DNA Demethylase ROS1 Interferes with DNA Methylation and Activates Stress Response Genes in Plants Infected with Beet Severe Curly Top Virus
Abstract
1. Introduction
2. Results
2.1. Symptoms of BSCTV-Infected Arabidopsis Thaliana Col-0 and ros1 Plants
2.2. Increased Expression of Stress Response Genes Is Mediated by C2 in BSCTV-Infected and C2 Transgenic Plants
2.3. The Repressor of Silencing ROS1 Is Involved in Regulating the Expression of the Stress Response Genes in BSCTV-Infected Plants
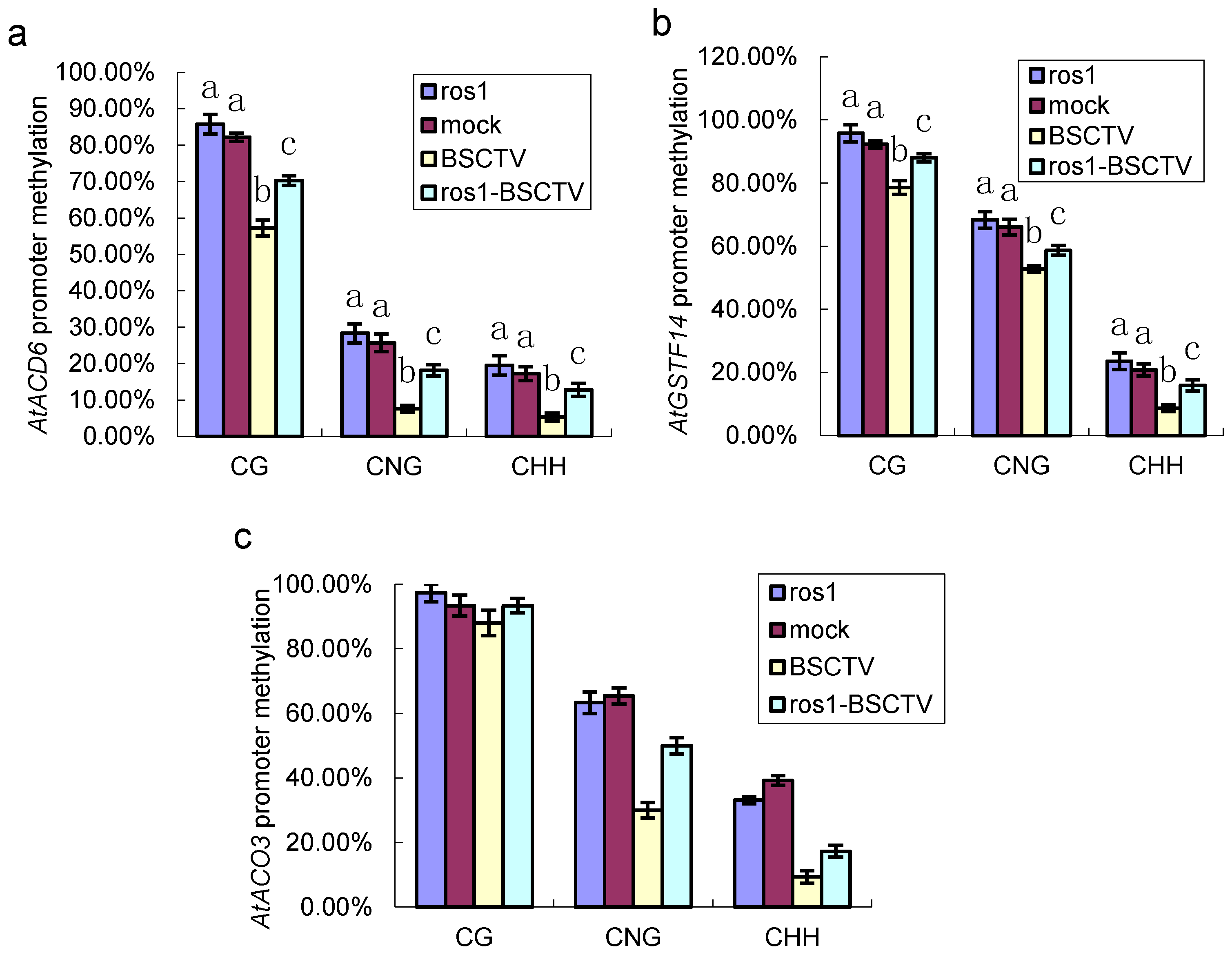
2.4. Increased Gene Expression Is Correlated with ROS1-Mediated DNA Demethylation in the Promoters of Stress Response Genes
3. Discussion
3.1. ROS1 Is Involved in the Regulation of Stress Response Gene Expression
3.2. DNA Demethylation in the Promoters of Stress Response Genes in BSCTV-Infected Plants Is Partially Dependent on ROS1
4. Materials and Methods
4.1. Plant Growth Conditions
4.2. Agrobacterium Tumefaciens Inoculation
4.3. Bisulfite Sequencing
4.4. RT-qPCR Analysis
4.5. Southern Blotting Analysis and Northern Blotting Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baulcombe, D. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Meister, G.; Tuschl, T. Mechanisms of gene silencing by double-stranded RNA. Nature 2004, 431, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O. Post-transcriptional RNA silencing in plant-microbe interactions: A touch of robustness and versatility. Curr. Opin. Plant Biol. 2008, 11, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Llave, C. Virus-derived small interfering RNAs at the core of plant-virus interactions. Trends Plant Sci. 2010, 15, 701–707. [Google Scholar] [CrossRef]
- Brodersen, P.; Sakvarelidze-Achard, L.; Bruun-Rasm Ussen, M.; Dunoyer, P.; Yamamoto, Y.Y.; Sieburth, L.; Voinnet, O. Widespread translational inhibition by plant miRNAs and siRNAs. Science 2008, 320, 1185–1190. [Google Scholar] [CrossRef]
- Voinnet, O. Induction and suppression of RNA silencing: Insights from viral infections. Nat. Rev. Genet. 2005, 6, 206–220. [Google Scholar] [CrossRef]
- Yang, L.P.; Fang, Y.Y.; An, C.P.; Dong, L.; Zhang, Z.H.; Chen, H.; Xie, Q.; Guo, H.S. C2-mediated decrease in DNA methylation, accumulation of siRNAs, and increase in expression for genes involved in defense pathways in plants infected with beet severe curly top virus. Plant J. 2013, 73, 910–917. [Google Scholar] [CrossRef]
- Wang, H.; Buckley, K.J.; Yang, X.J.; Buchmann, R.C.; Bisaro, D.M. Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J. Virol. 2005, 79, 7410–7418. [Google Scholar] [CrossRef]
- Raja, P.; Sanville, B.C.; Buchmann, R.C.; Bisaro, D.M. Viral genome methylation as an epigenetic defense against geminiviruses. J. Virol. 2008, 82, 8997–9007. [Google Scholar] [CrossRef]
- Qi, Y.J.; Denli, A.M.; Hannon, G.J. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 2005, 19, 421–428. [Google Scholar] [CrossRef]
- Vanitharani, R.; Chellappan, P.; Fauquet, C.M. Geminiviruses and RNA silencing. Trends Plant Sci. 2005, 10, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Meyer, P. DNA methylation systems and targets in plants. FEBS Lett. 2011, 585, 2008–2015. [Google Scholar] [CrossRef] [PubMed]
- Simon, S.A.; Meyers, B.C. Small RNA-mediated epigenetic modifications in plants. Curr. Opin. Plant Biol. 2011, 14, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.M.; Zhu, J.K. RNA-directed DNA methylation. Curr. Opin. Plant Biol. 2011, 14, 142–147. [Google Scholar] [CrossRef]
- Yang, L.P.; Xu, Y.N.; Liu, Y.Q.; Meng, D.W.; Jin, T.C.; Zhou, X.F. HC-Pro viral suppressor from tobacco vein banding mosaic virus interferes with DNA methylation and activates the salicylic acid pathway. Virology 2016, 497, 244–250. [Google Scholar] [CrossRef]
- Wang, B.; Yang, X.; Wang, Y.; Xie, Y.; Zhou, X. Tomato yellow leaf curl Virus V2 interacts with host histone deacetylase6 to suppress methylation-mediated transcriptional gene silencing in plants. J. Virol. 2018, 92, 10–1128. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Y.; Gong, Q.; Ismayil, A.; Yuan, Y.; Lian, B.; Jia, Q.; Han, M.; Deng, H.; Hong, Y.; et al. Geminiviral V2 protein suppresses transcriptional gene silencing through interaction with AGO4. J. Virol. 2019, 93, 10–1128. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Chen, H.; Huang, X.H.; Xia, R.; Zhao, Q.Z.; Lai, J.B.; Teng, K.L.; Li, Y.; Liang, L.M.; Du, Q.S.; et al. BSCTV C2 Attenuates the Degradation of SAMDC1 to Suppress DNA Methylation-Mediated Gene Silencing in Arabidopsis. Plant Cell 2011, 23, 273–288. [Google Scholar] [CrossRef]
- Yang, L.P.; Meng, D.W.; Wang, Y.; Wu, Y.J.; Lang, C.J.; Jin, T.C.; Zhou, X.F. The viral suppressor HCPro decreases DNA methylation and activates auxin biosynthesis genes. Virology 2020, 546, 133–140. [Google Scholar] [CrossRef]
- Gehring, M.; Jin, H.H.; Hsieh, T.F.; Penterman, J.; Choi, Y.; Harada, J.J.; Goldberg, R.B.; Fischer, R.L. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 2006, 124, 495–506. [Google Scholar] [CrossRef]
- Ortega-Galisteo, A.P.; Morales-Ruiz, T.; Ariza, R.R.; Roldán-Arjona, T. Arabidopsis Demeter-Like proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol. Biol. 2008, 67, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Ponferrada-Marin, M.I.; Roldan-Arjona, T.; Ariza, R.R. Demethylationinitiated by ROS1 glycosylase involves random sliding along DNA. Nucleic. Acids Res. 2012, 40, 11554–11562. [Google Scholar] [CrossRef] [PubMed]
- Schumann, U.; Lee, J.; Kazan, K.; Ayliffe, M.; Wang, M.B. DNA-demethylase regulated genes show methylation-independent spatiotemporal expression patterns. Front. Plant Sci. 2017, 8, 1449. [Google Scholar] [CrossRef] [PubMed]
- Le, T.N.; Schumann, U.; Smith, N.A.; Tiwari, S.; Au, P.; Zhu, Q.H.; Taylor, J.; Kazan, K.; Llewellyn, D.J.; Zhang, R. DNA demethylases target promoter transposable elements to positively regulate stress responsive genes in Arabidopsis. Genome Biol. 2014, 15, 458. [Google Scholar] [CrossRef]
- Dowen, R.H.; Pelizzola, M.; Schmitz, R.J.; Lister, R.; Ecker, J.R. Widespread dynamicDNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. USA 2012, 109, 2183–2191. [Google Scholar] [CrossRef]
- Yu, A.; Lepere, G.; Jay, F.; Wang, J.; Bapaume, L.; Wang, Y.; Abraham, A.L.; Penterman, J.; Fischer, R.L.; Voinnet, O.; et al. Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc. Natl. Acad. Sci. USA 2013, 110, 2389–2394. [Google Scholar] [CrossRef]
- Liu, R.; Lang, Z. The mechanism and function of active DNA demethylation in plants. J. Integr. Plant Biol. 2019, 62, 148–159. [Google Scholar] [CrossRef]
- Kim, J.S.; Lim, J.Y.; Shin, H.; Kim, B.G.; Yoo, S.D.; Kim, W.T.; Huh, J.H. ROS1-dependent DNA demethylation is required for ABA-inducible NIC3 expression. Plant Physiol. 2019, 179, 1810–1821. [Google Scholar] [CrossRef]
- Yang, L.P.; Lang, C.J.; Wu, Y.J.; Meng, D.W.; Yang, T.B.; Li, D.Q.; Jin, T.C.; Zhou, X.F. ROS1-mediated decrease in DNA methylation and increase in expression of defense genes and stress response genes in Arabidopsis thaliana due to abiotic stresses. BMC Plant Biol. 2022, 22, 104. [Google Scholar] [CrossRef]
- Ascencio-Ibáñez, J.T.; Sozzani, R.; Lee, T.J.; Chu, T.M.; Wolfinger, R.D.; Cella, R.; Hanley-Bowdoin, L. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008, 148, 436–454. [Google Scholar] [CrossRef]
- Marrs, K.A. The functions and regulation of glutathione stransferases in plants, Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 127–158. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.P.; Davis, B.G.; Edwardset, R. Functional divergence in the glutathione transferase superfamily in plants. J. Biol. Chem. 2002, 277, 30859–30869. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Ghanashyam, C.; Bhattacharjee, A. Comprehensive expression analysis suggests overlapping and specific roles of rice glutathione S-transferase genes during development and stress responses. BMC Genom. 2010, 11, 73. [Google Scholar] [CrossRef]
- Nianiou-Obeidat, I.; Madesis, P.; Kissoudis, C.; Voulgari, G.; Chronopoulou, E.; Tsaftaris, A.; Labrou, N.E. Plant glutathione transferase-mediated stress tolerance: Functions and biotechnological applications. Plant Cell Rep. 2017, 36, 791–805. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, Z.; Teng, K.; Lai, J.; Zhang, Y.; Huang, Y.; Li, Y.; Liang, L.; Wang, Y.; Chu, C.; et al. Up-regulation of LSB1/GDU3 affects geminivirus infection by activating the salicylic acid pathway. Plant J. 2010, 62, 12–23. [Google Scholar] [CrossRef]
- Zhang, H.; Lang, Z.; Zhu, J.K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef]
- Deniz, Ö.; Frost, M.F.; Branco, M.R. Regulation of transposable elements by DNA modifications. Nat. Rev. Genet. 2019, 20, 417–431. [Google Scholar] [CrossRef]
- Gong, Z.Z.; Morales-Ruiz, T.; Ariza, R.R.; Roldán-Arjona, T.; David, L.; Zhu, J.K. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 2002, 111, 803–814. [Google Scholar] [CrossRef]
- Agius, F.; Kapoor, A.; Zhu, J.K. Role of the “Arabidopsis” DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc. Natl. Acad. Sci. USA 2006, 103, 11796–11801. [Google Scholar] [CrossRef]
- Duan, C.G.; Wang, X.G.; Xie, S.J.; Li, P.; Miki, D.; Tang, K.; Hsu, C.C.; Lei, M.; Zhong, Y.; Hou, Y.J.; et al. A pair of transposon-derived proteins function in a histone acetyltransferase complex for active DNA demethylation. Cell Res. 2017, 27, 226–240. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, T.; Li, Y.; Sun, X.; Li, Y.; Xiao, Z.; Wang, W.; Yu, J.; Yang, L. DNA Demethylase ROS1 Interferes with DNA Methylation and Activates Stress Response Genes in Plants Infected with Beet Severe Curly Top Virus. Int. J. Mol. Sci. 2025, 26, 2807. https://doi.org/10.3390/ijms26062807
Jin T, Li Y, Sun X, Li Y, Xiao Z, Wang W, Yu J, Yang L. DNA Demethylase ROS1 Interferes with DNA Methylation and Activates Stress Response Genes in Plants Infected with Beet Severe Curly Top Virus. International Journal of Molecular Sciences. 2025; 26(6):2807. https://doi.org/10.3390/ijms26062807
Chicago/Turabian StyleJin, Taicheng, Yushuo Li, Xu Sun, Yidi Li, Zhuyi Xiao, Weiyan Wang, Jiaxue Yu, and Liping Yang. 2025. "DNA Demethylase ROS1 Interferes with DNA Methylation and Activates Stress Response Genes in Plants Infected with Beet Severe Curly Top Virus" International Journal of Molecular Sciences 26, no. 6: 2807. https://doi.org/10.3390/ijms26062807
APA StyleJin, T., Li, Y., Sun, X., Li, Y., Xiao, Z., Wang, W., Yu, J., & Yang, L. (2025). DNA Demethylase ROS1 Interferes with DNA Methylation and Activates Stress Response Genes in Plants Infected with Beet Severe Curly Top Virus. International Journal of Molecular Sciences, 26(6), 2807. https://doi.org/10.3390/ijms26062807



