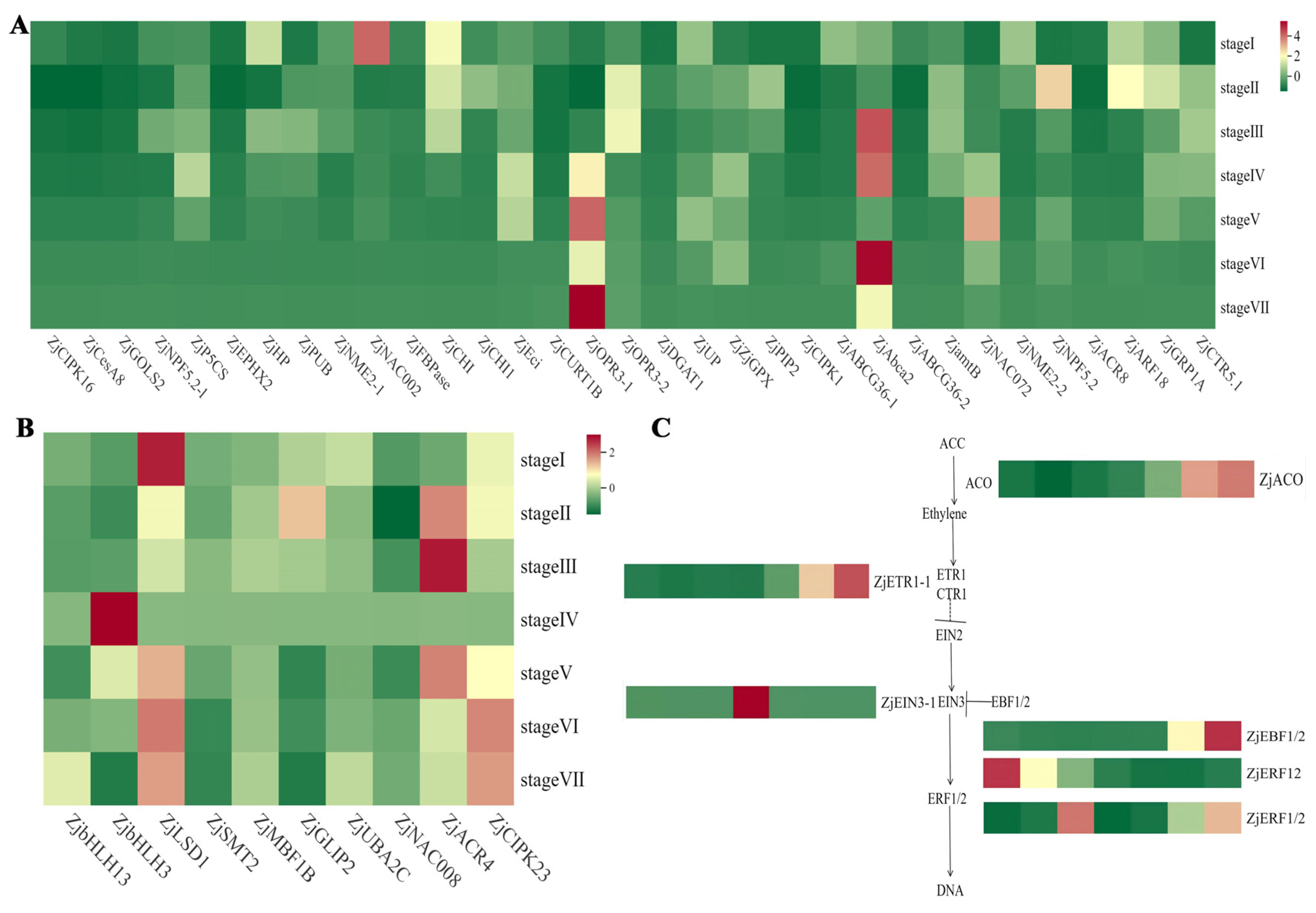
2.4. Significant Differences in the Expression of Differentially Expressed Genes Involved in the ABA Signaling Pathway in Z. jujuba Mill. cv. Goutou Fruits at Different Ripening Stages
2.5. Expression Analysis of Differentially Expressed Genes Related to the Ethylene Signaling Pathway in Z. jujuba Mill. cv. Goutou Fruits at Different Ripening Stages
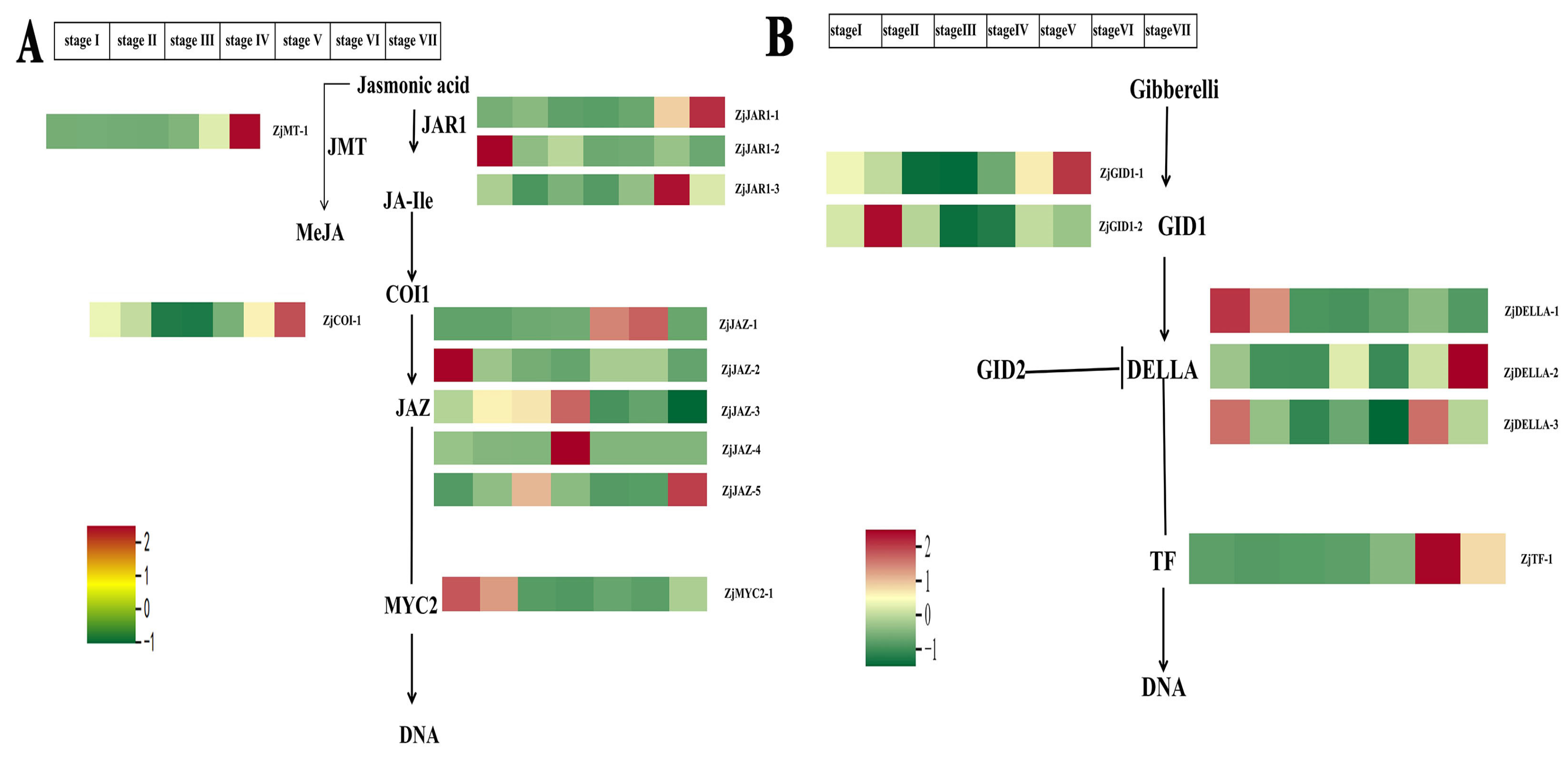
2.6. Expression Analysis of DEGs Regulating the Jasmonic Acid and Gibberellic Acid Signaling Pathway in Z. jujuba Mill. cv. Goutou Fruits at Different Ripening Stages
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Transcriptome Sequencing and Functional Annotations
4.3. Metal Element Analysis
4.4. Determination of Ursolic Acid, Oleanolic Acid, and Betulinic Acid Content
4.5. Real-Time Quantitative PCR (RT-qPCR) Analysis
4.6. Data Processing and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pan, F.; Zhao, X.; Liu, F.; Luo, Z.; Chen, S.; Liu, Z.; Zhao, Z.; Liu, M.; Wang, L. Triterpenoids in Jujube: A Review of Composition, Content Diversity, Pharmacological Effects, Synthetic Pathway, and Variation during Domestication. Plants 2023, 12, 1501. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Chen, Q.S.; Kong, X.B.; Han, L.X.; Zhang, Q.; Li, Q.J.; Hao, B.; Zhao, X.; Liu, L.; Wan, H.; et al. Heavy metal contamination and risk assessment in winter jujube (Ziziphus jujuba Mill. cv. Dongzao). Food Chem. Toxicol. 2023, 174, 113645. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.H.; Calderone, L.A.; Pan, L.Y.; Trent, Q.; Pandelia, M. The Fe and Zn cofactor dilemma. Biochim. Biophys. Acta Proteins Proteom 2023, 1871, 140931. [Google Scholar] [CrossRef]
- Costa, M.I.; Sarmento-Ribeiro, A.B.; Gonçalves, A.C. Zinc: From Biological Functions to Therapeutic Potential. Int. J. Mol. Sci. 2023, 24, 4822. [Google Scholar] [CrossRef]
- Zhu, M.; Yu, J.; Wang, R.; Zeng, Y.; Kang, L.; Chen, Z. Nano-calcium alleviates the cracking of nectarine fruit and improves fruit quality. Plant Physiol. Biochem. 2023, 196, 370–380. [Google Scholar] [CrossRef]
- Wen, C.; Zhang, Z.; Shi, Q.; Yue, R.; Li, X. Metabolite and Gene Expression Analysis Underlying Temporal and Spatial Accumulation of Pentacyclic Triterpenoids in Jujube. Genes 2022, 13, 823. [Google Scholar] [CrossRef] [PubMed]
- Sakna, S.T.; Maghraby, Y.R.; Abdelfattah, M.S.; Farag, M.A. Phytochemical diversity and pharmacological effects of triterpenes from genus Ziziphus: A comprehensive review. Phytochem. Rev. 2023, 22, 1611–1636. [Google Scholar] [CrossRef]
- Zhong, Y.; Xun, W.; Wang, X.; Tian, S.; Zhang, Y.; Li, D.; Zhou, Y.; Qin, Y.; Zhang, B.; Zhao, G.; et al. Root-secreted bitter triterpene modulates the rhizosphere microbiota to improve plant fitness. Nat. Plants 2022, 8, 887–896. [Google Scholar] [CrossRef]
- Ji, Y.; Xu, M.; Wang, A. Recent Advances in The Regulation of Climacteric Fruit Ripening: Hormone, Transcription Factor and Epigenetic Modifications. Front. Agric. Sci. Eng. 2021, 8, 314–334. [Google Scholar] [CrossRef]
- Wang, Y.W.; Nambeesan, S.U. Ethylene promotes fruit ripening initiation by downregulating photosynthesis, enhancing abscisic acid and suppressing jasmonic acid in blueberry (Vaccinium ashei). BMC Plant Biol. 2024, 24, 418. [Google Scholar] [CrossRef]
- Zhai, X.; Lan, D.; Jia, M.; Gan, Z.; Chen, C.; Zeng, X.; Chen, J.; Kai, W. Characterization and expression analysis of key abscisic acid signal transduction genes during kiwifruit development. Sci. Hortic. 2023, 309, 111672. [Google Scholar] [CrossRef]
- Mou, W.; Li, D.; Bu, J.; Jiang, Y.; Khan, Z.U.; Luo, Z.; Mao, L.; Ying, T. Comprehensive Analysis of ABA Effects on Ethylene Biosynthesis and Signaling during Tomato Fruit Ripening. PLoS ONE 2016, 11, e0154072. [Google Scholar] [CrossRef]
- Bai, Z.Q.; Zu, H.; Wang, R.; Gao, X.; Zou, T.; Chen, G.L.; Wu, J. Molecular role of ethylene in fruit ripening of Ziziphus jujube Mill. Plant Signal. Behav. 2020, 15, 1834749. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Kang, C.; Zhang, S.; Li, X. Transcript analyses reveal a comprehensive role of abscisic acid in modulating fruit ripening in Chinese jujube. BMC Plant Biol. 2019, 19, 189. [Google Scholar] [CrossRef]
- Barickman, T.C.; Kopsell, D.A.; Sams, C.E. Applications of Abscisic Acid and Increasing Concentrations of Calcium Affect the Partitioning of Mineral Nutrients between Tomato Leaf and Fruit Tissue. Horticulturae 2019, 5, 49. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, S.; Yin, J.; Li, C.; Zhan, Y.; Xiao, J.; Liang, T.; Li, X. Molecular cloning and promoter analysis of squalene synthase and squalene epoxidase genes from Betula platyphylla. Protoplasma 2016, 253, 1347–1363. [Google Scholar] [CrossRef]
- Paul, P.; Singh, S.K.; Patra, B.; Liu, X.; Pattanaik, S.; Yuan, L. Mutually Regulated AP2/ERF Gene Clusters Modulate Biosynthesis of Specialized Metabolites in Plants. Plant Physiol. 2020, 182, 840–856. [Google Scholar] [CrossRef]
- Salazar, J.; Zapata, P.; Silva, C.; González, M.; Pacheco, I.; Bastías, M.; Meneses, C.; Jorquera, C.; Moreno, I.; Shinya, P.; et al. Transcriptome analysis and postharvest behavior of the kiwifruit ‘Actinidia deliciosa’ reveal the role of ethylene-related phytohormones during fruit ripening. Tree Genet. Genomes 2021, 17, 8. [Google Scholar] [CrossRef]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic Acid Signaling Pathway in Plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef]
- Zhao, Y.; Song, C.; Brummell, D.A.; Qi, S.; Lin, Q.; Duan, Y. Jasmonic acid treatment alleviates chilling injury in peach fruit by promoting sugar and ethylene metabolism. Food Chem. 2020, 338, 128005. [Google Scholar] [CrossRef]
- Cao, Y.; Liu, L.; Ma, K.; Wang, W.; Lv, H.; Gao, M.V.; Wang, X.; Zhang, X.; Ren, S.; Zhang, N.; et al. The jasmonate-induced bHLH gene SlJIG functions in terpene biosynthesis and resistance to insects and fungus. J. Integr. Plant Biol. 2022, 64, 1102–1115. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.Y.; Shi, X.C.; Liu, F.Q.; Laborda, P. Effects of exogenous methyl jasmonate on quality and preservation of postharvest fruits: A review. Food Chem. 2021, 353, 129482. [Google Scholar] [CrossRef]
- Kuhn, N.; Ponce, C.; Arellano, M.; Time, A.; Sagredo, B.; Donoso, J.M.; Meisel, L.A. Gibberellic Acid Modifies the Transcript Abundance of ABA Pathway Orthologs and Modulates Sweet Cherry (Prunus avium) Fruit Ripening in Early- and Mid-Season Varieties. Plants 2020, 9, 1796. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wu, T.; Liu, J.; Cong, L.; Zhu, Y.; Zhai, R.; Yang, C.; Wang, Z.; Ma, F.; Xu, L. PbGA20ox2 Regulates Fruit Set and Induces Parthenocarpy by Enhancing GA4 Content. Front. Plant Sci. 2020, 11, 113. [Google Scholar] [CrossRef]
- Liu, Y.; Lei, X.; Guo, Y.; Yao, S.; Zeng, K. Effects of methionine treatment on storage quality and antioxidant activity of postharvest jujube fruit. J. Integr. Agric. 2023, 22, 2893–2904. [Google Scholar] [CrossRef]
- Dou, J.-F.; Kou, X.-H.; Wu, C.-E.; Fan, G.-J.; Li, T.-T.; Li, X.-J.; Zhou, D.-D.; Yan, Z.-C.; Zhu, J.-P. Recent advances and development of postharvest management research for fresh jujube fruit: A review. Sci. Hortic. 2023, 310, 111769. [Google Scholar] [CrossRef]
- Fenn, M.A.; Giovannoni, J.J. Phytohormones in fruit development and maturation. Plant J. 2021, 105, 446–458. [Google Scholar] [CrossRef]
- Yang, Y.; Hao, C.; Du, J.; Xu, L.; Guo, Z.; Li, D.; Cai, H.; Guo, H.; Li, L. The carboxy terminal transmembrane domain of SPL7 mediates interaction with RAN1 at the endoplasmic reticulum to regulate ethylene signaling in Arabidopsis. New Phytol. 2022, 236, 878–892. [Google Scholar] [CrossRef]
- Li, J.; Liang, C.; Liu, X.; Huai, B.; Chen, J.; Yao, Q.; Qin, Y.; Liu, Z.; Luo, X. Effect of Zn and NAA co-treatment on the occurrence of creasing fruit and the peel development of ‘Shatangju’ mandarin. Sci. Hortic. 2016, 201, 230–237. [Google Scholar] [CrossRef]
- Wang, Z.G.; Yu, Y.H.; Guo, D.L. Advances in ROS promoting fruit development and ripening. Acta Agric. Zhejiangensis 2020, 32, 2103–2110. [Google Scholar] [CrossRef]
- Li, Y.; Lin, Y.; Jia, B.; Chen, G.; Shi, H.N.; Xu, R.; Li, X.; Tang, J.; Tang, Q.; Zhang, G.; et al. Transcriptome Analysis Reveals Molecular Mechanisms Underlying Methyl Jasmonate-mediated Biosynthesis of Protopanaxadiol-type Saponins in Panax notoginseng Leaves. J. Plant Biol. 2022, 65, 29–41. [Google Scholar] [CrossRef]
- Liu, Y.; Tang, L.; Wang, C.; Li, J. NAA and 6-BA promote accumulation of oleanolic acid by JA regulation in Achyranthes bidentata Bl. PLoS ONE 2020, 15, e0229490. [Google Scholar] [CrossRef]
- Kou, X.H.; Yang, S.; Chai, L.P.; Wu, C.E.; Zhou, J.Q.; Liu, Y.F.; Xue, Z. Abscisic acid and fruit ripening: Multifaceted analysis of the effect of abscisic acid on fleshy fruit ripening. Sci. Hortic. 2021, 281, 109999. [Google Scholar] [CrossRef]
- Wu, W.; Cao, S.F.; Shi, L.Y.; Chen, W.; Yin, X.R.; Yang, Z.F. Abscisic acid biosynthesis, metabolism and signaling in ripening fruit. Front. Plant Sci. 2023, 14, 1279031. [Google Scholar] [CrossRef]
- Liu, Y.D.; Tang, M.F.; Liu, M.C.; Su, D.; Chen, J.; Gao, Y.S.; Bouzayen, M.; Li, Z. The Molecular Regulation of Ethylene in Fruit Ripening. Small Methods 2020, 4, 1900485. [Google Scholar] [CrossRef]
- Zhang, Z.; Huang, J.; Li, X. Transcript analyses of ethylene pathway genes during ripening of Chinese jujube fruit. J. Plant Physiol. 2018, 224, 1–10. [Google Scholar] [CrossRef]
- Huang, W.; Hu, N.; Xiao, Z.; Qiu, Y.; Yang, Y.; Yang, J.; Mao, X.; Wang, Y.; Li, Z.; Guo, H. A molecular framework of ethylene-mediated fruit growth and ripening processes in tomato. Plant Cell 2022, 34, 3280–3300. [Google Scholar] [CrossRef]
- Li, T.; Xu, Y.; Zhang, L.; Ji, Y.; Tan, D.; Yuan, H.; Wang, A. The Jasmonate-Activated Transcription Factor MdMYC2 Regulates ETHYLENE RESPONSE FACTOR and Ethylene Biosynthetic Genes to Promote Ethylene Biosynthesis during Apple Fruit Ripening. Plant Cell 2017, 29, 1316–1334. [Google Scholar] [CrossRef]
- Ding, X.; Wang, B.; Gong, Y.; Yan, X.; Chen, X.; Zhong, Y.; Zhao, Z. Exogenous Methyl-Jasmonate (MeJA) Improves ‘Ruixue’ Apple Fruit Quality by Regulating Cell Wall Metabolism. Foods 2024, 13, 1594. [Google Scholar] [CrossRef]
- Premathilake, A.T.; Ni, J.; Shen, J.; Bai, S.; Teng, Y. Transcriptome analysis provides new insights into the transcriptional regulation of methyl jasmonate-induced flavonoid biosynthesis in pear calli. BMC Plant Biol. 2020, 20, 388. [Google Scholar] [CrossRef]
- Xi, H.; He, Y.; Chen, H. Functional Characterization of SmbHLH13 in Anthocyanin Biosynthesis and Flowering in Eggplant. Hortic. Plant J. 2021, 7, 73–80. [Google Scholar] [CrossRef]
- Hu, Y.N.; Sun, H.L.; Han, Z.Y.; Wang, S.; Wang, T.; Li, Q.; Tian, J.; Wang, Y.; Zhang, X.; Xu, X.; et al. ERF4 affects fruit ripening by acting as a JAZ interactor between ethylene and jasmonic acid hormone signaling pathways. Hortic. Plant J. 2022, 8, 689–699. [Google Scholar] [CrossRef]
- Lorenzo, O.; Piqueras, R.; Sánchez-Serrano, J.J.; Solano, R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 2003, 15, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Zhao, Y.; Tao, R.; Yin, L.; Gao, L.; Strid, A.; Qian, M.; Li, J.; Li, Y.; Shen, J.; et al. Ethylene mediates the branching of the jasmonate-induced flavonoid biosynthesis pathway by suppressing anthocyanin biosynthesis in red Chinese pear fruits. Plant Biotechnol. J. 2020, 18, 1223–1240. [Google Scholar] [CrossRef]
- Ali, A.; Zhao, Q.Y.; Zhang, H.M.; Zhang, S.; He, L.; Wang, F.D.; Gao, J.W. The fundamental role of DELLA protein and regulatory mechanism during plant growth and development. Not. Bot. Horti Agrobot. Cluj-Napoca 2021, 49, 12561. [Google Scholar] [CrossRef]
- Xue, H.D.; Gao, X.; He, P.; Xiao, G.H. Origin, evolution, and molecular function of DELLA proteins in plants. Crop J. 2022, 10, 287–299. [Google Scholar] [CrossRef]
- Wu, M.; Liu, K.; Li, H.; Li, Y.; Zhu, Y.; Su, D.; Zhang, Y.; Deng, H.; Wang, Y.; Liu, M. Gibberellins involved in fruit ripening and softening by mediating multiple hormonal signals in tomato. Hortic. Res. 2023, 11, uhad275. [Google Scholar] [CrossRef]
- Nagai, K.; Mori, Y.; Ishikawa, S.; Furuta, T.; Gamuyao, R.; Niimi, Y.; Hobo, T.; Fukuda, M.; Kojima, M.; Takebayashi, Y.; et al. Antagonistic regulation of the gibberellic acid response during stem growth in rice. Nature 2020, 584, 109–114. [Google Scholar] [CrossRef]
- Liu, M.J.; Zhao, J.; Cai, Q.L.; Liu, G.C.; Wang, J.R.; Zhao, Z.H.; Liu, P.; Dai, L.; Yan, G.; Wang, W.-J.; et al. The complex jujube genome provides insights into fruit tree biology. Nat. Commun. 2021, 5, 5315. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, S.; Zhang, Y.; Shi, B.; Wang, R.; Zhang, Z.; Wu, J.; Bai, Z.; Chen, G. Transcriptome Screening of Hormone-Regulated Genes Related to Fruit Development in Zizyphus jujuba Mill. cv. Goutou Fruits at Different Ripening Stages. Int. J. Mol. Sci. 2025, 26, 3476. https://doi.org/10.3390/ijms26083476
Luo S, Zhang Y, Shi B, Wang R, Zhang Z, Wu J, Bai Z, Chen G. Transcriptome Screening of Hormone-Regulated Genes Related to Fruit Development in Zizyphus jujuba Mill. cv. Goutou Fruits at Different Ripening Stages. International Journal of Molecular Sciences. 2025; 26(8):3476. https://doi.org/10.3390/ijms26083476
Chicago/Turabian StyleLuo, Shuting, Yusen Zhang, Beibei Shi, Rui Wang, Ziyan Zhang, Jiawen Wu, Zhenqing Bai, and Guoliang Chen. 2025. "Transcriptome Screening of Hormone-Regulated Genes Related to Fruit Development in Zizyphus jujuba Mill. cv. Goutou Fruits at Different Ripening Stages" International Journal of Molecular Sciences 26, no. 8: 3476. https://doi.org/10.3390/ijms26083476
APA StyleLuo, S., Zhang, Y., Shi, B., Wang, R., Zhang, Z., Wu, J., Bai, Z., & Chen, G. (2025). Transcriptome Screening of Hormone-Regulated Genes Related to Fruit Development in Zizyphus jujuba Mill. cv. Goutou Fruits at Different Ripening Stages. International Journal of Molecular Sciences, 26(8), 3476. https://doi.org/10.3390/ijms26083476




