Identification of R2R3-MYB Gene Family and Functional Analysis of Responses of S22 Subfamily to Abiotic Stresses in Dandelion (Taraxacum mongolicum Hand.-Mazz.)
Abstract
1. Introduction
2. Results
2.1. Identification, Physicochemical Properties, and Chromosomal Mapping of 130 TmR2R3-MYB TFs in Dandelions
2.2. Gene Structure and Sequence Analysis of TmR2R3-MYBs
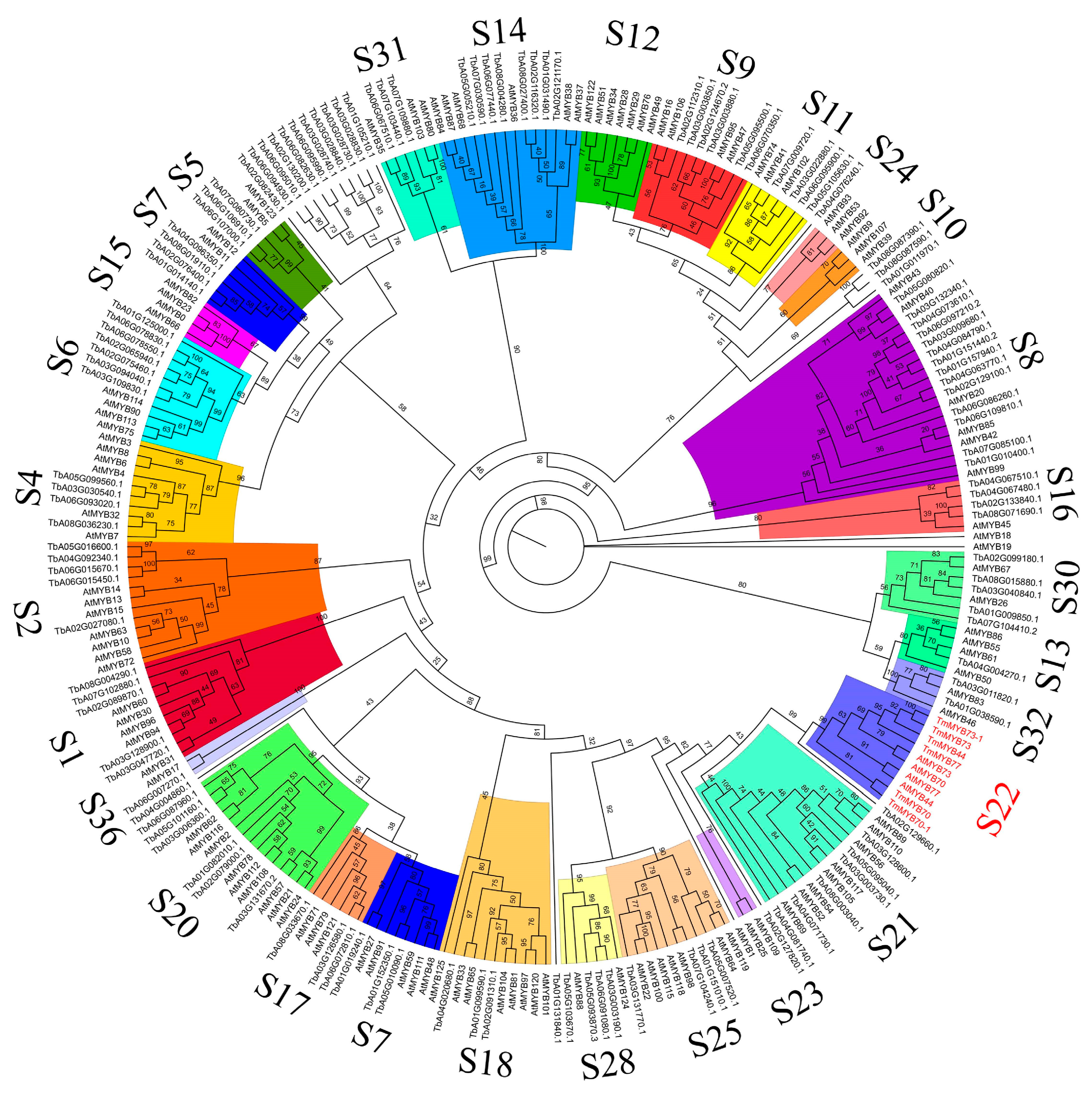
2.3. Phylogenetic Analysis of TmR2R3-MYBs
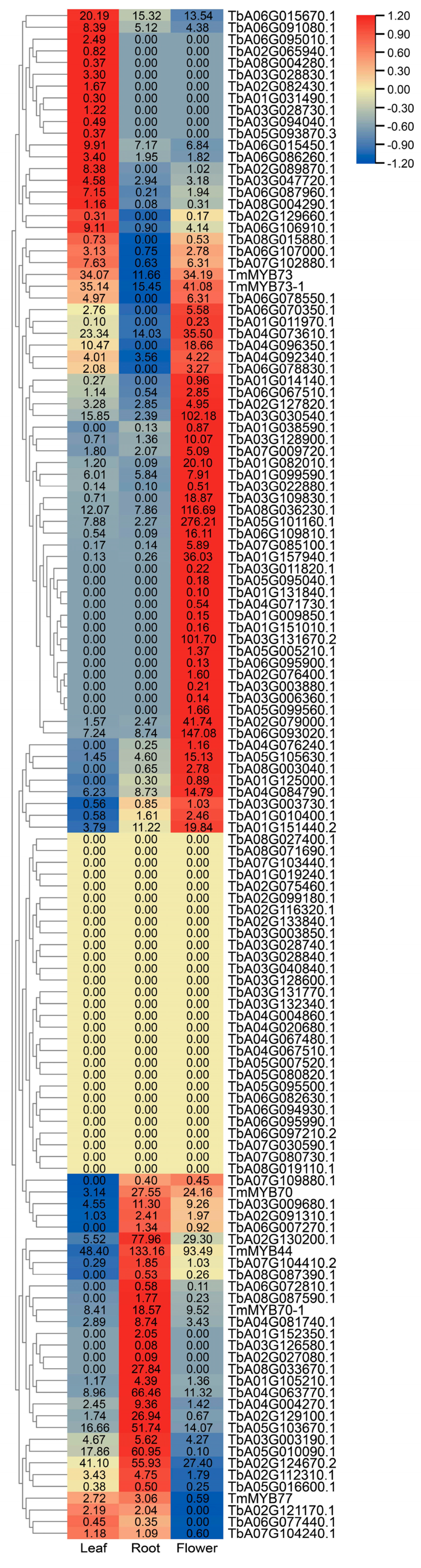
2.4. Expression Profiles of TmR2R3-MYBs in Different Tissues
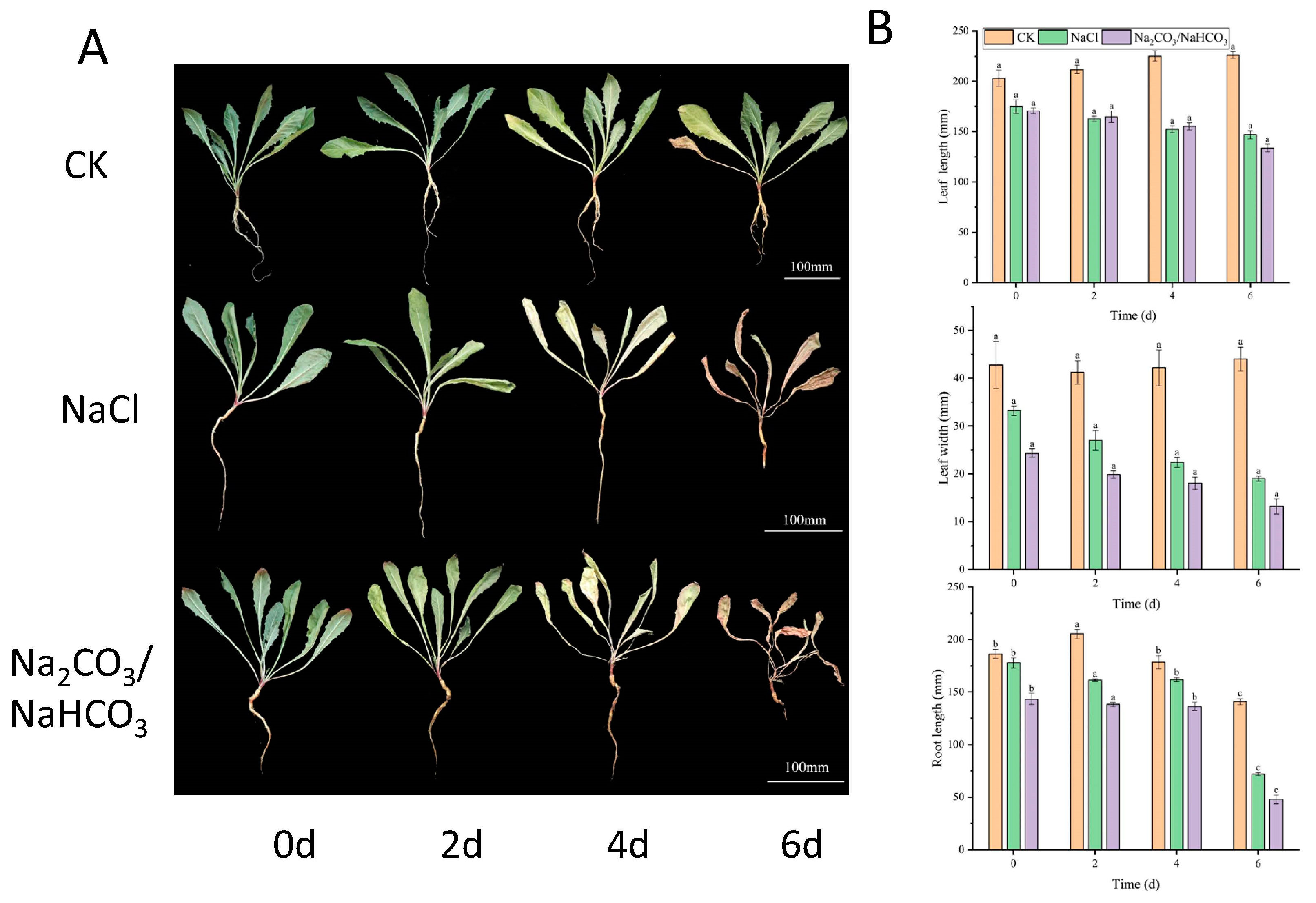
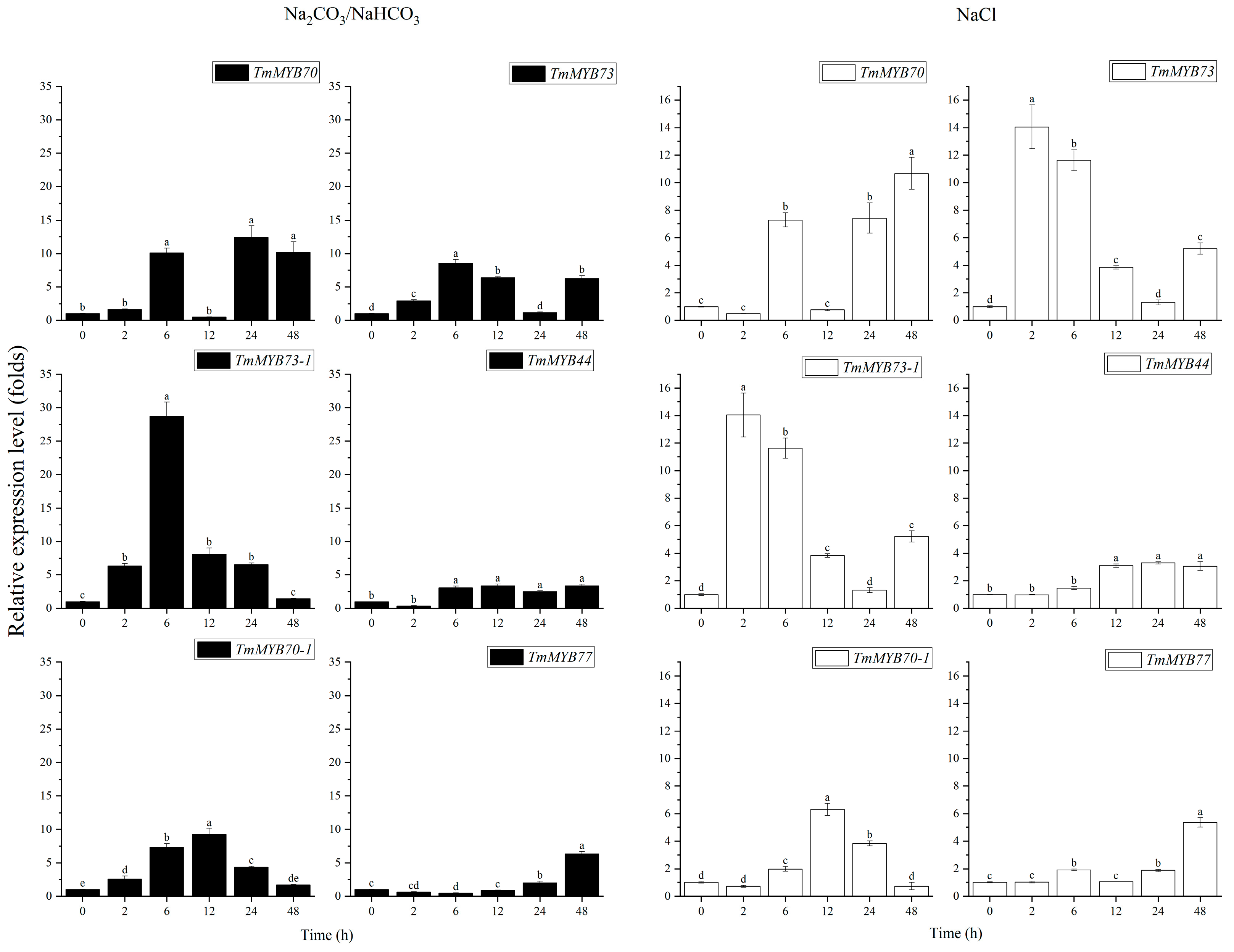
2.5. Expression Profiles of S22 Subfamily Members After Subjecting Dandelions to Different Saline–Alkaline Stress Treatments
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Identification of TmR2R3-MYB Gene Family
4.3. Analysis of TmR2R3-MYBs’ Physicochemical Properties and Conserved Sequence
4.4. Phylogenetic Analysis of TmMYBs
4.5. Analysis of Dandelion Tissue-Specific Expression Patterns
4.6. RNA Extraction and Analysis of Gene Expression Pattern
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, X.; Li, Y.; Huang, Y.; Shen, J.; Xu, H.; Li, K. Integrative analysis of the metabolome and transcriptome reveals the mechanism of polyphenol biosynthesis in Taraxacum mongolicum. Front. Plant Sci. 2024, 15, 1418585. [Google Scholar]
- Lee, Y.S.; Kim, J.; Woo, S.; Park, J.Y.; Park, H.-S.; Shim, H.; Choi, H.-I.; Kang, J.H.; Lee, T.J.; Sung, S.H.; et al. Assessing the genetic and chemical diversity of Taraxacum species in the Korean Peninsula. Phytochemistry 2021, 181, 112576. [Google Scholar] [PubMed]
- Wei, S.; Zhou, Q.; Wang, X. Identification of weed plants excluding the uptake of heavy metals. Environ. Int. 2005, 31, 829–834. [Google Scholar] [PubMed]
- Wei, S.; Zhou, Q.; Mathews, S. A newly found cadmium accumulator-Taraxacum mongolicum. J. Hazard. Mater. 2008, 159, 544–547. [Google Scholar] [CrossRef]
- Wu, Z.; Meng, R.; Feng, W.; Wongsnansilp, T.; Li, Z.J.; Lu, X.L.; Wang, X.P. Study of Dandelion (Taraxacum mongolicum Hand.-Mazz.) Salt Response and Caffeic Acid Metabolism under Saline Stress by Transcriptome Analysis. Genes 2024, 15, 220. [Google Scholar] [CrossRef]
- Yue, X.; Zhong, L.; Ye, M.; Luan, Y.; Zhang, Q.; Wang, Q. Taraxacum mongolicum polysaccharide promotes white adipocyte browning by regulating miR-134-3p via Akt/GSK-3β signalling. Int. J. Biol. Macromol. 2024, 257, 128296. [Google Scholar]
- Xue, H.; Nima, L.; Wang, S.; Tan, J. Ultrasound assisted hot water extraction of polysaccharides from Taraxacum mongolicum: Optimization, purification, structure characterization, and antioxidant activity. J. Food Sci. 2024, 89, 2827–2842. [Google Scholar]
- Liu, Y.; Shi, Y.; Zou, J.; Zhang, X.; Zhai, B.; Guo, D.; Sun, J.; Luan, F. Extraction, purification, structural features, biological activities, modifications, and applications from Taraxacum mongolicum polysaccharides: A review. Int. J. Biol. Macromol. 2024, 259, 129193. [Google Scholar]
- Kim, Y.H.; Choo, S.J.; Ryoo, I.J.; Ahn, J.S.; Yoo, I.D. Eudesmanolides from Taraxacum mongolicum and their inhibitory effects on the production of nitric oxide. Arch. Pharm. Res. 2011, 34, 37–41. [Google Scholar] [CrossRef]
- Jia, Y.Y.; Guan, R.F.; Wu, Y.H.; Yu, X.P.; Lin, W.Y.; Zhang, Y.Y.; Liu, T.; Zhao, J.; Shi, S.Y.; Zhao, Y. Taraxacum mongolicum extract exhibits a protective effect on hepatocytes and an antiviral effect against hepatitis B virus in animal and human cells. Mol. Med. Rep. 2014, 9, 1381–1387. [Google Scholar] [CrossRef]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Frangedakis, E.; Yelina, N.E.; Billakurthi, K.; Hua, L.; Schreier, T.; Dickinson, P.J.; Tomaselli, M.; Haseloff, J.; Hibberd, J.M. MYB-related transcription factors control chloroplast biogenesis. Cell 2024, 187, 4859–4876.e22. [Google Scholar] [CrossRef] [PubMed]
- Gong, Q.; Zhou, M.; Li, X.; Guo, Y. Transcription factor MYB8 regulates iron deficiency stress response in Arabidopsis. Plant Sci. 2024, 340, 111973. [Google Scholar] [CrossRef]
- Li, Y.; Tian, B.; Wang, Y.; Wang, J.; Zhang, H.; Wang, L.; Sun, G.; Yu, Y.; Zhang, H. The transcription factor MYB37 positively regulates photosynthetic inhibition and oxidative damage in Arabidopsis leaves under salt stress. Front. Plant Sci. 2022, 13, 943153. [Google Scholar] [CrossRef]
- Kim, D.; Jeon, S.J.; Yanders, S.; Park, S.C.; Kim, H.S.; Kim, S. MYB3 plays an important role in lignin and anthocyanin biosynthesis under salt stress condition in Arabidopsis. Plant Cell Rep. 2022, 41, 1549–1560. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Qiu, T.; Yue, J.; Guo, N.; He, Y.; Han, X.; Wang, Q.; Jia, P.; Wang, H.; Li, M. Arabidopsis ADF1 is regulated by MYB73 and is involved in response to salt stress affecting actin filament organization. Plant Cell Physiol. 2021, 62, 1387–1395. [Google Scholar] [CrossRef]
- Wang, C.; Lei, J.; Jin, X.; Chai, S.; Jiao, C.; Yang, X.; Wang, L. A Sweet Potato MYB Transcription Factor IbMYB330 Enhances Tolerance to Drought and Salt Stress in Transgenic Tobacco. Genes 2024, 15, 693. [Google Scholar] [CrossRef]
- Hu, J.; Zou, S.; Huang, J.; Huan, X.; Jin, X.; Zhou, L.; Zhao, K.; Han, Y.; Wang, S. PagMYB151 facilitates proline accumulation to enhance salt tolerance of poplar. BMC Genom. 2023, 24, 345. [Google Scholar] [CrossRef]
- Ren, C.; Li, Z.; Song, P.; Wang, Y.; Liu, W.; Zhang, L.; Li, X.; Li, W.; Han, D. Overexpression of a Grape MYB Transcription Factor Gene VhMYB2 Increases Salinity and Drought Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2023, 24, 10743. [Google Scholar] [CrossRef]
- Zhou, Z.; Wei, X.; Lan, H. CgMYB1, an R2R3-MYB transcription factor, can alleviate abiotic stress in an annual halophyte Chenopodium glaucum. Plant Physiol. Biochem. 2023, 196, 484–496. [Google Scholar] [CrossRef]
- Hu, D.; Cui, R.; Wang, K.; Yang, Y.; Wang, R.; Zhu, H.; He, M.; Fan, Y.; Wang, L.; Wang, L.; et al. The Myb73-GDPD2-GA2ox1 transcriptional regulatory module confers phosphate deficiency tolerance in soybean. Plant Cell 2024, 36, 2176–2200. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yang, Z.; Ding, Y.; Liu, L.; Han, X.; Zhan, J.; Wei, X.; Diao, Y.; Qin, W.; Wang, P.; et al. Over-expression of an R2R3 MYB Gene, GhMYB73, increases tolerance to salt stress in transgenic Arabidopsis. Plant Sci. 2019, 286, 28–36. [Google Scholar] [PubMed]
- Gu, K.D.; Zhang, Q.Y.; Yu, J.Q.; Wang, J.H.; Zhang, F.J.; Wang, C.K.; Zhao, Y.W.; Sun, C.H.; You, C.X.; Hu, D.G.; et al. R2R3-MYB Transcription Factor MdMYB73 Confers Increased Resistance to the Fungal Pathogen Botryosphaeria dothidea in Apples via the Salicylic Acid Pathway. J. Agric. Food Chem. 2021, 69, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wen, J.; Xia, Y.; Zhang, L.; Du, H. Evolution and functional diversification of R2R3-MYB transcription factors in plants. Hortic. Res. 2022, 9, uhac058. [Google Scholar] [CrossRef]
- Jia, B.-B.; Zhou, X.-N.; Ding, S.-L.; Shi, Z.-Q.; Hao, L.-J.; Xu, J.; Guo, W. Effects of arbuscular mycorrhizal fungi inoculation on growth and salt ion accumulation of sunflower under different saline-alkali stresses. J. S. China Agric. Univ. (Nat. Sci. Ed.) 2021, 42, 45–54. [Google Scholar]
- Lv, H.-Y.; Liang, Z.-W.; Lv, B.-S.; Yin, H.-B. Responses of rice (Oryza sativa L.) mutant ML04 and its wild type seedling growth to saline-alkaline stress. Res. Crops 2015, 16, 1–14. [Google Scholar] [CrossRef]
- Wang, Z.; Li, X.; Yao, X.; Ma, J.; Lu, K.; An, Y.; Sun, Z.; Wang, Q.; Zhou, M.; Qin, L.; et al. MYB44 regulates PTI by promoting the expression of EIN2 and MPK3/6 in Arabidopsis. Plant Commun. 2023, 4, 100628. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, S.; Xu, L.; Chu, S.; Yan, X.; Lin, L.; Wen, J.; Zheng, B.; Chen, S.; Li, Q. Transcription factor PagMYB31 positively regulates cambium activity and negatively regulates xylem development in poplar. Plant Cell 2024, 36, 1806–1828. [Google Scholar] [CrossRef]
- Su, T.; Liu, H.; Wu, Y.; Wang, J.; He, F.; Li, H.; Li, S.; Wang, L.; Li, L.; Cao, J.; et al. Soybean hypocotyl elongation is regulated by a MYB33-SWEET11/21-GA2ox8c module involving long-distance sucrose transport. Plant Biotechnol. J. 2024, 22, 2859–2872. [Google Scholar]
- Han, Y.; Yang, R.; Zhang, X.; Wang, Q.; Wang, Y.; Li, Y.; Prusky, D.; Bi, Y. MYB24, MYB144, and MYB168 positively regulate suberin biosynthesis at potato tuber wounds during healing. Plant J. 2024, 119, 1239–1257. [Google Scholar] [CrossRef]
- Nagaoka, S.; Takano, T. Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J. Exp. Bot. 2003, 54, 2231–2237. [Google Scholar] [PubMed]
- Qin, Y.; Wang, M.; Tian, Y.; He, W.; Han, L.; Xia, G. Over-expression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis. Mol. Biol. Rep. 2012, 39, 7183–7192. [Google Scholar] [PubMed]
- Xu, R.; Wang, Y.; Zheng, H.; Lu, W.; Wu, C.; Huang, J.; Yan, K.; Yang, G.; Zheng, C. Salt-induced transcription factor MYB74 is regulated by the RNA-directed DNA methylation pathway in Arabidopsis. J. Exp. Bot. 2015, 66, 5997–6008. [Google Scholar] [PubMed]
- Ganesan, G.; Sankararamasubramanian, H.; Harikrishnan, M.; Ashwin, G.; Parida, A. A MYB transcription factor from the grey mangrove is induced by stress and confers NaCl tolerance in tobacco. J. Exp. Bot. 2012, 63, 4549–4561. [Google Scholar]
- Zhang, F.; Zhang, X.; Wan, W.; Zhu, X.; Shi, M.; Zhang, L.; Yang, F.; Jin, S. MYB4 in Lilium pumilum affects plant saline-alkaline tolerance. Plant Signal. Behav. 2024, 19, 2370724. [Google Scholar]
- Wang, F.; Yang, F.; Zhu, D.; Saniboere, B.; Zhou, B.; Peng, D. MYB44 plays key roles in regulating plant responses to abiotic and biotic stress, metabolism, and development. J. Plant Biochem. Biotechnol. 2024, 33, 462–473. [Google Scholar]
- Zhang, W.; Zhang, N.; Qin, Q.; Zhang, X.; Zhang, J.; Yang, T.; Zhang, Y.; Dong, J.; Che, D. RmMYB44 Confers Resistance to Chilling, Drought, and Salt Stress in Both Rosa multiflora and Tobacco. Agriculture 2024, 14, 1212. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Cheong, J.J. H2A.Z-containing nucleosomes are evicted to activate AtMYB44 transcription in response to salt stress. Biochem. Biophys. Res. Commun. 2018, 499, 1039–1043. [Google Scholar]
- Sharifi Alishah, M.; Darvishzadeh, R.; Ahmadabadi, M.; Piri Kashtiban, Y.; Hasanpur, K. Identification of differentially expressed genes in salt-tolerant oilseed sunflower (Helianthus annuus L.) genotype by RNA sequencing. Mol. Biol. Rep. 2022, 49, 3583–3596. [Google Scholar] [CrossRef]
- He, Y.; Li, W.; Lv, J.; Jia, Y.; Wang, M.; Xia, G. Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 1511–1522. [Google Scholar]
- Shin, R.; Burch, A.Y.; Huppert, K.A.; Tiwari, S.B.; Murphy, A.S.; Guilfoyle, T.J.; Schachtman, D.P. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 2007, 19, 2440–2453. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Zhao, Y.; Gao, J.; Xiang, C.; Zhu, J.K. The ABA receptor PYL9 together with PYL8 plays an important role in regulating lateral root growth. Sci. Rep. 2016, 6, 27177. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Xing, L.; Wang, X.; Hou, Y.J.; Gao, J.; Wang, P.; Duan, C.G.; Zhu, X.; Zhu, J.K. The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci. Signal. 2014, 7, ra53. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef]
- Qin, B.; Fan, S.L.; Yu, H.Y.; Lu, Y.X.; Wang, L.F. HbMYB44, a Rubber Tree MYB Transcription Factor with Versatile Functions in Modulating Multiple Phytohormone Signaling and Abiotic Stress Responses. Front. Plant Sci. 2022, 13, 893896. [Google Scholar] [CrossRef]
- Larralde, M.; Zeller, G. PyHMMER: A Python library binding to HMMER for efficient sequence analysis. Bioinformatics 2023, 39, btad214. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using ClustalW and ClustalX, 2008/09/17, ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2003; Chapter 2; pp. 2.3.1–2.3.22. [Google Scholar]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, L.; Fan, S.; Qin, B.; Wang, J.; Wang, L.; Liu, S. Identification of R2R3-MYB Gene Family and Functional Analysis of Responses of S22 Subfamily to Abiotic Stresses in Dandelion (Taraxacum mongolicum Hand.-Mazz.). Int. J. Mol. Sci. 2025, 26, 3422. https://doi.org/10.3390/ijms26073422
Lu L, Fan S, Qin B, Wang J, Wang L, Liu S. Identification of R2R3-MYB Gene Family and Functional Analysis of Responses of S22 Subfamily to Abiotic Stresses in Dandelion (Taraxacum mongolicum Hand.-Mazz.). International Journal of Molecular Sciences. 2025; 26(7):3422. https://doi.org/10.3390/ijms26073422
Chicago/Turabian StyleLu, Liangruinan, Songle Fan, Bi Qin, Jingang Wang, Lifeng Wang, and Shizhong Liu. 2025. "Identification of R2R3-MYB Gene Family and Functional Analysis of Responses of S22 Subfamily to Abiotic Stresses in Dandelion (Taraxacum mongolicum Hand.-Mazz.)" International Journal of Molecular Sciences 26, no. 7: 3422. https://doi.org/10.3390/ijms26073422
APA StyleLu, L., Fan, S., Qin, B., Wang, J., Wang, L., & Liu, S. (2025). Identification of R2R3-MYB Gene Family and Functional Analysis of Responses of S22 Subfamily to Abiotic Stresses in Dandelion (Taraxacum mongolicum Hand.-Mazz.). International Journal of Molecular Sciences, 26(7), 3422. https://doi.org/10.3390/ijms26073422




