Genome-Wide Identification and Evolution-Profiling Analysis of Heat Shock Protein Gene Family in Poaceae Barnhart
Abstract
1. Introduction
2. Results
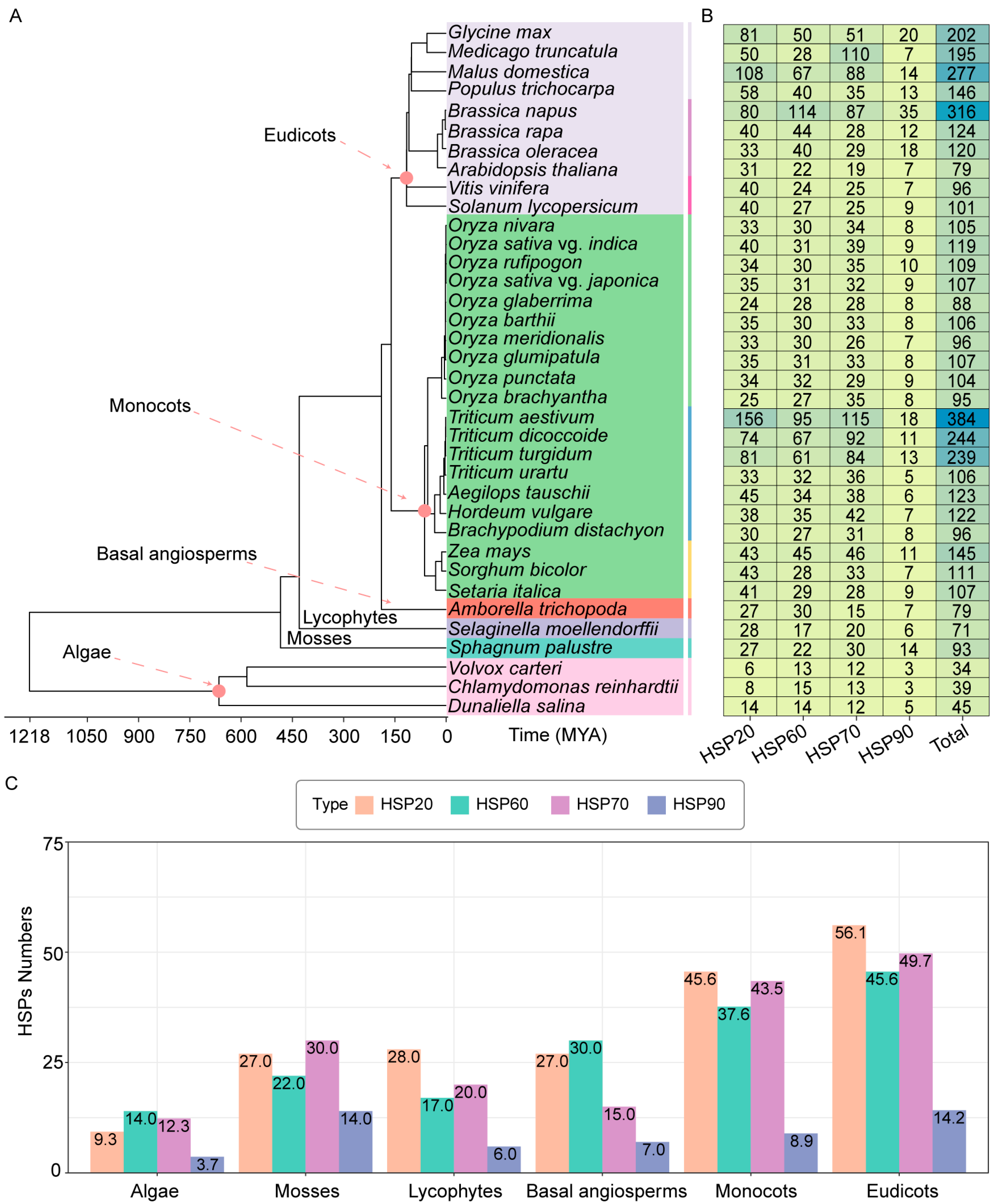
2.1. The Number of HSPs Is Increased in Higher Plants
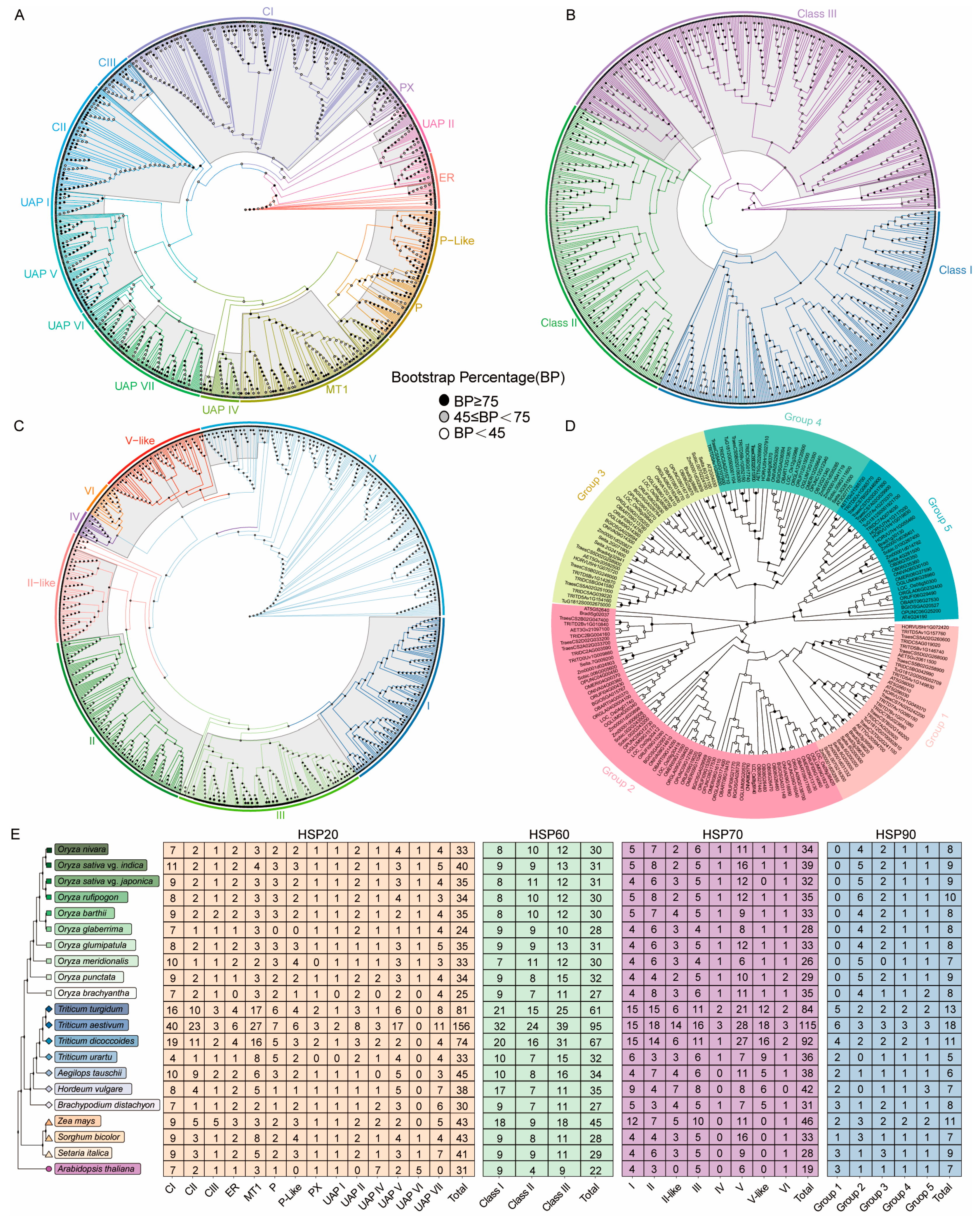
2.2. Phylogenetic Analysis of HSPs in Poaceae
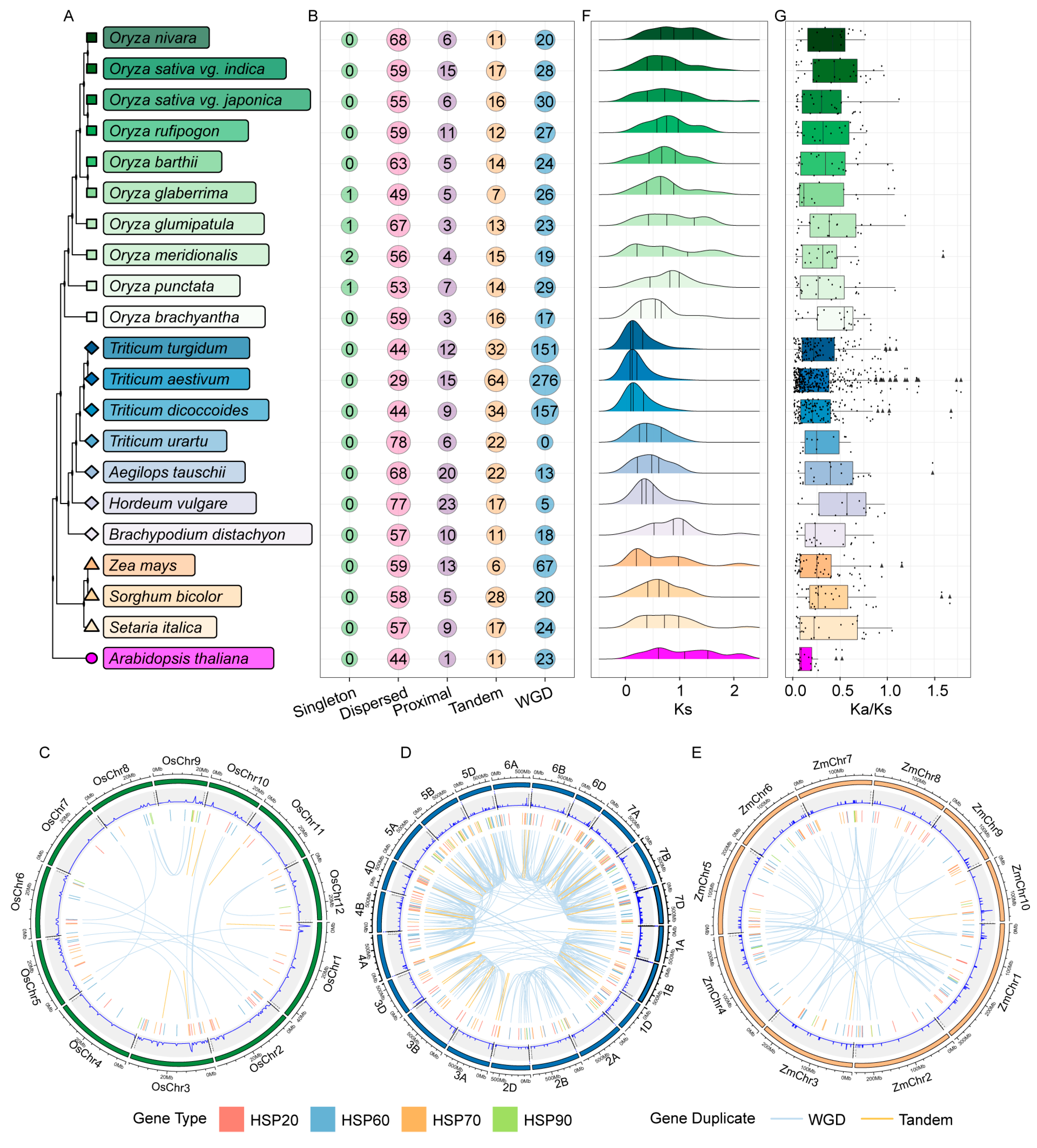
2.3. Gene Duplication Events Lead to the Expansion of HSP Genes
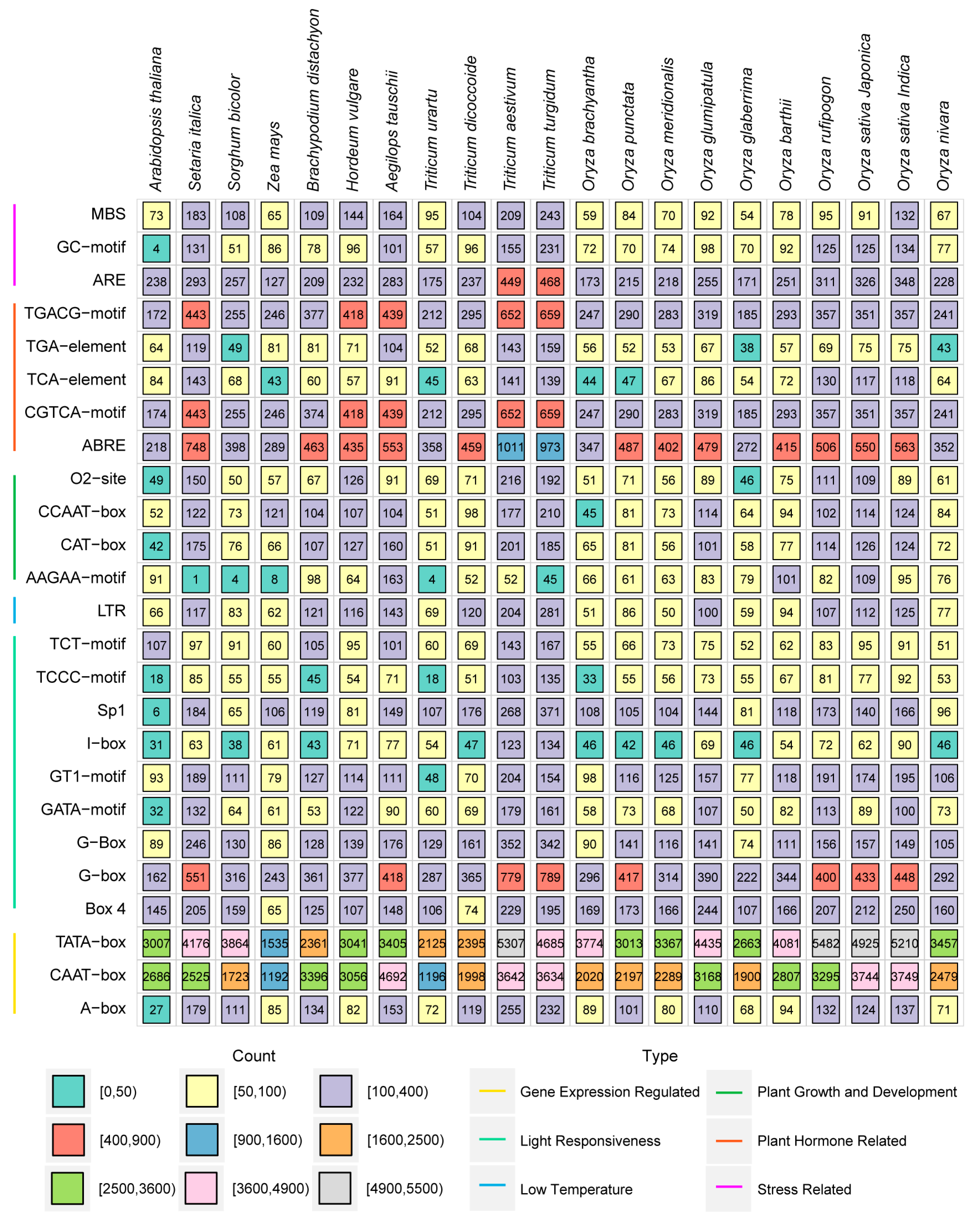
2.4. Cis-Acting Element Analysis of HSP Genes in Poaceae
2.5. Functional Conservation Among HSP Members
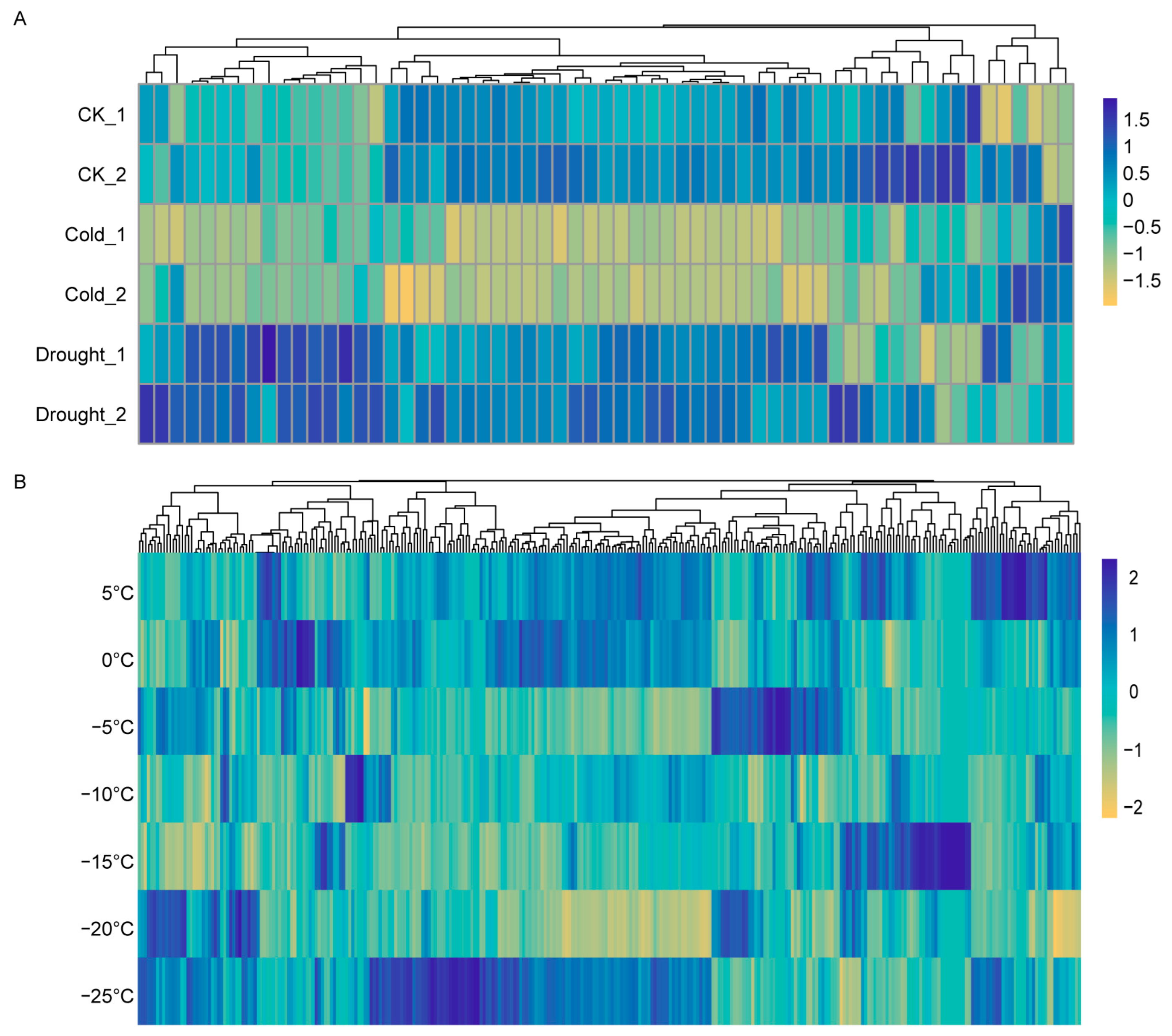
2.6. Transcriptomic Analysis of HSP Expression in Maize and Wheat Under Cold Stress Using RNA-Seq
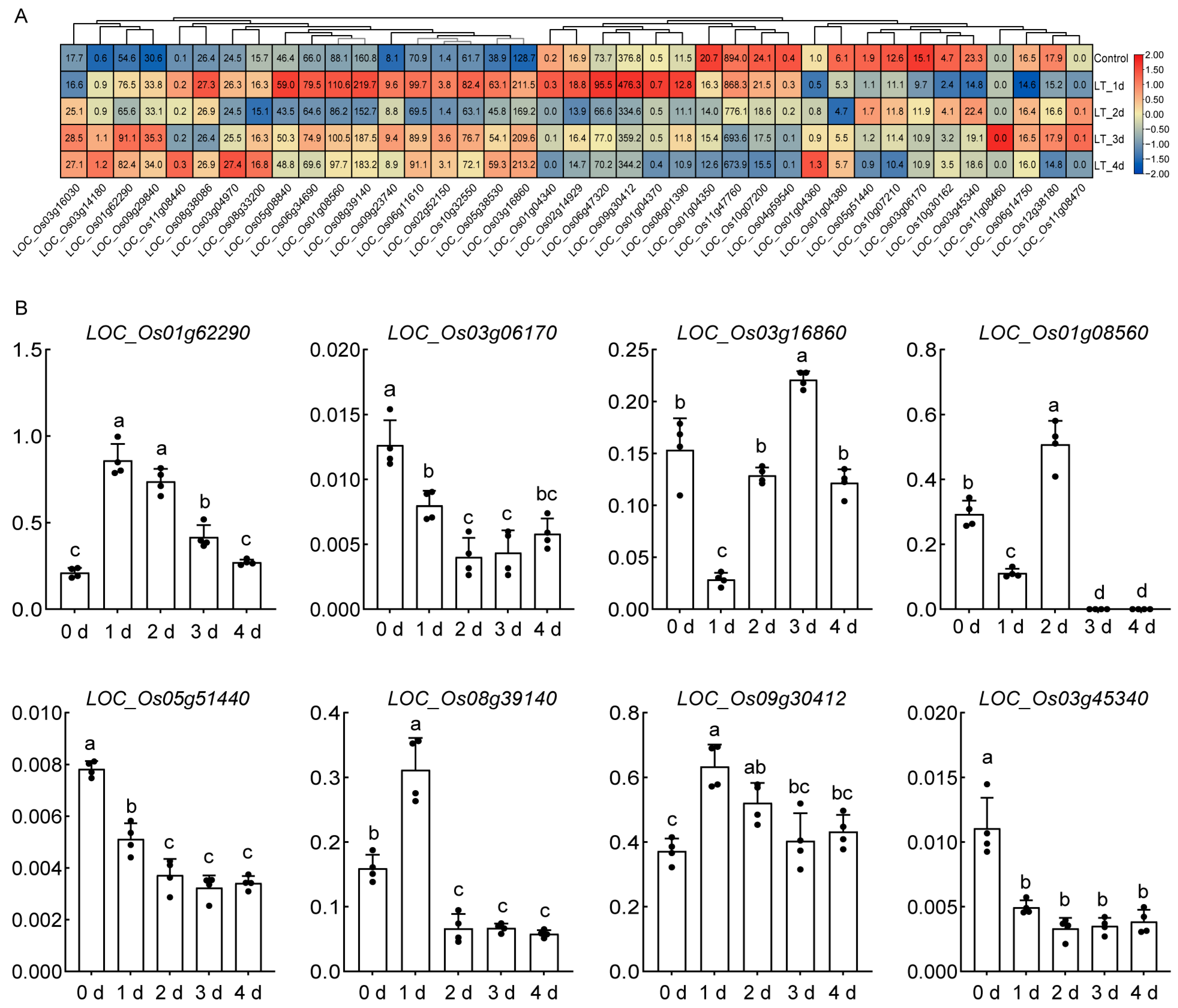
2.7. Expression Patterns of OsHSP Under Cold Stress
3. Discussion
4. Materials and Methods
4.1. Acquisition of Plant Genomic Data and Genome-Wide Identification of HSPs
4.2. Phylogenetic Analysis of HSP Genes
4.3. Chromosomal Distribution, Gene Duplication, Ka/Ks, and Synteny Analysis
4.4. Analysis of Cis-Acting Element HSP Genes
4.5. GO Functional Annotation
4.6. Expression Pattern Analysis of HSP Genes Using RNA-seq Data
4.7. RNA Isolation and Expression Analysis of OsHSP Genes by RT-qPCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Ismail, A.M.; Horie, T. Genomics, Physiology, and Molecular Breeding Approaches for Improving Salt Tolerance. Annu. Rev. Plant Biol. 2017, 68, 405–434. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.K.; Bohnert, H.J. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009, 29, 185–212. [Google Scholar] [CrossRef]
- Zhu, J.-K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003, 6, 441–445. [Google Scholar] [CrossRef]
- Kasuga, M.; Liu, Q.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999, 17, 287–291. [Google Scholar] [CrossRef]
- Repo, T.; Mononen, K.; Alvila, L.; Pakkanen, T.T.; Hänninen, H. Cold acclimation of pedunculate oak (Quercus robur L.) at its northernmost distribution range. Environ. Exp. Bot. 2008, 63, 59–70. [Google Scholar] [CrossRef]
- Jha, U.C.; Bohra, A.; Jha, R. Breeding approaches and genomics technologies to increase crop yield under low-temperature stress. Plant Cell Rep. 2017, 36, 1–35. [Google Scholar] [CrossRef]
- Dangi, A.K.; Sharma, B.; Khangwal, I.; Shukla, P. Combinatorial interactions of biotic and abiotic stresses in plants and their molecular mechanisms: Systems biology approach. Mol. Biotechnol. 2018, 60, 636–650. [Google Scholar] [CrossRef]
- Swindell, W.R.; Huebner, M.; Weber, A.P. Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genom. 2007, 8, 125. [Google Scholar] [CrossRef]
- Timperio, A.M.; Egidi, M.G.; Zolla, L. Proteomics applied on plant abiotic stresses: Role of heat shock proteins (HSP). J. Proteom. 2008, 71, 391–411. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhan, C.; Huang, B. Heat shock proteins in association with heat tolerance in grasses. Int. J. Proteom. 2011, 2011, 529648. [Google Scholar] [CrossRef] [PubMed]
- Al-Whaibi, M.H. Plant heat-shock proteins: A mini review. J. King Saud Univ. Sci. 2011, 23, 139–150. [Google Scholar] [CrossRef]
- Ritossa, F. A new puffing pattern induced by temperature shock and DNP in Drosophila. Experientia 1962, 18, 571–573. [Google Scholar] [CrossRef]
- Lindquist, S.; Craig, E.A. The heat-shock proteins. Annu. Rev. Genet. 1988, 22, 631–677. [Google Scholar] [CrossRef]
- Vierling, E. The roles of heat shock proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1991, 42, 579–620. [Google Scholar] [CrossRef]
- Boston, R.S.; Viitanen, P.V.; Vierling, E. Molecular chaperones and protein folding in plants. Post-Transcr. Control Gene Expr. Plants 1996, 32, 191–222. [Google Scholar]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]
- Waters, E.R. The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 2013, 64, 391–403. [Google Scholar] [CrossRef]
- Sun, W.; Van Montagu, M.; Verbruggen, N. Small heat shock proteins and stress tolerance in plants. Biochim. Biophys. Acta (BBA)-Gene Struct. Expr. 2002, 1577, 1–9. [Google Scholar] [CrossRef]
- Charng, Y.-Y.; Liu, H.-C.; Liu, N.-Y.; Hsu, F.-C.; Ko, S.-S. Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol. 2006, 140, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Basha, E.; O’Neill, H.; Vierling, E. Small heat shock proteins and α-crystallins: Dynamic proteins with flexible functions. Trends Biochem. Sci. 2012, 37, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Matas, M.A.; Nunez, P.; Soto, A.; Allona, I.; Casado, R.; Collada, C.; Guevara, M.A.; Aragoncillo, C.; Gomez, L. Protein cryoprotective activity of a cytosolic small heat shock protein that accumulates constitutively in chestnut stems and is up-regulated by low and high temperatures. Plant Physiol. 2004, 134, 1708–1717. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.K.; Choi, Y.J. A nuclear-localized HSP70 confers thermoprotective activity and drought-stress tolerance on plants. Biotechnol. Lett. 2009, 31, 597–606. [Google Scholar] [CrossRef]
- Fandino, L.B.; Riemann, L.; Steward, G.F.; Long, R.A.; Azam, F. Variations in bacterial community structure during a dinoflagellate bloom analyzed by DGGE and 16S rDNA sequencing. Aquat. Microb. Ecol. 2001, 23, 119–130. [Google Scholar] [CrossRef]
- Tukaj, S.; Bisewska, J.; Roeske, K.; Tukaj, Z. Time-and dose-dependent induction of HSP70 in Lemna minor exposed to different environmental stressors. Bull. Environ. Contam. Toxicol. 2011, 87, 226–230. [Google Scholar] [CrossRef]
- Aparicio, F.; Thomas, C.L.; Lederer, C.; Niu, Y.; Wang, D.; Maule, A.J. Virus induction of heat shock protein 70 reflects a general response to protein accumulation in the plant cytosol. Plant Physiol. 2005, 138, 529–536. [Google Scholar] [CrossRef]
- Jungkunz, I.; Link, K.; Vogel, F.; Voll, L.M.; Sonnewald, S.; Sonnewald, U. AtHsp70-15-deficient Arabidopsis plants are characterized by reduced growth, a constitutive cytosolic protein response and enhanced resistance to TuMV. Plant J. 2011, 66, 983–995. [Google Scholar] [CrossRef]
- Shirasu, K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 2009, 60, 139–164. [Google Scholar] [CrossRef]
- Kadota, Y.; Shirasu, K. The HSP90 complex of plants. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2012, 1823, 689–697. [Google Scholar] [CrossRef]
- Wang, G.F.; Wei, X.; Fan, R.; Zhou, H.; Wang, X.; Yu, C.; Dong, L.; Dong, Z.; Wang, X.; Kang, Z. Molecular analysis of common wheat genes encoding three types of cytosolic heat shock protein 90 (Hsp90): Functional involvement of cytosolic Hsp90s in the control of wheat seedling growth and disease resistance. New Phytol. 2011, 191, 418–431. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hamada, S.; Fujiwara, M.; Zhu, T.; Thao, N.P.; Wong, H.L.; Krishna, P.; Ueda, T.; Kaku, H.; Shibuya, N. The Hop/Sti1-Hsp90 chaperone complex facilitates the maturation and transport of a PAMP receptor in rice innate immunity. Cell Host Microbe 2010, 7, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Mishra, D.; Shekhar, S.; Singh, D.; Chakraborty, S.; Chakraborty, N. Heat shock proteins and abiotic stress tolerance in plants. In Regulation of Heat Shock Protein Responses; Springer International Publishing: Cham, Switzerland, 2018; pp. 41–69. [Google Scholar]
- Krishna, P.; Gloor, G. The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperones 2001, 6, 238. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.-L.; Wang, J.-S.; Liu, H.-C.; Chen, R.-W.; Meyer, Y.; Barakat, A.; Delseny, M. Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones 2001, 6, 201. [Google Scholar] [CrossRef]
- Scharf, K.-D.; Siddique, M.; Vierling, E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing α-crystallin domains (Acd proteins). Cell Stress Chaperones 2001, 6, 225. [Google Scholar] [CrossRef]
- Ouyang, Y.; Chen, J.; Xie, W.; Wang, L.; Zhang, Q. Comprehensive sequence and expression profile analysis of Hsp20 gene family in rice. Plant Mol. Biol. 2009, 70, 341–357. [Google Scholar] [CrossRef]
- Xu, Z.-S.; Li, Z.-Y.; Chen, Y.; Chen, M.; Li, L.-C.; Ma, Y.-Z. Heat shock protein 90 in plants: Molecular mechanisms and roles in stress responses. Int. J. Mol. Sci. 2012, 13, 15706–15723. [Google Scholar] [CrossRef]
- Sarkar, N.K.; Kundnani, P.; Grover, A. Functional analysis of Hsp70 superfamily proteins of rice (Oryza sativa). Cell Stress Chaperones 2013, 18, 427–437. [Google Scholar] [CrossRef]
- Muthusamy, S.K.; Dalal, M.; Chinnusamy, V.; Bansal, K.C. Genome-wide identification and analysis of biotic and abiotic stress regulation of small heat shock protein (HSP20) family genes in bread wheat. J. Plant Physiol. 2017, 211, 100–113. [Google Scholar] [CrossRef]
- Lu, Y.; Zhao, P.; Zhang, A.; Ma, L.; Xu, S.; Wang, X. Alternative splicing diversified the heat response and evolutionary strategy of conserved heat shock protein 90s in hexaploid wheat (Triticum aestivum L.). Front. Genet. 2020, 11, 577897. [Google Scholar] [CrossRef]
- Lai, D.-L.; Yan, J.; Fan, Y.; Li, Y.; Ruan, J.-J.; Wang, J.-Z.; Fan, Y.; Cheng, X.-B.; Cheng, J.-P. Genome-wide identification and phylogenetic relationships of the Hsp70 gene family of Aegilops tauschii, wild emmer wheat (Triticum dicoccoides) and bread wheat (Triticum aestivum). 3 Biotech. 2021, 11, 301. [Google Scholar] [CrossRef] [PubMed]
- Noureddine, J.; Mu, B.; Hamidzada, H.; Mok, W.L.; Bonea, D.; Nambara, E.; Zhao, R. Knockout of endoplasmic reticulum-localized molecular chaperone HSP90.7 impairs seedling development and cellular auxin homeostasis in Arabidopsis. Plant J. 2024, 119, 218–236. [Google Scholar] [CrossRef] [PubMed]
- Queitsch, C.; Sangster, T.A.; Lindquist, S. Hsp90 as a capacitor of phenotypic variation. Nature 2002, 417, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.-Y.; Juan, Y.-T.; Hsu, Y.-H.; Wu, S.-H.; Liao, H.-T.; Fung, R.W.; Charng, Y.-Y. Interplay between Heat Shock Proteins HSP101 and HSA32 Prolongs Heat Acclimation Memory Posttranscriptionally in Arabidopsis. Plant Physiol. 2013, 161, 2075–2084. [Google Scholar] [CrossRef]
- Gross, B.L.; Olsen, K.M. Genetic perspectives on crop domestication. Trends Plant Sci. 2010, 15, 529–537. [Google Scholar] [CrossRef]
- Meyer, R.S.; Purugganan, M.D. Evolution of crop species: Genetics of domestication and diversification. Nat. Rev. Genet. 2013, 14, 840–852. [Google Scholar] [CrossRef]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 2019, 20, 1–23. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Freeling, M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 2009, 60, 433–453. [Google Scholar] [CrossRef]
- Holland, P.W. Evolution of homeobox genes. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Bjornsdottir, G.; Myers, L.C. Minimal components of the RNA polymerase II transcription apparatus determine the consensus TATA box. Nucleic Acids Res. 2008, 36, 2906–2916. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zhou, X.; Cao, Y.; Zhou, M.; McNeil, D.; Liang, S.; Yang, C. RNA-seq Analysis of Cold and Drought Responsive Transcriptomes of Zea mays ssp. mexicana L. Front. Plant Sci. 2017, 8, 136. [Google Scholar] [CrossRef]
- Tian, Y.; Peng, K.; Lou, G.; Ren, Z.; Sun, X.; Wang, Z.; Xing, J.; Song, C.; Cang, J. Transcriptome analysis of the winter wheat Dn1 in response to cold stress. BMC Plant Biol. 2022, 22, 277. [Google Scholar] [CrossRef]
- Qu, Z.; Jia, Y.; Duan, Y.; Chen, H.; Wang, X.; Zheng, H.; Liu, H.; Wang, J.; Zou, D.; Zhao, H. Integrated Isoform Sequencing and Dynamic Transcriptome Analysis Reveals Diverse Transcripts Responsible for Low Temperature Stress at Anther Meiosis Stage in Rice. Front. Plant Sci. 2021, 12, 795834. [Google Scholar] [CrossRef]
- Bondino, H.G.; Valle, E.M.; Ten Have, A. Evolution and functional diversification of the small heat shock protein/α-crystallin family in higher plants. Planta 2012, 235, 1299–1313. [Google Scholar] [CrossRef]
- Jiang, L.; Hu, W.; Qian, Y.; Ren, Q.; Zhang, J. Genome-wide identification, classification and expression analysis of the Hsf and Hsp70 gene families in maize. Gene 2021, 770, 145348. [Google Scholar] [CrossRef]
- Kumar, D.; Chattopadhyay, S. Glutathione modulates the expression of heat shock proteins via the transcription factors BZIP10 and MYB21 in Arabidopsis. J. Exp. Bot. 2018, 69, 3729–3743. [Google Scholar] [CrossRef]
- Milioni, D.; Hatzopoulos, P. Genomic organization of hsp90 gene family in Arabidopsis. Plant Mol. Biol. 1997, 35, 955–961. [Google Scholar] [CrossRef]
- Takahashi, A.; Casais, C.; Ichimura, K.; Shirasu, K. HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2003, 100, 11777–11782. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhang, X.; Cheng, Y.; Takano, T.; Liu, S. rHsp90 gene expression in response to several environmental stresses in rice (Oryza sativa L.). Plant Physiol. Biochem. PPB 2006, 44, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Lukatkin, A.S.; Brazaitytė, A.; Bobinas, Č.; Duchovskis, P. Chilling injury in chilling-sensitive plants: A review. Zemdirb.-Agric. 2012, 99, 111–124. [Google Scholar]
- Hashimoto, M.; Komatsu, S. Proteomic analysis of rice seedlings during cold stress. Proteomics 2007, 7, 1293–1302. [Google Scholar] [CrossRef]
- Kosová, K.r.; Vítámvás, P.; Planchon, S.b.; Renaut, J.; Vanková, R.; Prášil, I.T. Proteome analysis of cold response in spring and winter wheat (Triticum aestivum) crowns reveals similarities in stress adaptation and differences in regulatory processes between the growth habits. J. Proteome Res. 2013, 12, 4830–4845. [Google Scholar] [CrossRef]
- Bae, M.S.; Cho, E.J.; Choi, E.Y.; Park, O.K. Analysis of the Arabidopsis nuclear proteome and its response to cold stress. Plant J. 2003, 36, 652–663. [Google Scholar] [CrossRef]
- Reddy, R.K.; Chaudhary, S.; Patil, P.; Krishna, P. The 90 kDa heat shock protein (hsp90) is expressed throughout Brassica napus seed development and germination. Plant Sci. 1998, 131, 131–137. [Google Scholar] [CrossRef]
- Jin, Y.; Zhang, C.; Yang, H.; Yang, Y.; Huang, C.; Tian, Y.; Lu, X. Proteomic analysis of cold stress responses in tobacco seedlings. Afr. J. Biotechnol. 2011, 10, 18991–19004. [Google Scholar]
- Kollipara, K.P.; Saab, I.N.; Wych, R.D.; Lauer, M.J.; Singletary, G.W. Expression profiling of reciprocal maize hybrids divergent for cold germination and desiccation tolerance. Plant Physiol. 2002, 129, 974–992. [Google Scholar] [CrossRef]
- Vítámvás, P.; Prášil, I.T.; Kosová, K.; Planchon, S.; Renaut, J. Analysis of proteome and frost tolerance in chromosome 5A and 5B reciprocal substitution lines between two winter wheats during long-term cold acclimation. Proteomics 2012, 12, 68–85. [Google Scholar] [CrossRef]
- Hlaváčková, I.; Vítámvás, P.; Šantrůček, J.; Kosová, K.; Zelenková, S.; Prášil, I.T.; Ovesná, J.; Hynek, R.; Kodíček, M. Proteins involved in distinct phases of cold hardening process in frost resistant winter barley (Hordeum vulgare L.) cv Luxor. Int. J. Mol. Sci. 2013, 14, 8000–8024. [Google Scholar] [CrossRef] [PubMed]
- Degand, H.; Faber, A.; Dauchot, N.; Mingeot, D.; Watillon, B.; Van Cutsem, P.; Morsomme, P.; Boutry, M. Proteomic analysis of chicory root identifies proteins typically involved in cold acclimation. Proteomics 2009, 9, 2903–2907. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.L.; Heazlewood, J.L.; Day, D.A.; Millar, A.H. Differential impact of environmental stresses on the pea mitochondrial proteome. Mol. Cell. Proteom. 2005, 4, 1122–1133. [Google Scholar] [CrossRef]
- Renaut, J.; Lutts, S.; Hoffmann, L.; Hausman, J.-F. Responses of poplar to chilling temperatures: Proteomic and physiological aspects. Plant Biol. 2004, 7, 81–90. [Google Scholar] [CrossRef]
- Cui, S.; Huang, F.; Wang, J.; Ma, X.; Cheng, Y.; Liu, J. A proteomic analysis of cold stress responses in rice seedlings. Proteomics 2005, 5, 3162–3172. [Google Scholar] [CrossRef]
- Balbuena, T.S.; Salas, J.J.; Martinez-Force, E.; Garces, R.; Thelen, J.J. Proteome analysis of cold acclimation in sunflower. J. Proteome Res. 2011, 10, 2330–2346. [Google Scholar] [CrossRef]
- Shirasu, K.; Schulze-Lefert, P. Regulators of cell death in disease resistance. Plant Mol. Biol. 2000, 44, 371–385. [Google Scholar] [CrossRef]
- Tang, S.; Ling, Q.; Ma, Q.; Cheng, Y.; Mei, P.; Miao, Y.; Pan, Y.; Jia, Y.; Wu, M.; Yong, X.; et al. LrHSP17.2 Plays an Important Role in Abiotic Stress Responses by Regulating ROS Scavenging and Stress-Related Genes in Lilium regale. Plants 2024, 13, 2416. [Google Scholar] [CrossRef]
- Kumar, B.; Verma, S.K.; Singh, H.P. Effect of temperature on seed germination parameters in Kalmegh (Andrographis paniculata Wall. ex Nees.). Ind. Crops Prod. 2011, 34, 1241–1244. [Google Scholar] [CrossRef]
- Vollenweider, P.; Günthardt-Goerg, M.S. Diagnosis of abiotic and biotic stress factors using the visible symptoms in foliage. Environ. Pollut. 2005, 137, 455–465. [Google Scholar] [CrossRef]
- Cleland, E.E.; Chuine, I.; Menzel, A.; Mooney, H.A.; Schwartz, M.D. Shifting plant phenology in response to global change. Trends Ecol. Evol. 2007, 22, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Rilkey, G.J. Effects of high temperature on protein synthesis during germination of maize (Zea mays L.). Planta 1981, 151, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Akman, Z. Comparison of high temperature tolerance in maize, rice and sorghum seeds by plant growth regulators. J. Anim. Vet. Adv. 2009, 8, 358–361. [Google Scholar]
- Sung, D.Y.; Vierling, E.; Guy, C.L. Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol. 2001, 126, 789–800. [Google Scholar] [CrossRef]
- Wang, Y.X.; Yu, T.F.; Wang, C.X.; Wei, J.T.; Zhang, S.X.; Liu, Y.W.; Chen, J.; Zhou, Y.B.; Chen, M.; Ma, Y.Z.; et al. Heat shock protein TaHSP17.4, a TaHOP interactor in wheat, improves plant stress tolerance. Int. J. Biol. Macromol. 2023, 246, 125694. [Google Scholar] [CrossRef]
- Yang, Z.; Du, H.; Xing, X.; Li, W.; Kong, Y.; Li, X.; Zhang, C. A small heat shock protein, GmHSP17.9, from nodule confers symbiotic nitrogen fixation and seed yield in soybean. Plant Biotechnol. J. 2022, 20, 103–115. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Tang, H.; Bowers, J.E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A.H. Synteny and collinearity in plant genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z. KaKs_Calculator 3.0: Calculating Selective Pressure on Coding and Non-coding Sequences. Genom. Proteom. Bioinform. 2022, 20, 536–540. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, X.; Liu, Y.; Yu, X.; Cai, Y.; Hou, L.; Zhang, J.; Yang, H. Genome-Wide Identification and Evolution-Profiling Analysis of Heat Shock Protein Gene Family in Poaceae Barnhart. Int. J. Mol. Sci. 2025, 26, 4269. https://doi.org/10.3390/ijms26094269
Huang X, Liu Y, Yu X, Cai Y, Hou L, Zhang J, Yang H. Genome-Wide Identification and Evolution-Profiling Analysis of Heat Shock Protein Gene Family in Poaceae Barnhart. International Journal of Molecular Sciences. 2025; 26(9):4269. https://doi.org/10.3390/ijms26094269
Chicago/Turabian StyleHuang, Xiaoyi, Yue Liu, Xiao Yu, Yajun Cai, Lingyu Hou, Jingyuan Zhang, and Hongchun Yang. 2025. "Genome-Wide Identification and Evolution-Profiling Analysis of Heat Shock Protein Gene Family in Poaceae Barnhart" International Journal of Molecular Sciences 26, no. 9: 4269. https://doi.org/10.3390/ijms26094269
APA StyleHuang, X., Liu, Y., Yu, X., Cai, Y., Hou, L., Zhang, J., & Yang, H. (2025). Genome-Wide Identification and Evolution-Profiling Analysis of Heat Shock Protein Gene Family in Poaceae Barnhart. International Journal of Molecular Sciences, 26(9), 4269. https://doi.org/10.3390/ijms26094269





