AcMYB176-Regulated AcCHS5 Enhances Salt Tolerance in Areca catechu by Modulating Flavonoid Biosynthesis and Reactive Oxygen Species Scavenging
Abstract
1. Introduction
2. Results
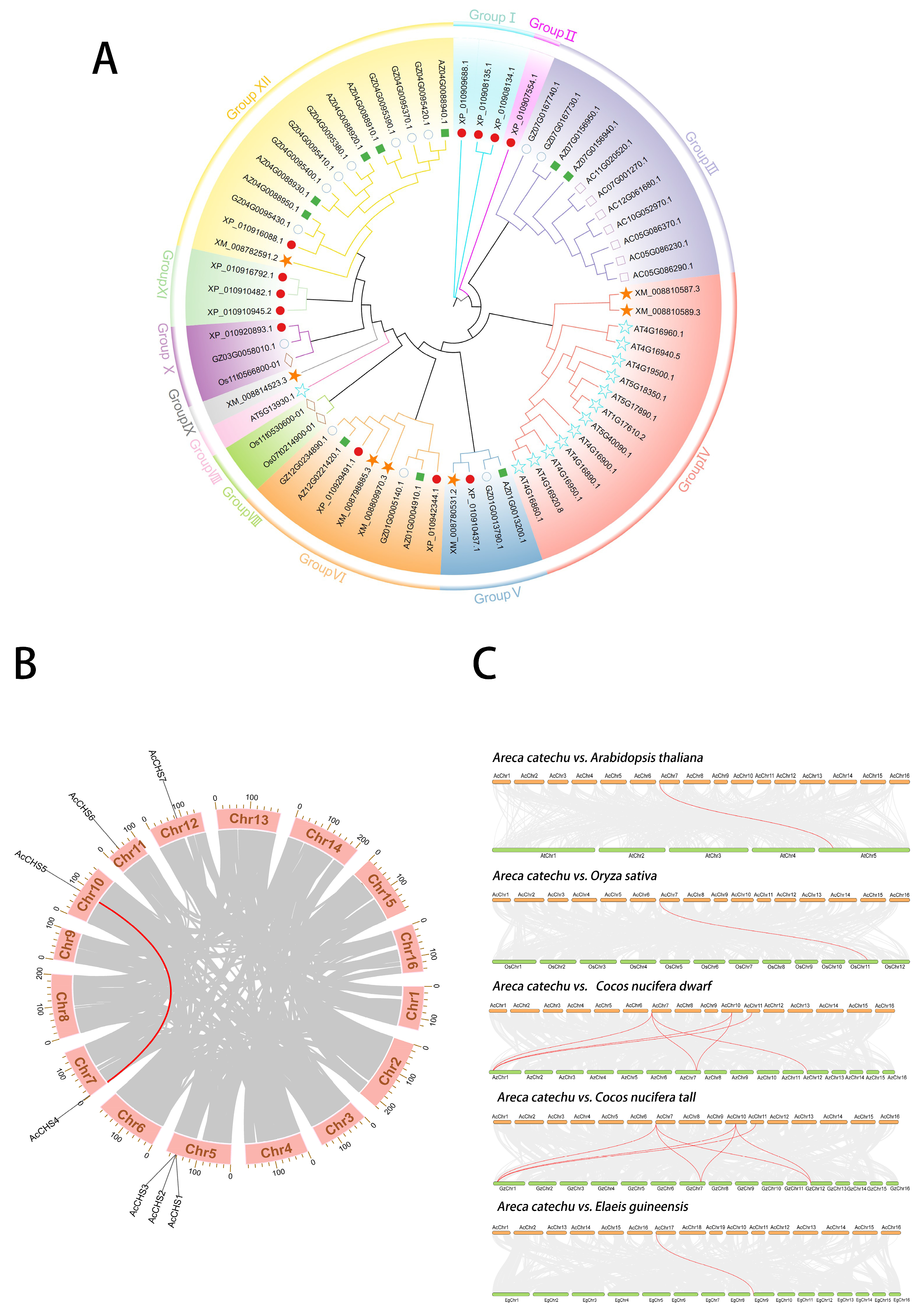
2.1. Phylogenetic Analysis of CHS Family
2.2. Chromosomal Localization and Intra- and Interspecific Collinearity Analyses of A. catechu CHS Family
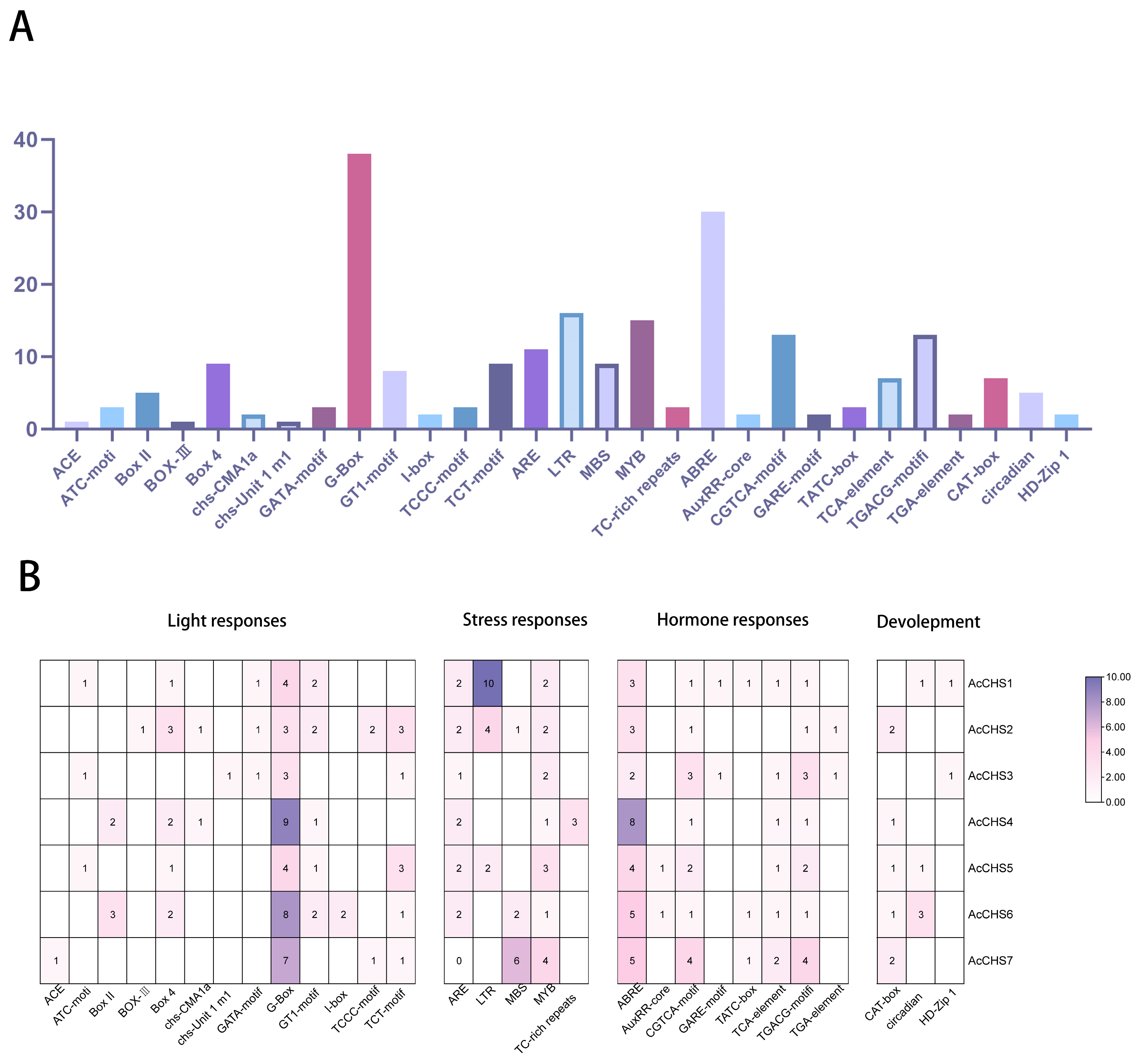
2.3. Cis-Acting Element Analysis of Areca CHS Family Members
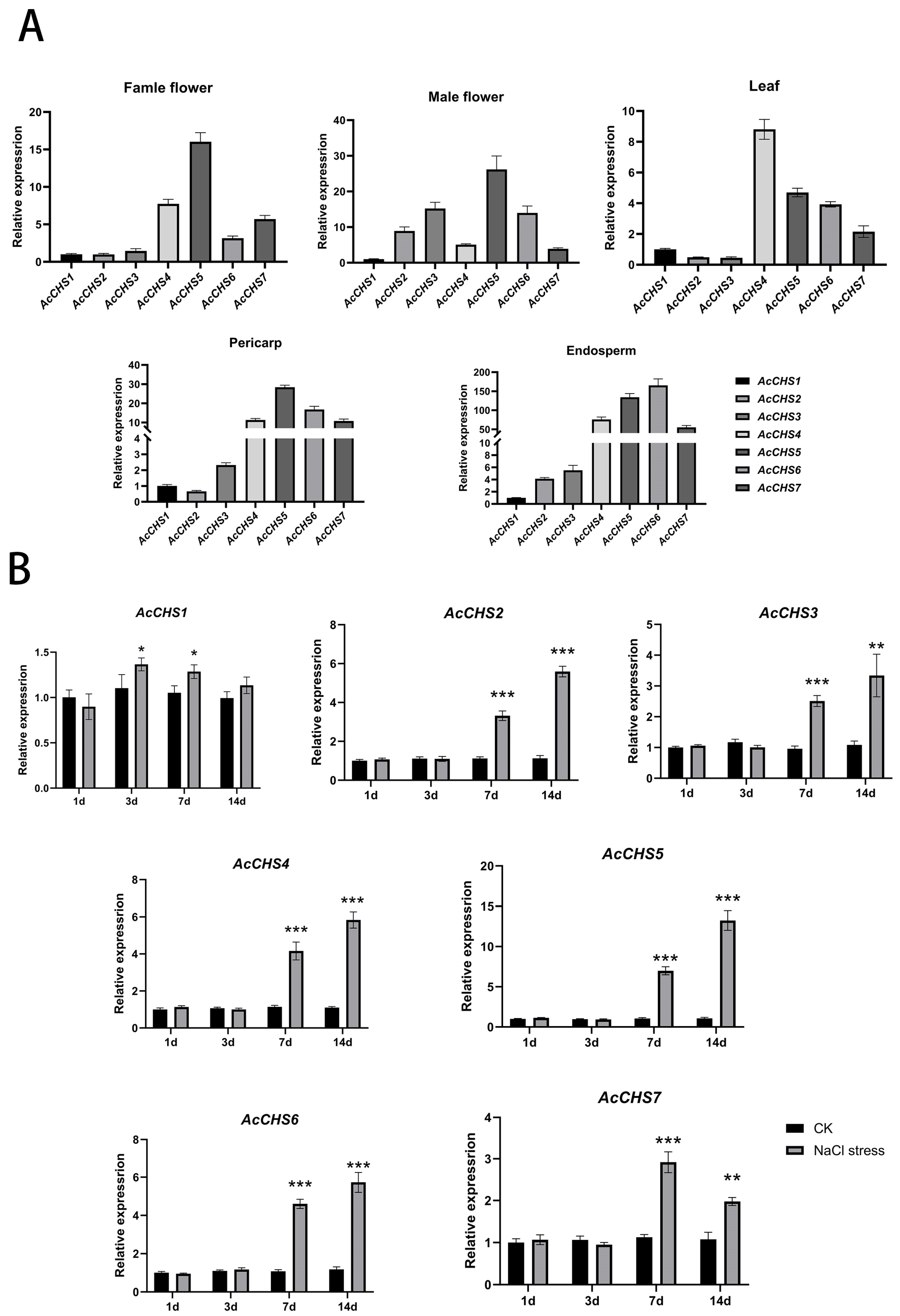
2.4. Expression Profiling of A. catechu CHS Genes
2.5. Expression Patterns Under Salt Stress
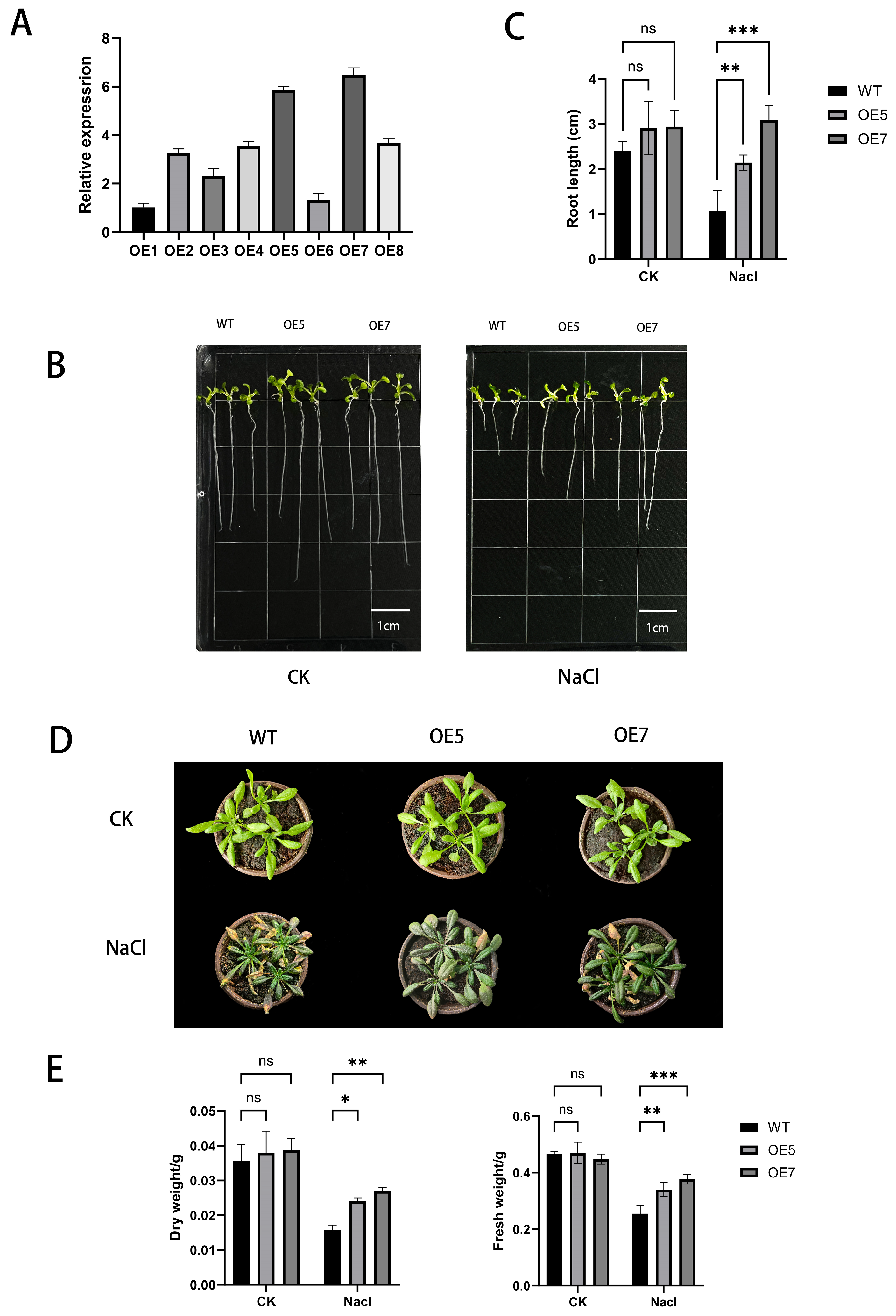
2.6. Identification of Transgenic A. thaliana
2.7. Phenotypic Evaluation of A. thaliana Under Salt Stress
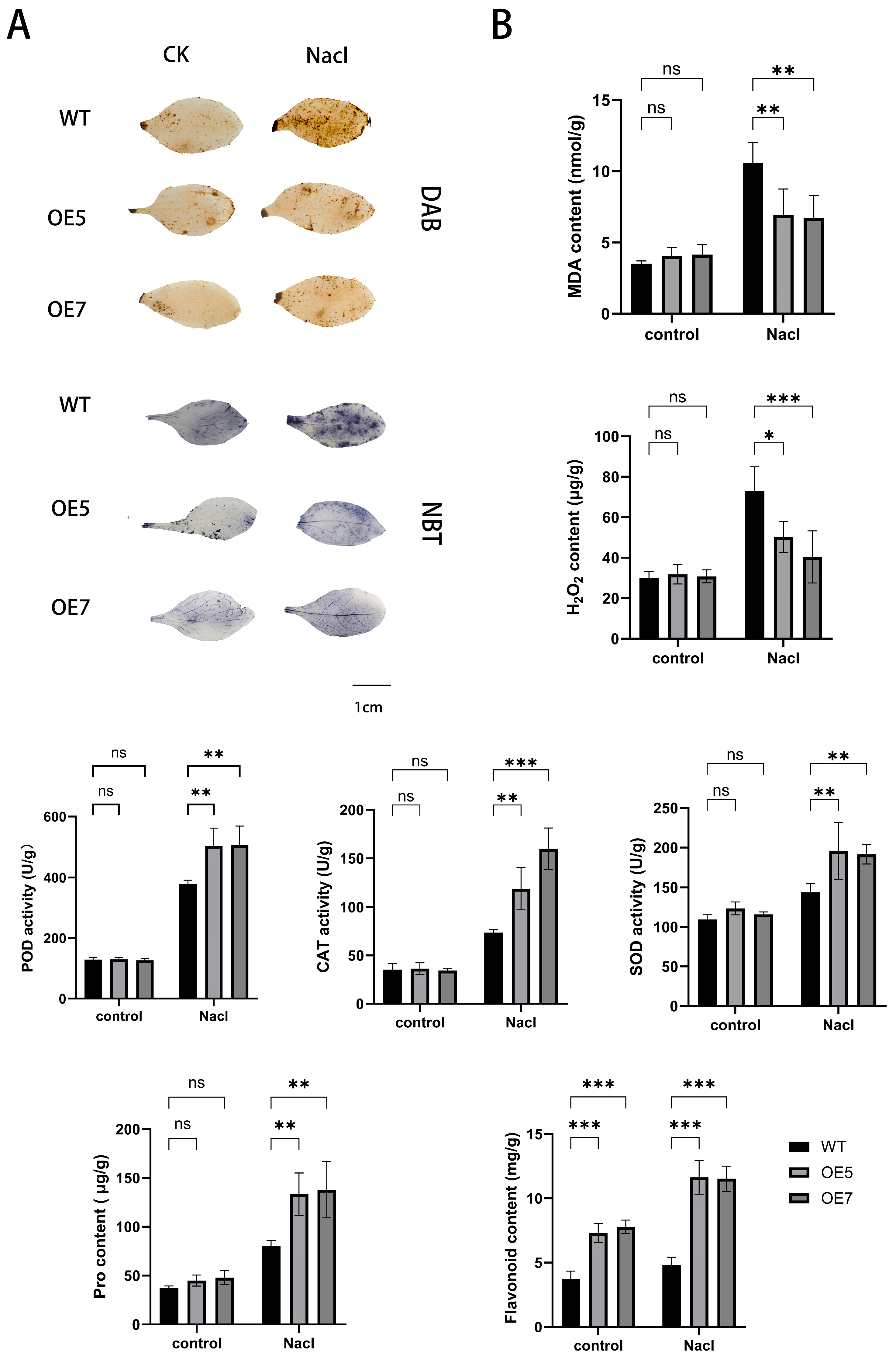
2.8. Detection of Stress-Resistant Physiological Indicators in A. thaliana Under Various Stress Treatments
2.9. Correlation Analysis of MYB Transcription Factors and Flavonoid Biosynthesis Genes in A. catechu
2.10. Yeast One-Hybrid Assay Validation
2.11. Dual-Luciferase Assay Results Analysis
3. Discussion
4. Materials and Methods
4.1. Phylogenetic Analysis of the CHS Gene Family in A. catechu
4.2. Inter- and Intra-Species Collinearity Analysis of the CHS Gene Family in A. catechu
4.3. Cis-Acting Element Analysis of the CHS Gene Family in A. catechu
4.4. Expression Pattern Analysis of CHS Genes in A. catechu
4.5. Construction of AcCHS5 Overexpression Vector and Generation of Transgenic A. thaliana
4.6. Phenotypic Observation of A. thaliana Under Salt Stress Treatment
4.7. Detection of Stress-Resistant Physiological Indices in A. thaliana Under Salt Stress
4.8. NBT Staining and DAB Staining
4.9. Correlation Analysis of MYB Transcription Factors and Flavonoid Biosynthesis Genes in A. catechu
4.10. Yeast One-Hybrid Vector Construction
4.11. Screening of Bait Yeast Strain Sensitivity to AbA
4.12. Yeast One-Hybrid Interaction Assay
4.13. Dual-Luciferase Reporter Assay (LUC)
4.14. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, N.; Haldar, S.; Saikia, R. Root Exudation as a Strategy for Plants to Deal with Salt Stress: An Updated Review. Environ. Exp. Bot. 2023, 216, 105518. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Gupta, B.; Huang, B. Mechanism of Salinity Tolerance in Plants: Physiological, Biochemical, and Molecular Characterization. Int. J. Genom. 2014, 2014, 701596. [Google Scholar] [CrossRef]
- Tebini, M.; Chieb, M.; Luu, D.T.; Dailly, H.; Lutts, S.; Ahmed, H.B.; Chalh, A. Assessment of Salt Stress Effects on Antioxidant Levels and Membrane Transport Protein in Amaranthus caudatus. J. Plant Growth Regul. 2025, 39, 105519. [Google Scholar] [CrossRef]
- Taylor, L.P.; Grotewold, E. Flavonoids as Developmental Regulators. Int. J. Mol. Sci. 2005, 8, 317–323. [Google Scholar] [CrossRef]
- Chen, L.; Guo, H.; Li, Y.; Chen, H. Chalcone Synthase EaCHS1 from Eupatorium adenophorum Functions in Salt Stress Tolerance in Tobacco. Plant Cell Rep. 2015, 34, 885–894. [Google Scholar] [CrossRef]
- Baldoni, E.; Genga, A.; Cominelli, E. Plant MYB Transcription Factors: Their Role in Drought Response Mechanisms. Int. J. Mol. Sci. 2015, 16, 15811–15851. [Google Scholar] [CrossRef]
- Liu, L.; Si, L.; Zhang, L.; Li, X.; Li, Y.; Wang, X.; Zhao, Q.; Gao, Y.; Zhang, J.; Liu, G. Metabolomics and Transcriptomics Analysis Revealed the Response Mechanism of Alfalfa to Combined Cold and Saline-Alkali Stress. Plant J. 2024, 119, 1900–1919. [Google Scholar] [CrossRef]
- Wang, X.; Yang, Y.; Zhang, H. Analysis of the CHS Gene Family Reveals Its Functional Responses to Hormones, Salinity, and Drought Stress in Moso Bamboo (Phyllostachys edulis). Plants 2025, 14, 161. [Google Scholar] [CrossRef]
- Chen, S.; Wu, F.; Li, Y.; Qian, Y.; Pan, X.; Li, F.; Yang, A. NtMYB4 and NtCHS1 Are Critical Factors in the Regulation of Flavonoid Biosynthesis and Are Involved in Salinity Responsiveness. Front. Plant Sci. 2019, 10, 178. [Google Scholar] [CrossRef]
- Hou, Q.; Li, S.; Shang, C.; Wen, Z.; Cai, X.; Hong, Y.; Qiao, G. Genome-Wide Characterization of Chalcone Synthase Genes in Sweet Cherry and Functional Characterization of CpCHS1 under Drought Stress. Front. Plant Sci. 2022, 13, 989959. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Li, K.; Li, Y.; Zhao, X.; Wang, L. MYB Transcription Factors as Regulators of Secondary Metabolism in Plants. Biology 2020, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, G.; Tang, Q.; Song, W.; Gao, Q.; Xiang, G.; Zhai, C. EbMYBP1, a R2R3-MYB Transcription Factor, Promotes Flavonoid Biosynthesis in Erigeron breviscapus. Front. Plant Sci. 2022, 13, 946827. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Misra, P.; Trivedi, P.K. Constitutive Expression of Arabidopsis MYB Transcription Factor, AtMYB11, in Tobacco Modulates Flavonoid Biosynthesis in Favor of Flavonol Accumulation. Plant Cell Rep. 2015, 34, 1515–1528. [Google Scholar] [CrossRef]
- Jian, Y.; Gao, C.; Shang, Y.; Qin, J.; Duan, S.; Bian, C.; Li, G. Metabolomic and Transcriptomic Analyses Reveal MYB-Related Genes Involved in Drought Resistance in Grafted Potatoes via the Flavonoid Pathway. Plant Stress 2024, 14, 100665. [Google Scholar] [CrossRef]
- Ravaglia, D.; Espley, R.V.; Henry-Kirk, R.A.; Andreotti, C.; Ziosi, V.; Hellens, R.P.; Allan, A.C. Transcriptional Regulation of Flavonoid Biosynthesis in Nectarine (Prunus persica) by a Set of R2R3 MYB Transcription Factors. BMC Plant Biol. 2013, 13, 68. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, F.; Cheng, S.; Liao, Y. Characterization and Functional Analysis of a MYB Gene (GbMYBFL) Related to Flavonoid Accumulation in Ginkgo biloba. Genes Genom. 2018, 40, 49–61. [Google Scholar] [CrossRef]
- Li, X.W.; Li, J.W.; Zhai, Y.; Zhao, Y.; Zhao, X.; Zhang, H.J.; Wang, Q.Y. A R2R3-MYB Transcription Factor, GmMYB12B2, Affects the Expression Levels of Flavonoid Biosynthesis Genes Encoding Key Enzymes in Transgenic Arabidopsis Plants. Gene 2013, 532, 72–79. [Google Scholar] [CrossRef]
- Wang, C.; Yin, Z.; Luo, R.; Qian, J.; Fu, C.; Wang, Y.; Xie, Y.; Liu, Z.; Qiu, Z.; Pei, H. Spatiotemporal Evolution and Impact Mechanisms of Areca Palm Plantations in China (1987–2022). Forests 2024, 15, 1679. [Google Scholar] [CrossRef]
- Ansari, A.; Mahmood, T.; Bagga, P.; Ahsan, F.; Shamim, A.; Ahmad, S.; Shariq, M.; Parveen, S. Areca catechu: A Phytopharmacological Legwork. Food Front. 2021, 2, 163–183. [Google Scholar] [CrossRef]
- Lai, J.; Li, C.; Zhang, Y.; Wu, Z.; Li, W.; Zhang, Z.; Yang, J. Integrated Transcriptomic and Metabolomic Analyses Reveal the Molecular and Metabolic Basis of Flavonoids in Areca catechu L. J. Agric. Food Chem. 2023, 71, 4851–4862. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, R.K. Does Salinity Stress Amend the Morphology and Physiological Appearance of Young Betel Nut (Areca catechu L.) Seedlings? J. Plant. Crops 2016, 44, 129–131. [Google Scholar]
- Fu, Z.W.; Feng, Y.R.; Gao, X.; Ding, F.; Li, J.H.; Yuan, T.T.; Lu, Y.T. Salt Stress-Induced Chloroplastic Hydrogen Peroxide Stimulates pdTPI Sulfenylation and Methylglyoxal Accumulation. Plant Cell 2023, 35, 1593–1616. [Google Scholar] [CrossRef] [PubMed]
- An, Q.; Jiang, Y.; Zhou, G. Genome-Wide Investigation of MYB Gene Family in Areca catechu and Potential Roles of AcDF in Transgenic Arabidopsis. Mol. Biol. Rep. 2024, 51, 1–13. [Google Scholar] [CrossRef]
- Ahmad, S.; Ali, S.; Shah, A.Z.; Khan, A.; Faria, S. Chalcone Synthase (CHS) Family Genes Regulate the Growth and Response of Cucumber (Cucumis sativus L.) to Botrytis cinerea and Abiotic Stresses. Plant Stress 2023, 8, 100159. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, S.; Liu, X.; Shang, J.; Zhang, A.; Zhu, Z.; Zha, D. Chalcone Synthase (CHS) Family Members Analysis from Eggplant (Solanum melongena L.) in the Flavonoid Biosynthetic Pathway and Expression Patterns in Response to Heat Stress. PLoS ONE 2020, 15, e0226537. [Google Scholar] [CrossRef]
- Isıyel, M.; İlhan, E.; Kasapoğlu, A.G.; Muslu, S.; Öner, B.M.; Aygören, A.S.; Yıldırım, E. Identification and Characterization of Phaseolus vulgaris CHS Genes in Response to Salt and Drought Stress. Genet. Resour. Crop Evol. 2024, 72, 271–293. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Z.; Vang, S.; Yu, J.; Wong, G.K.S.; Wang, J. Correlation Between Ka/Ks and Ks Is Related to Substitution Model and Evolutionary Lineage. J. Mol. Evol. 2009, 68, 414–423. [Google Scholar] [CrossRef]
- Walther, D.; Brunnemann, R.; Selbig, J. The Regulatory Code for Transcriptional Response Diversity and Its Relation to Genome Structural Properties in A. thaliana. PLoS Genet. 2007, 3, e11. [Google Scholar] [CrossRef]
- Sun, Y.; Oh, D.H.; Duan, L.; Ramachandran, P.; Ramirez, A.; Bartlett, A.; Dinneny, J.R. Divergence in the ABA Gene Regulatory Network Underlies Differential Growth Control. Nat. Plants 2022, 8, 549–560. [Google Scholar] [CrossRef]
- Dao, T.T.H.; Linthorst, H.J.M.; Verpoorte, R. Chalcone Synthase and Its Functions in Plant Resistance. Phytochem. Rev. 2011, 10, 397–412. [Google Scholar] [CrossRef] [PubMed]
- Lai, G.T.; Que, Q.X.; Pan, R.; Liu, Y.X.; Wang, Q.; Lai, P.F.; Lai, C.C. Response of Chalcone Synthase Gene (CHS) to Different Light Quality and Transcription Factor Regulation in Vitis davidii. Biotechnol. Bull. 2022, 38, 129. [Google Scholar] [CrossRef]
- Pan, L.; Ma, J.; Li, J.; Yin, B.; Fu, C. Advances of Salt Stress-Responsive Transcription Factors in Plants. Chin. J. Biotechnol. 2022, 38, 50–65. [Google Scholar] [CrossRef]
- Acosta-Motos, J.R.; Ortuño, M.F.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.J.; Hernandez, J.A. Plant Responses to Salt Stress: Adaptive Mechanisms. Agronomy 2017, 7, 18. [Google Scholar] [CrossRef]
- von der Mark, C.; Ivanov, R.; Eutebach, M.; Maurino, V.G.; Bauer, P.; Brumbarova, T. Reactive Oxygen Species Coordinate the Transcriptional Responses to Iron Availability in Arabidopsis. J. Exp. Bot. 2021, 72, 2181–2195. [Google Scholar] [CrossRef]
- Liu, Q.; Gu, X.; Zhang, Y.; Zhang, T.; Wang, Y.; Dhankher, O.P.; Yuan, H. Molecular and Metabolomics Analysis Reveals New Insight into the Mechanism Underlying Iris halophila Pall. IhCHS1-Mediated Regulation of Plant Salt Tolerance. Environ. Exp. Bot. 2025, 229, 106080. [Google Scholar] [CrossRef]
- Cardona, T.; Shao, S.; Nixon, P.J. Enhancing photosynthesis in plants: The light reactions. Essays Biochem. 2018, 62, 85–94. [Google Scholar] [CrossRef]
- Mira, L.; Fernandez, M.T.; Santos, M.; Rocha, R.; Florêncio, M.H.; Jennings, K.R. Interactions of flavonoids with iron and copper ions: A mechanism for their antioxidant activity. Free Radic. Res. 2002, 36, 1199–1208. [Google Scholar] [CrossRef]
- Kobori, M.; Takahashi, Y.; Akimoto, Y.; Sakurai, M.; Matsunaga, I.; Nishimuro, H.; Ohnishi-Kameyama, M. Chronic high intake of quercetin reduces oxidative stress and induces expression of the antioxidant enzymes in the liver and visceral adipose tissues in mice. J. Funct. Foods 2015, 15, 551–560. [Google Scholar] [CrossRef]
- Li, M.; Wang, W.; Wang, Y.; Guo, L.; Liu, Y.; Jiang, X. Duplicated Chalcone Synthase (CHS) Genes Modulate Flavonoid Production in Tea Plants in Response to Light Stress. J. Integr. Agric. 2024, 23, 1940–1955. [Google Scholar] [CrossRef]
- Yong, Y.; Zhang, Y.; Lyu, Y. A MYB-Related Transcription Factor from Lilium lancifolium L. (LlMYB3) Is Involved in Anthocyanin Biosynthesis Pathway and Enhances Multiple Abiotic Stress Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 3195. [Google Scholar] [CrossRef]
- Li, B.; Fan, R.; Guo, S.; Wang, P.; Zhu, X.; Fan, Y.; Song, C.P. The Arabidopsis MYB Transcription Factor, MYB111 Modulates Salt Responses by Regulating Flavonoid Biosynthesis. Environ. Exp. Bot. 2019, 166, 103807. [Google Scholar] [CrossRef]

| Species | Seq1 | Seq2 | Ka | Ks | Ka/Ks |
|---|---|---|---|---|---|
| Ac vs. Az | AC07G001270.1 | AZ01G0004910.1 | 0.019008 | 0.34062 | 0.055803 |
| Ac vs. Az | AC07G001270.1 | AZ07G0156950.1 | 0.060103 | 1.375058 | 0.043709 |
| Ac vs. Az | AC07G001270.1 | AZ12G0221420.1 | 0.012157 | 0.249037 | 0.048816 |
| Ac vs. Az | AC10G052970.1 | AZ01G0004910.1 | 0.053033 | 0.889335 | 0.059633 |
| Ac vs. Az | AC10G052970.1 | AZ07G0156950.1 | 0.011962 | 0.180102 | 0.066416 |
| Ac vs. Az | AC11G020520.1 | AZ01G0004910.1 | 0.077037 | 0.263941 | 0.291871 |
| Ac vs. Gz | AC07G001270.1 | GZ01G0005140.1 | 0.019008 | 0.34062 | 0.055803 |
| Ac vs. Gz | AC07G001270.1 | GZ07G0167730.1 | 0.069142 | 1.231382 | 0.05615 |
| Ac vs. Gz | AC07G001270.1 | GZ12G0234890.1 | 0.012157 | 0.249037 | 0.048816 |
| Ac vs. Gz | AC10G052970.1 | GZ01G0005140.1 | 0.053033 | 0.889335 | 0.059633 |
| Ac vs. Gz | AC10G052970.1 | GZ07G0167730.1 | 0.014559 | 0.191921 | 0.07586 |
| Ac vs. Gz | AC10G052970.1 | GZ12G0234890.1 | 0.057815 | 1.070942 | 0.053985 |
| Ac vs. Gz | AC11G020520.1 | GZ01G0005140.1 | 0.077037 | 0.263941 | 0.291871 |
| Ac vs. At | AC07G001270.1 | AT5G13930.1 | 0.102025 | 2.071949 | 0.049241 |
| Ac vs. Os | AC07G001270.1 | Os11t0530600-01 | 0.102015 | 0.697797 | 0.146195 |
| Ac vs. Eg | AC07G001270.1 | XM_010931189.3 | 0.015307 | 0.211388 | 0.07241 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, Y.; Khan, N.M.; Ali, A.; Zhou, G.; Zhou, Y.; Li, P.; Wan, Y. AcMYB176-Regulated AcCHS5 Enhances Salt Tolerance in Areca catechu by Modulating Flavonoid Biosynthesis and Reactive Oxygen Species Scavenging. Int. J. Mol. Sci. 2025, 26, 3216. https://doi.org/10.3390/ijms26073216
Jiang Y, Khan NM, Ali A, Zhou G, Zhou Y, Li P, Wan Y. AcMYB176-Regulated AcCHS5 Enhances Salt Tolerance in Areca catechu by Modulating Flavonoid Biosynthesis and Reactive Oxygen Species Scavenging. International Journal of Molecular Sciences. 2025; 26(7):3216. https://doi.org/10.3390/ijms26073216
Chicago/Turabian StyleJiang, Yiqi, Noor Muhammad Khan, Akhtar Ali, Guangzhen Zhou, Yue Zhou, Panjing Li, and Yinglang Wan. 2025. "AcMYB176-Regulated AcCHS5 Enhances Salt Tolerance in Areca catechu by Modulating Flavonoid Biosynthesis and Reactive Oxygen Species Scavenging" International Journal of Molecular Sciences 26, no. 7: 3216. https://doi.org/10.3390/ijms26073216
APA StyleJiang, Y., Khan, N. M., Ali, A., Zhou, G., Zhou, Y., Li, P., & Wan, Y. (2025). AcMYB176-Regulated AcCHS5 Enhances Salt Tolerance in Areca catechu by Modulating Flavonoid Biosynthesis and Reactive Oxygen Species Scavenging. International Journal of Molecular Sciences, 26(7), 3216. https://doi.org/10.3390/ijms26073216





