BolANT3 Positively Regulates Indolic Glucosinolate Accumulation by Transcriptionally Activating BolCYP83B1 in Cabbage
Abstract
1. Introduction
2. Results
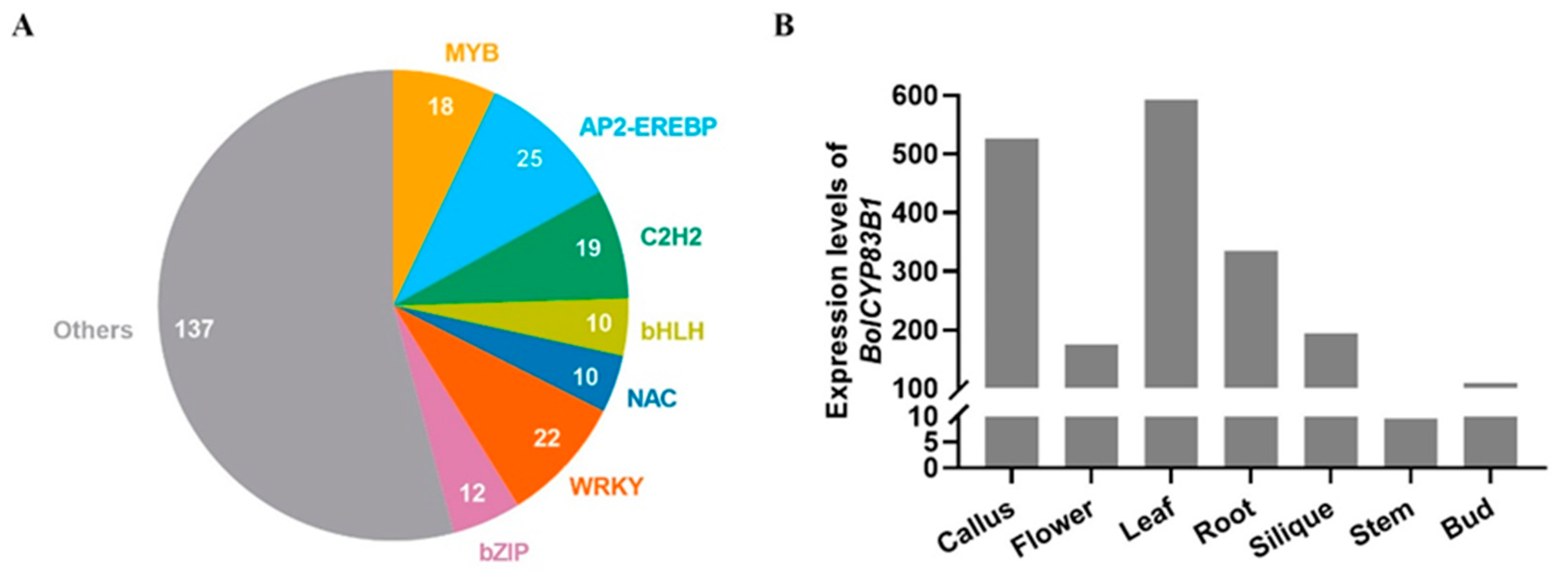
2.1. Transcriptional Regulation of CYP83B1 in Arabidopsis
2.2. Functional Validation of BolCYP83B1 in Cabbage
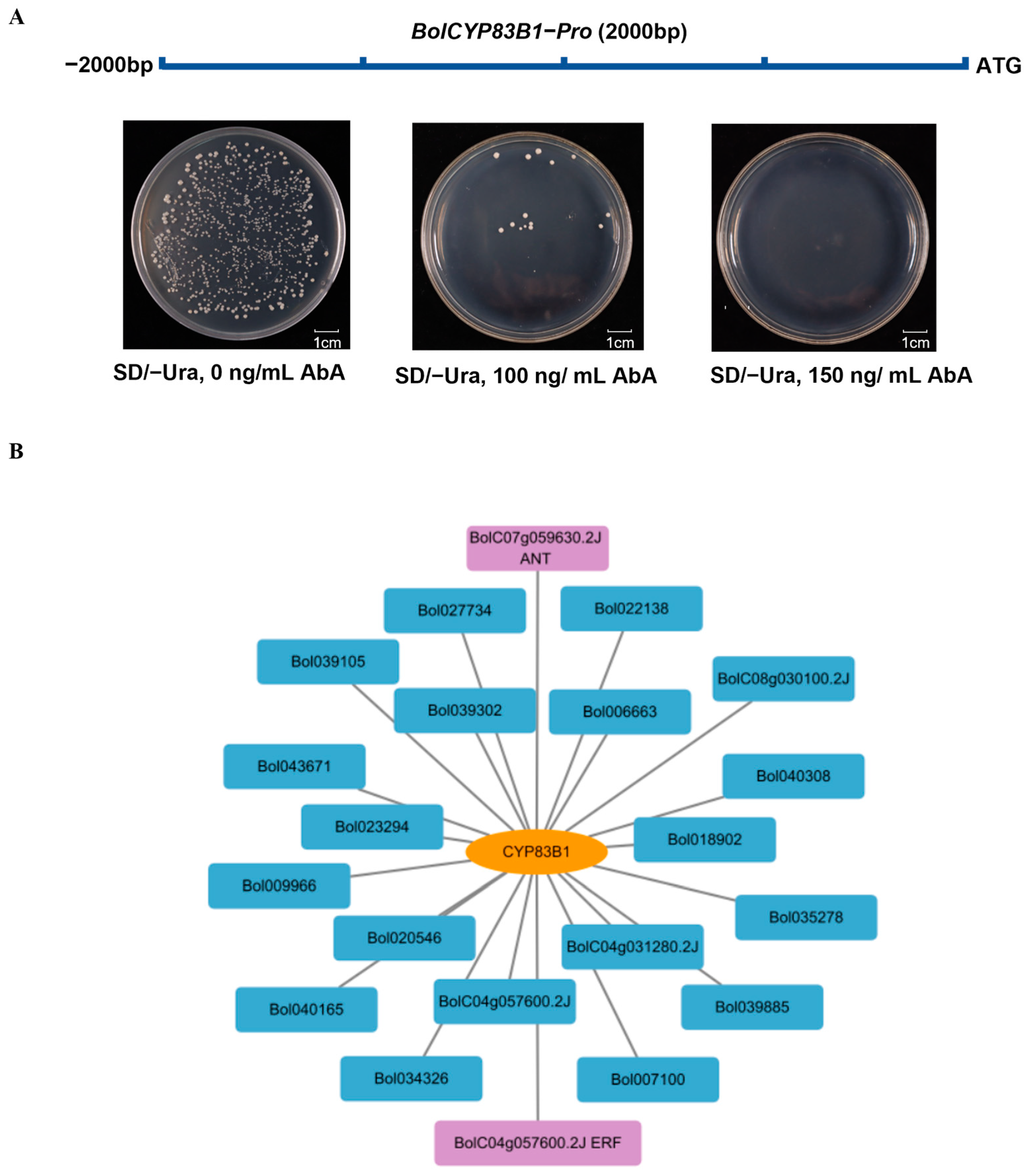
2.3. Screening of Upstream Regulators of BolCYP83B1
2.4. BolANT1 and BolANT3 Bind the Promoter of BolCYP83B1
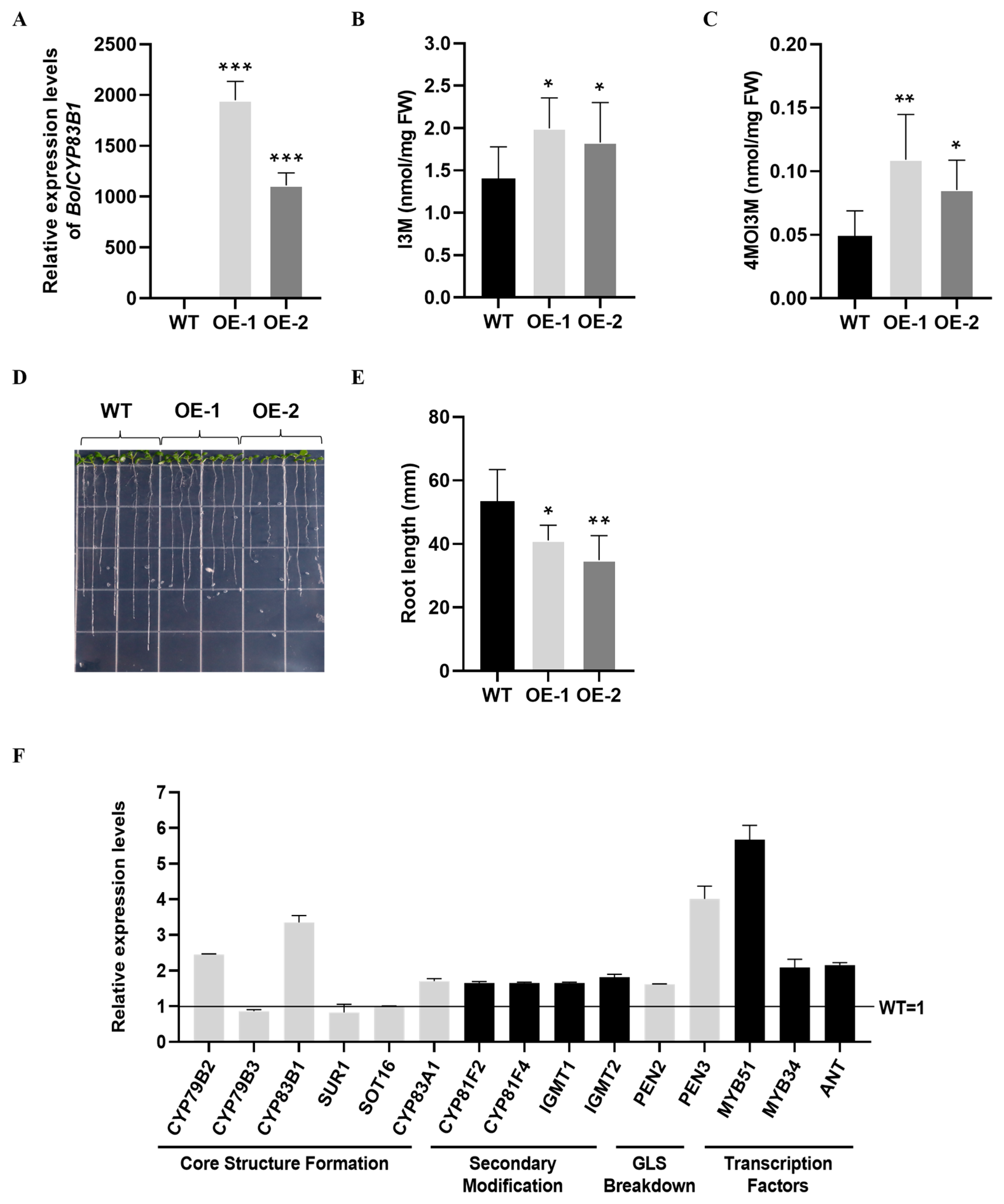
2.5. Overexpression of BolANT3 Induced Indolic Glucosinolate Accumulation in Cabbage
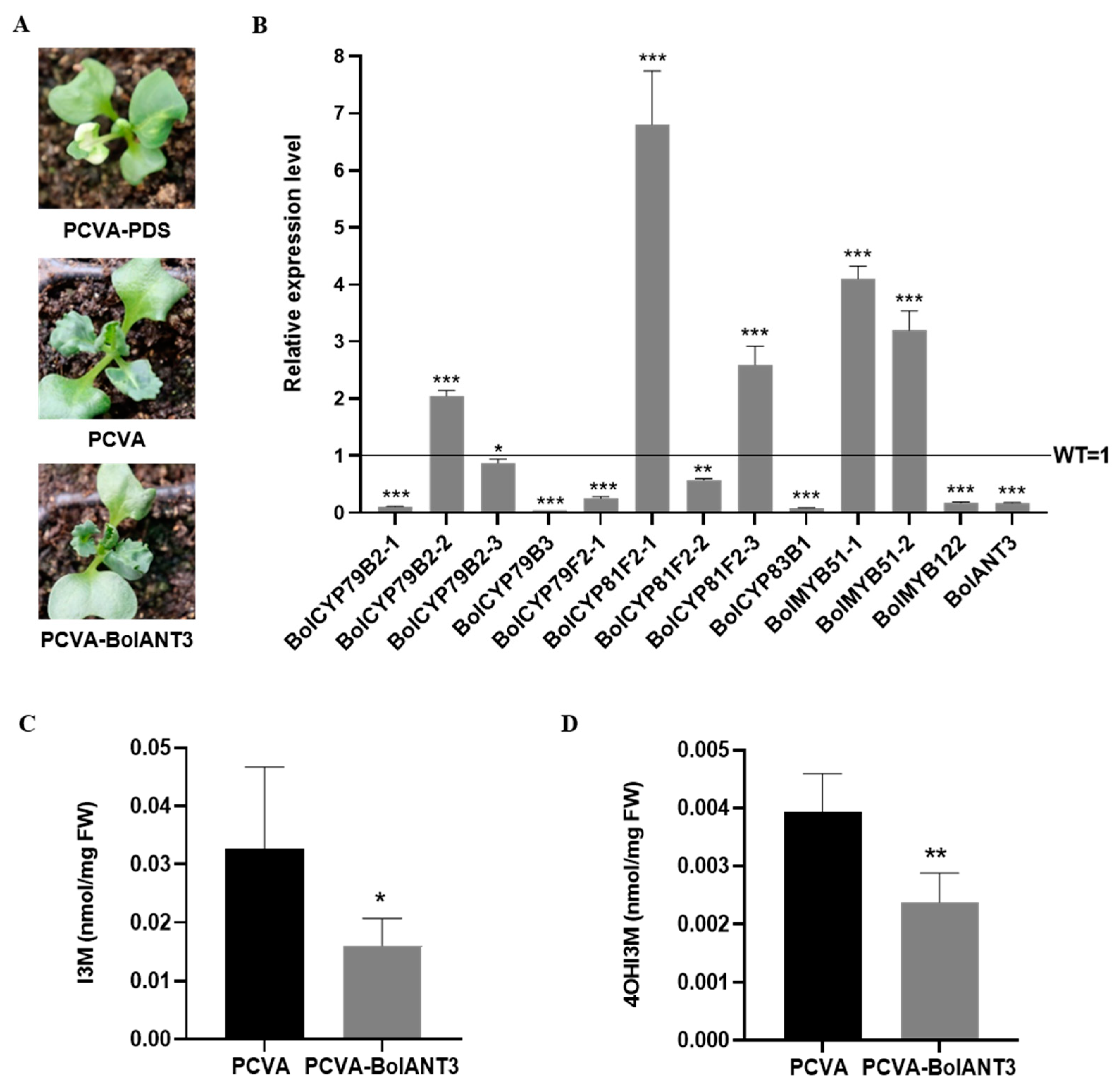
2.6. VIGS of BolANT3 Repressed Indolic Glucosinolate Accumulation in Cabbage
3. Discussion
3.1. Value of Transcriptional Regulation of Indolic Glucosinolates in Brassica Vegetables
3.2. The Transcriptional Regulation of Plant Defense and Development by the BolANT3-BolCYP83B1 Module
3.3. Disease Resistance, Nutrition Value, and Better Growth Phenotypes Achieved by Manipulating the Upstream Regulators of BolCYP83B1
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Gene Cloning and Sequence Analysis
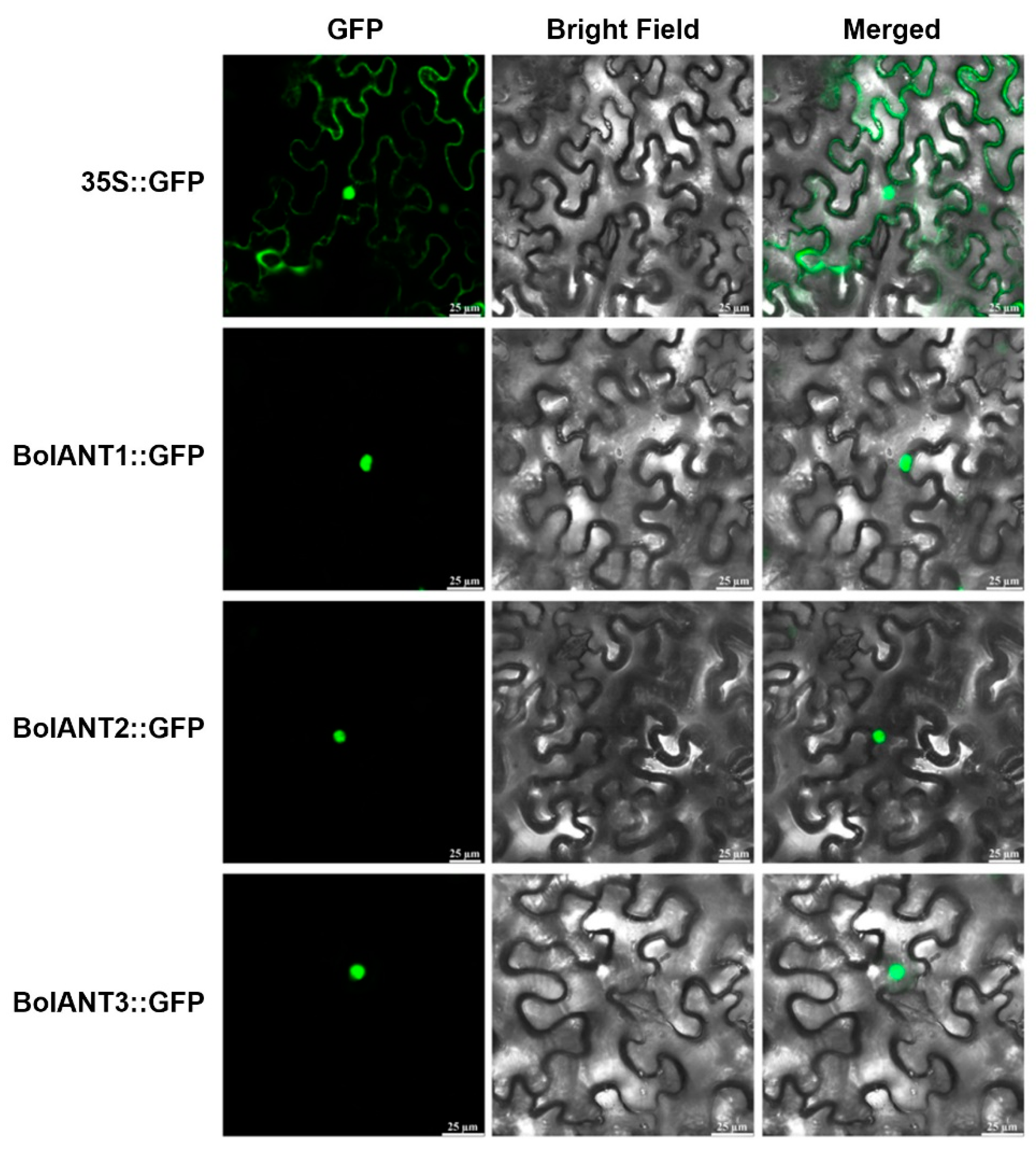
4.3. Subcellular Localization of BolANT Proteins
4.4. Yeast One-Hybrid (Y1H) Assay
4.5. Dual-Luciferase Reporter (LUC) Assay
4.6. Genetic Transformation of BolCYP83B1 and BolANT3
4.7. VIGS of BolANT3
4.8. Total Glucosinolate Extraction and HPLC Analysis
4.9. RNA Extraction and qPCR Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Erb, M.; Kliebenstein, D.J. Plant secondary metabolites as defenses, regulators, and primary metabolites: The blurred functional trichotomy. Plant Physiol. 2020, 184, 39–52. [Google Scholar] [CrossRef]
- Sønderby, I.E.; Geu-Flores, F.; Halkier, B.A. Biosynthesis of glucosinolates gene discovery and beyond. Trends Plant Sci. 2010, 15, 283–290. [Google Scholar] [CrossRef]
- Halkier, B.A.; Gershenzon, J. Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 2006, 57, 303–333. [Google Scholar] [CrossRef]
- Clay, N.K.; Adio, A.M.; Denoux, C.; Jander, G.; Ausubel, F.M. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 2009, 323, 95–101. [Google Scholar]
- Bednarek, P.; Pislewska-Bednarek, M.; Svatos, A.; Schneider, B.; Doubsky, J.; Mansurova, M.; Humphry, M.; Consonni, C.; Panstruga, R.; Sanchez-Vallet, A.; et al. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 2009, 323, 101–106. [Google Scholar]
- Katz, E.; Nisani, S.; Chamovitz, D.A. Indole-3-carbinol: A plant hormone combatting cancer. F1000Research 2018, 7, 689. [Google Scholar] [CrossRef]
- Stoewsand, G.S. Bioactive organosulfur phytochemicals in Brassica oleracea vegetables—A review. Food Chem. Toxicol. 1995, 33, 537–543. [Google Scholar] [CrossRef]
- Bekaert, M.; Edger, P.P.; Hudson, C.M.; Pires, J.C.; Conant, G.C. Metabolic and evolutionary costs of herbivory defense: Systems biology of glucosinolate synthesis. New Phytol. 2012, 196, 596–605. [Google Scholar]
- Shang, Y.; Ma, Y.; Zhou, Y.; Zhang, H.; Duan, L.; Chen, H.; Zeng, J.; Zhou, Q.; Wang, S.; Gu, W.; et al. Plant science. biosynthesis, regulation, and domestication of bitterness in cucumber. Science 2014, 346, 1084–1088. [Google Scholar] [CrossRef]
- Wang, H.; Ren, J.; Zhou, S.; Duan, Y.; Zhu, C.; Chen, C.; Liu, Z.; Zheng, Q.; Xiang, S.; Xie, Z.; et al. Molecular regulation of oil gland development and biosynthesis of essential oils in Citrus spp. Science 2024, 383, 659–666. [Google Scholar] [CrossRef]
- Frerigmann, H.; Gigolashvili, T. MYB34, MYB51 and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol. Plant 2014, 7, 814–828. [Google Scholar]
- Gigolashvili, T.; Berger, B.; Mock, H.P.; Müller, C.; Weisshaar, B.; Flügge, U.I. The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007, 50, 886–901. [Google Scholar] [CrossRef]
- Celenza, J.L.; Quiel, J.A.; Smolen, G.A.; Merrikh, H.; Silvestro, A.R.; Normanly, J.; Bender, J. The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 2005, 137, 253–262. [Google Scholar]
- Cai, C.X.; Yuan, W.X.; Miao, H.Y.; Deng, M.D.; Wang, M.Y.; Lin, J.Y.; Zeng, W.; Wang, Q.M. Functional characterization of BoaMYB51s as central regulators of indole glucosinolate biosynthesis in Brassica oleracea var. alboglabra Bailey. Front. Plant Sci. 2018, 9, 1599. [Google Scholar] [CrossRef]
- Wiggins, B.G.; Wang, Y.F.; Burke, A.; Grunberg, N.; Vlachaki Walker, J.M.; Dore, M.; Chahrour, C.; Pennycook, B.R.; Sanchez-Garrido, J.; Vernia, S.; et al. Endothelial sensing of AHR ligands regulates intestinal homeostasis. Nature 2023, 621, 821–829. [Google Scholar]
- Zeng, Q.; Liu, X.F.; Yan, X.M.; Zhang, J.H.; Li, C.; Yan, C.T.; Zhang, Y.F.; Kliebenstein, D.; Li, B.H. Novel Regulators and their epistatic networks in Arabidopsis’ defence responses to Alternaria alternata infection. Mol. Plant Pathol. 2025, 26, 70058. [Google Scholar]
- Tao, H.; Miao, H.; Chen, L.; Wang, M.; Xia, C.; Zeng, W.; Sun, B.; Zhang, F.; Zhang, S.; Li, C.; et al. WRKY33-mediated indolic glucosinolate metabolic pathway confers resistance against Alternaria brassicicola in Arabidopsis and Brassica crops. J. Integr. Plant Biol. 2022, 64, 1007–1019. [Google Scholar]
- Naur, P.; Petersen, B.L.; Mikkelsen, M.D.; Bak, S.; Rasmussen, H.; Olsen, C.E.; Halkier, B.A. CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol. 2003, 133, 63–72. [Google Scholar] [CrossRef]
- Hansen, C.H.; Du, L.; Naur, P.; Olsen, C.E.; Axelsen, K.B.; Hick, A.J.; Pickett, J.A.; Halkier, B.A. CYP83B1 is the oxime-metabolizing enzyme in the glucosinolate pathway in Arabidopsis. Biol. Chem. 2001, 276, 24790–24796. [Google Scholar] [CrossRef]
- Chen, L.; Zeng, Q.; Zhang, J.; Li, C.; Bai, X.; Sun, F.; Kliebenstein, D.J.; Li, B. Large-scale identification of novel transcriptional regulators of the aliphatic glucosinolate pathway in Arabidopsis. J. Exp. Bot. 2024, 75, 300–315. [Google Scholar] [CrossRef]
- Tang, M.; Li, B.; Zhou, X.; Bolt, T.; Li, J.J.; Cruz, N.; Gaudinier, A.; Ngo, R.; Clark-Wiest, C.; Kliebenstein, D.J.; et al. A genome-scale TF-DNA interaction network of transcriptional regulation of Arabidopsis primary and specialized metabolism. Mol. Syst. Biol. 2021, 17, e10625. [Google Scholar] [CrossRef]
- Li, B.; Gaudinier, A.; Tang, M.; Taylor-Teeples, M.; Nham, N.T.; Ghaffari, C.; Benson, D.S.; Steinmann, M.; Gray, J.A.; Brady, S.M.; et al. Promoter-based integration in plant defense regulation. Plant Physiol. 2014, 166, 1803–1820. [Google Scholar] [CrossRef]
- Barlier, I.; Kowalczyk, M.; Marchant, A.; Ljung, K.; Bhalerao, R.; Bennett, M.; Sandberg, G.; Bellini, C. The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc. Natl. Acad. Sci. USA 2000, 97, 14819–14824. [Google Scholar] [CrossRef]
- Glawischnig, E.; Hansen, B.G.; Olsen, C.E.; Halkier, B.A. Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 8245–82450. [Google Scholar] [CrossRef]
- Bai, X.; Zhang, R.; Zeng, Q.; Yang, W.; Fang, F.; Sun, Q.; Yan, C.; Li, F.; Liu, X.; Li, B. The RNA-binding protein BoRHON1 positively regulates the accumulation of aliphatic glucosinolates in cabbage. Int. J. Mol. Sci. 2024, 25, 5314. [Google Scholar] [CrossRef]
- Pan, Q.M.; Zhang, J.H.; Yan, C.T.; Khan, A.; Fei, S.M.; Lei, T.; Xu, Z.M.; Li, B.H.; Zhang, R.X.; Hui, M.X. Distribution of indolic glucosinolates in different developmental stages and tissues of 13 Varieties of Cabbage (Brassica oleracea L. var. capitata). Horticulturae 2023, 9, 867. [Google Scholar] [CrossRef]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A.; Zhao, M.; Ma, J.; Yu, J.; Huang, S.; et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014, 5, 3930. [Google Scholar] [CrossRef]
- Li, B.; Tang, M.; Caseys, C.; Nelson, A.; Zhou, M.; Zhou, X.; Brady, S.M.; Kliebenstein, D.J. Epistatic transcription factor networks differentially modulate Arabidopsis growth and defense. Genetics 2020, 214, 529–541. [Google Scholar] [CrossRef]
- Li, B.; Tang, M.; Nelson, A.; Caligagan, H.; Zhou, X.; Clark-Wiest, C.; Ngo, R.; Brady, S.M.; Kliebenstein, D.J. Network-guided discovery of extensive epistasis between transcription factors involved in aliphatic glucosinolate biosynthesis. Plant Cell 2018, 30, 178–195. [Google Scholar] [CrossRef]
- Muhich, A.J.; Agosto-Ramos, A.; Kliebenstein, D.J. The ease and complexity of identifying and using specialized metabolites for crop engineering. Emerg. Top. Life Sci. 2022, 6, 153–162. [Google Scholar] [CrossRef]
- Kliebenstein, D.J.; Osbourn, A. Making new molecules evolution of pathways for novel metabolites in plants. Curr. Opin. Plant Biol. 2012, 15, 415–423. [Google Scholar]
- Sanchez-Perez, R.; Pavan, S.; Mazzeo, R.; Moldovan, C.; Aiese Cigliano, R.; Del Cueto, J.; Ricciardi, F.; Lotti, C.; Ricciardi, L.; Dicenta, F.; et al. Mutation of a bHLH transcription factor allowed almond domestication. Science 2019, 364, 1095–1098. [Google Scholar]
- Schweizer, F.; Fernandez-Calvo, P.; Zander, M.; Diez-Diaz, M.; Fonseca, S.; Glauser, G.; Lewsey, M.G.; Ecker, J.R.; Solano, R.; Reymond, P. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 2013, 25, 3117–3132. [Google Scholar]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.M.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar]
- Boyes, D.C.; Zayed, A.M.; Ascenzi, R.; McCaskill, A.J.; Hoffman, N.E.; Davis, K.R.; Gorlach, J. Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 2001, 13, 1499–1510. [Google Scholar] [CrossRef]
- Sussman, M.R.; Amasino, R.M.; Young, J.C.; Krysan, P.J.; Austin-Phillips, S. The Arabidopsis knockout facility at the university of Wisconsin-Madison. Plant Physiol. 2000, 124, 1465–1467. [Google Scholar]
- Arabidopsis Genome, I. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar]
- Sønderby, I.E.; Hansen, B.G.; Bjarnholt, N.; Ticconi, C.; Halkier, B.A.; Kliebenstein, D.J. A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PLoS ONE 2007, 2, e1322. [Google Scholar] [CrossRef]
- Hirai, M.Y.; Sugiyama, K.; Sawada, Y.; Tohge, T.; Obayashi, T.; Suzuki, A.; Araki, R.; Sakurai, N.; Suzuki, H.; Aoki, K. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 6478–6483. [Google Scholar]
- Gigolashvili, T.; Yatusevich, R.; Berger, B.; Müller, C.; Flügge, U.I. The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant. J. 2007, 51, 247–261. [Google Scholar]
- Mabry, M.E.; Abrahams, R.S.; Al-Shehbaz, I.A.; Baker, W.J.; Barak, S.; Barker, M.S.; Barrett, R.L.; Beric, A.; Bhattacharya, S.; Carey, S.B.; et al. Complementing model species with model clades. Plant Cell 2024, 36, 1205–1226. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Jeong, C.W.; Nole-Wilson, S.; Krizek, B.A.; Wagner, D. AINTEGUMENTA and AINTEGUMENTA-LIKE6/PLETHORA3 induce LEAFY expression in response to auxin to promote the onset of flower formation in Arabidopsis. Plant Physiol. 2016, 170, 283–293. [Google Scholar] [CrossRef]
- Klucher, K.M.; Chow, H.; Reiser, L.; Fischer, R.L. The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 1996, 8, 137–153. [Google Scholar]
- Elliott, R.C.; Betzner, A.S.; Huttner, E.; Oakes, M.P.; Tucker, W.Q.; Gerentes, D.; Perez, P.; Smyth, D.R. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 1996, 8, 155–168. [Google Scholar]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Ahmed, S.S.; Gong, Z.-H.; Ji, J.-J.; Yin, Y.-X.; Xiao, H.-J.; Khan, M.A.; Rehman, A.; Ahmad, I. Construction of the intermediate vector pVBG2307 by incorporating vital elements of expression vectors pBI121 and pBI221. Genet. Mol. Res. 2012, 11, 3091–3104. [Google Scholar] [CrossRef]
- Xiao, Z.; Xing, M.; Liu, X.; Fang, Z.; Yang, L.; Zhang, Y.; Wang, Y.; Zhuang, M.; Lv, H. An efficient virus-induced gene silencing (VIGS) system for functional genomics in Brassicas using a cabbage leaf curl virus (CaLCuV)-based vector. Planta 2020, 252, 42. [Google Scholar] [CrossRef]
- Kliebenstein, D.J.; Kroymann, J.; Brown, P.; Figuth, A.; Pedersen, D.; Gershenzon, J.; Mitchell-Olds, T. Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol. 2001, 126, 811–825. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, C.; Yang, W.; Yan, X.; Liu, Y.; Zhang, J.; Bai, X.; Zeng, Q.; Liu, X.; Shao, D.; Li, B. BolANT3 Positively Regulates Indolic Glucosinolate Accumulation by Transcriptionally Activating BolCYP83B1 in Cabbage. Int. J. Mol. Sci. 2025, 26, 3415. https://doi.org/10.3390/ijms26073415
Yan C, Yang W, Yan X, Liu Y, Zhang J, Bai X, Zeng Q, Liu X, Shao D, Li B. BolANT3 Positively Regulates Indolic Glucosinolate Accumulation by Transcriptionally Activating BolCYP83B1 in Cabbage. International Journal of Molecular Sciences. 2025; 26(7):3415. https://doi.org/10.3390/ijms26073415
Chicago/Turabian StyleYan, Chengtai, Wenjing Yang, Xuemei Yan, Yao Liu, Jiahao Zhang, Xue Bai, Qi Zeng, Xifan Liu, Dengkui Shao, and Baohua Li. 2025. "BolANT3 Positively Regulates Indolic Glucosinolate Accumulation by Transcriptionally Activating BolCYP83B1 in Cabbage" International Journal of Molecular Sciences 26, no. 7: 3415. https://doi.org/10.3390/ijms26073415
APA StyleYan, C., Yang, W., Yan, X., Liu, Y., Zhang, J., Bai, X., Zeng, Q., Liu, X., Shao, D., & Li, B. (2025). BolANT3 Positively Regulates Indolic Glucosinolate Accumulation by Transcriptionally Activating BolCYP83B1 in Cabbage. International Journal of Molecular Sciences, 26(7), 3415. https://doi.org/10.3390/ijms26073415





