Genome-Wide Identification of the bHLH Gene Family in Magnolia sieboldii and Response of MsPIFs to Different Light Qualities
Abstract
1. Introduction
2. Results
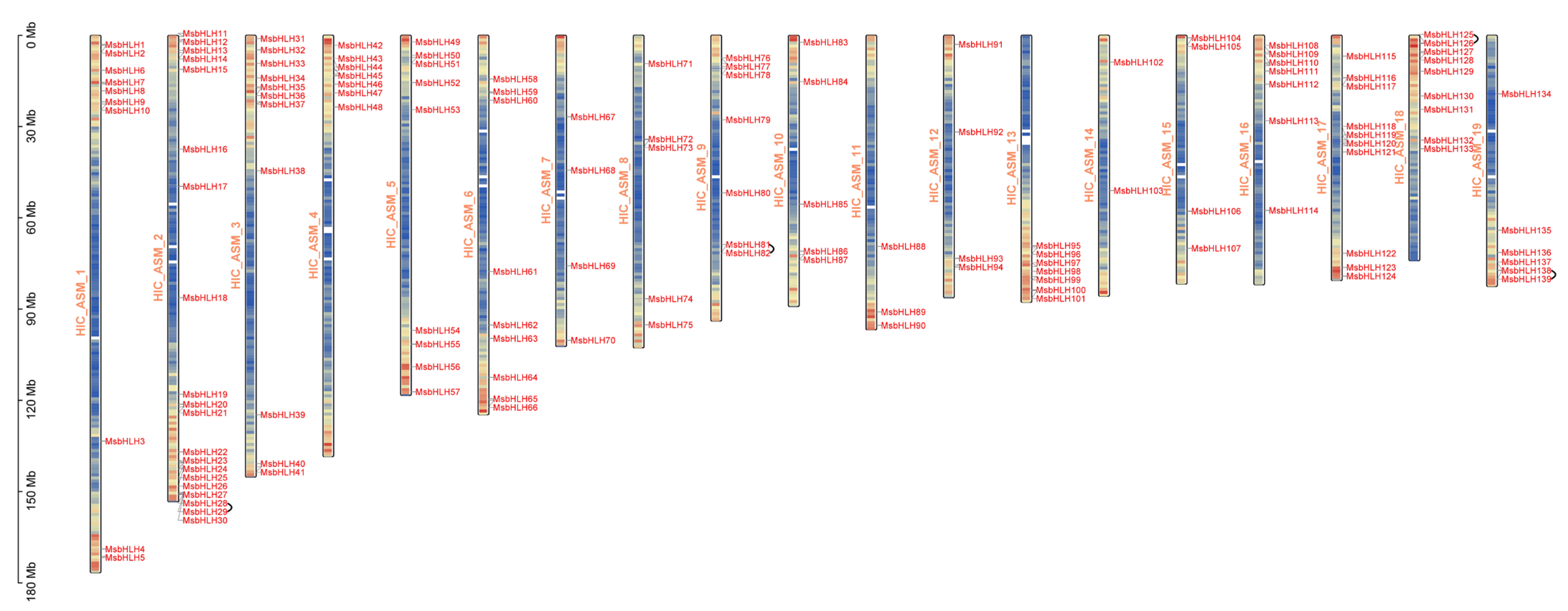
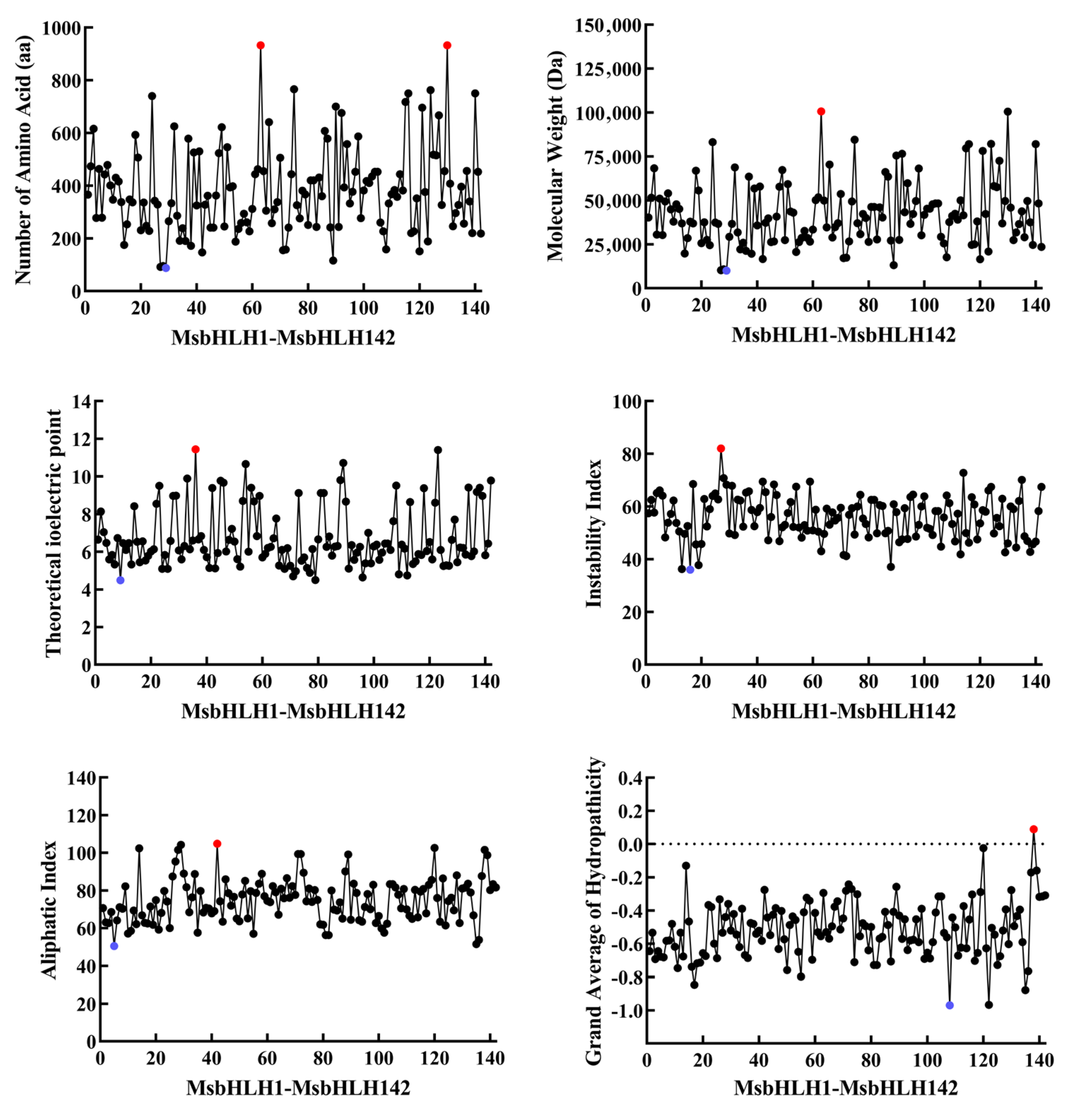
2.1. Whole-Genome Identification of bHLH TFs in M. sieboldii
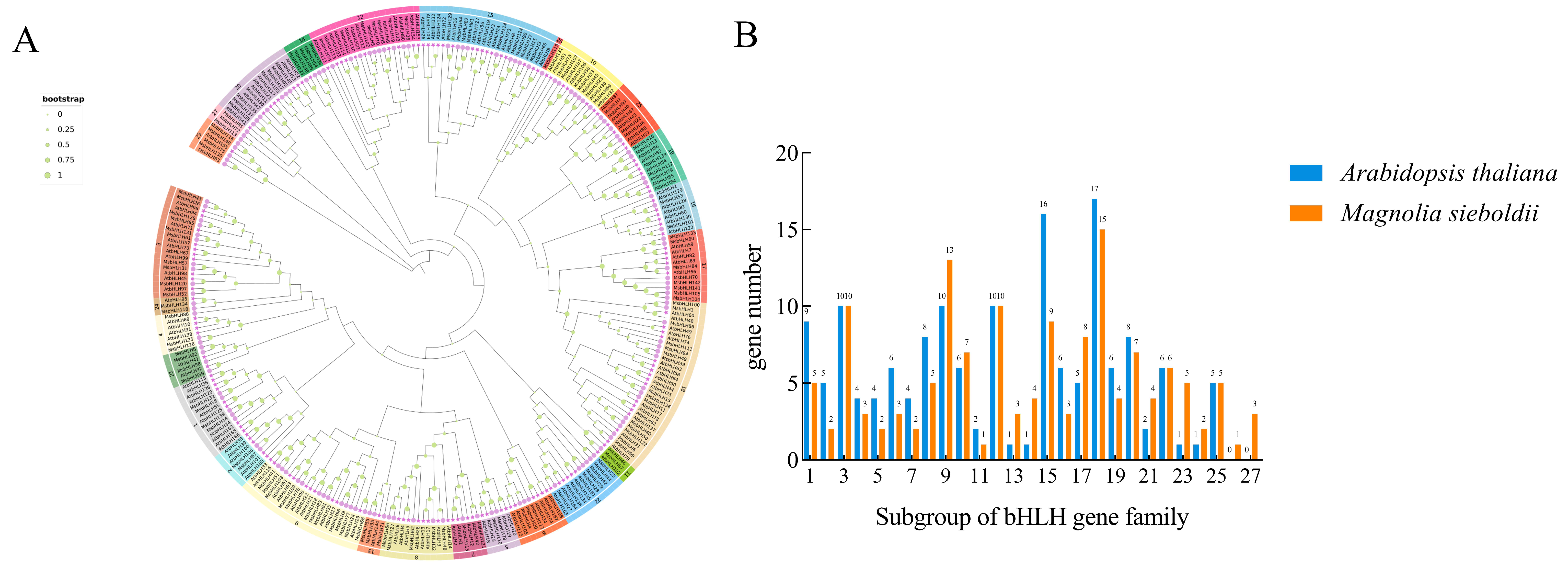
2.2. Phylogenetic Relationship Between AtbHLHs and MsbHLHs
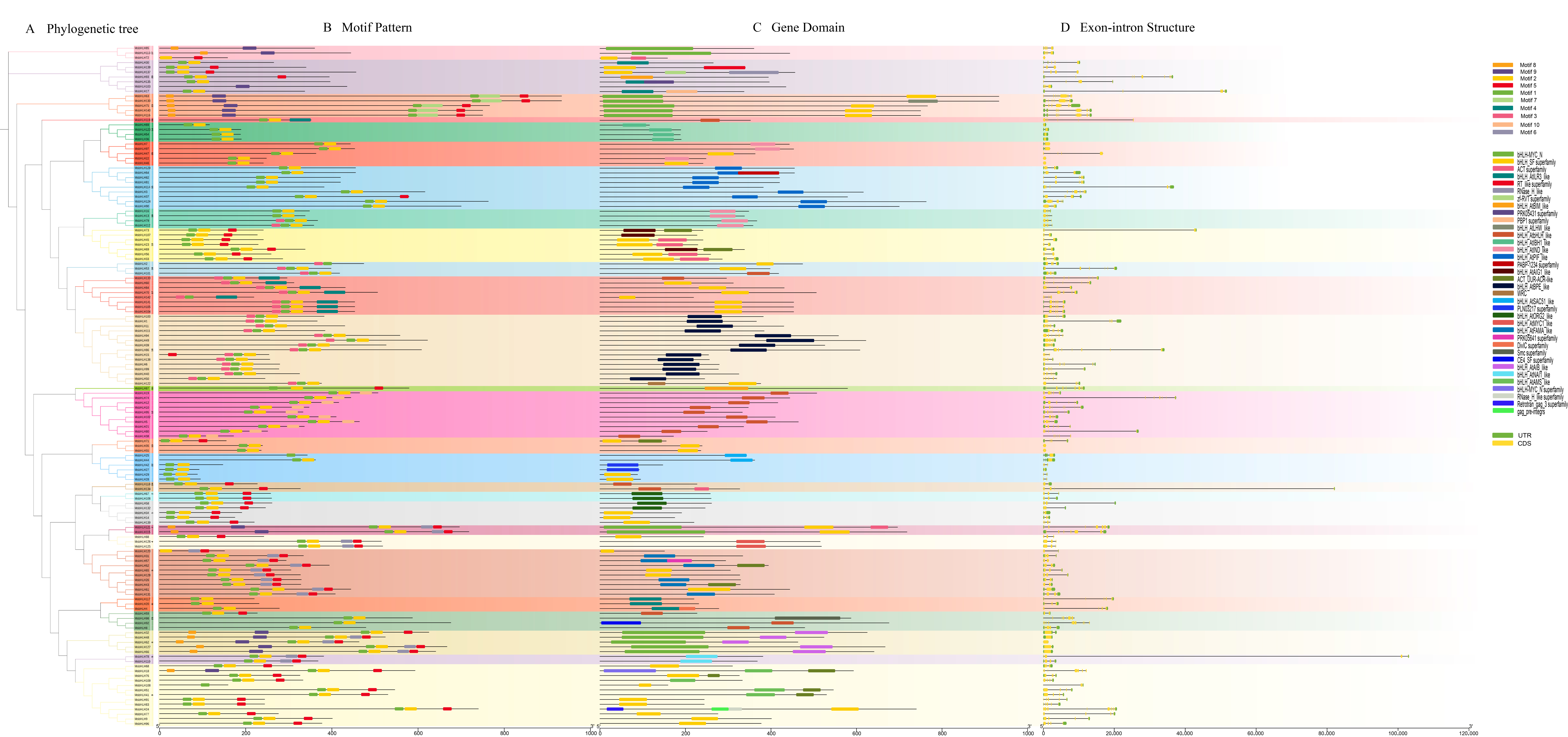
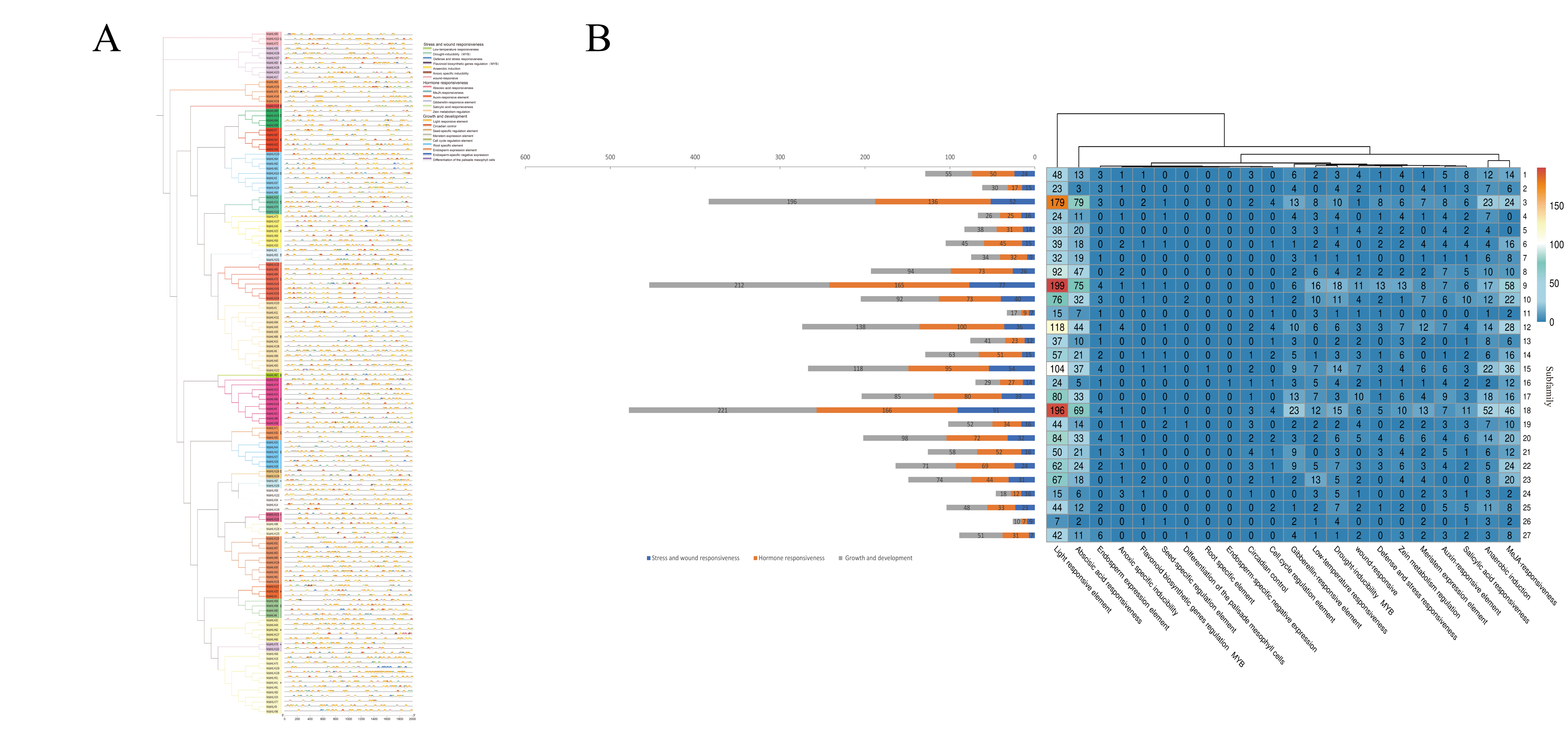
2.3. Gene Structure, Motif, and Intron–Exon Analysis
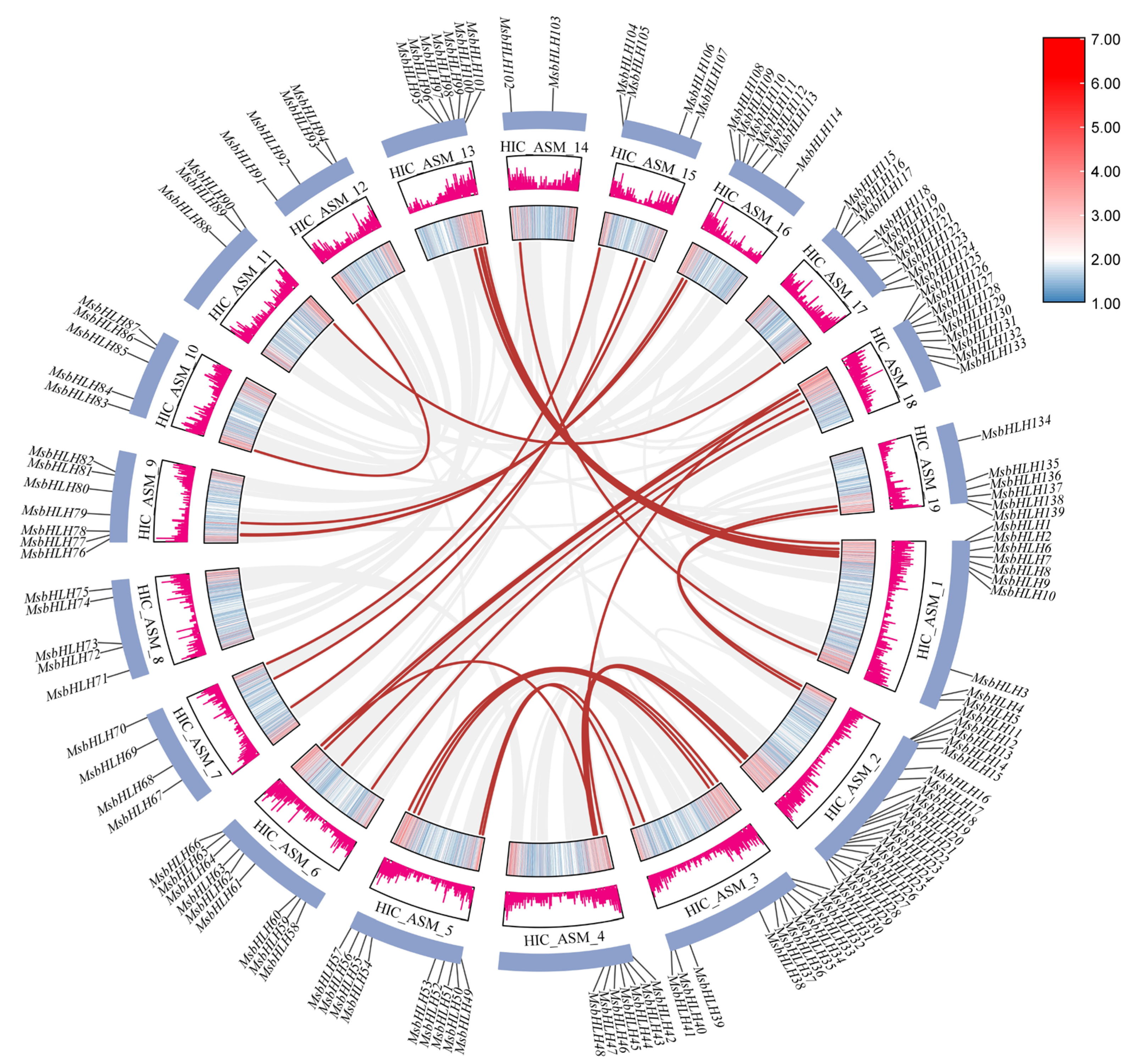
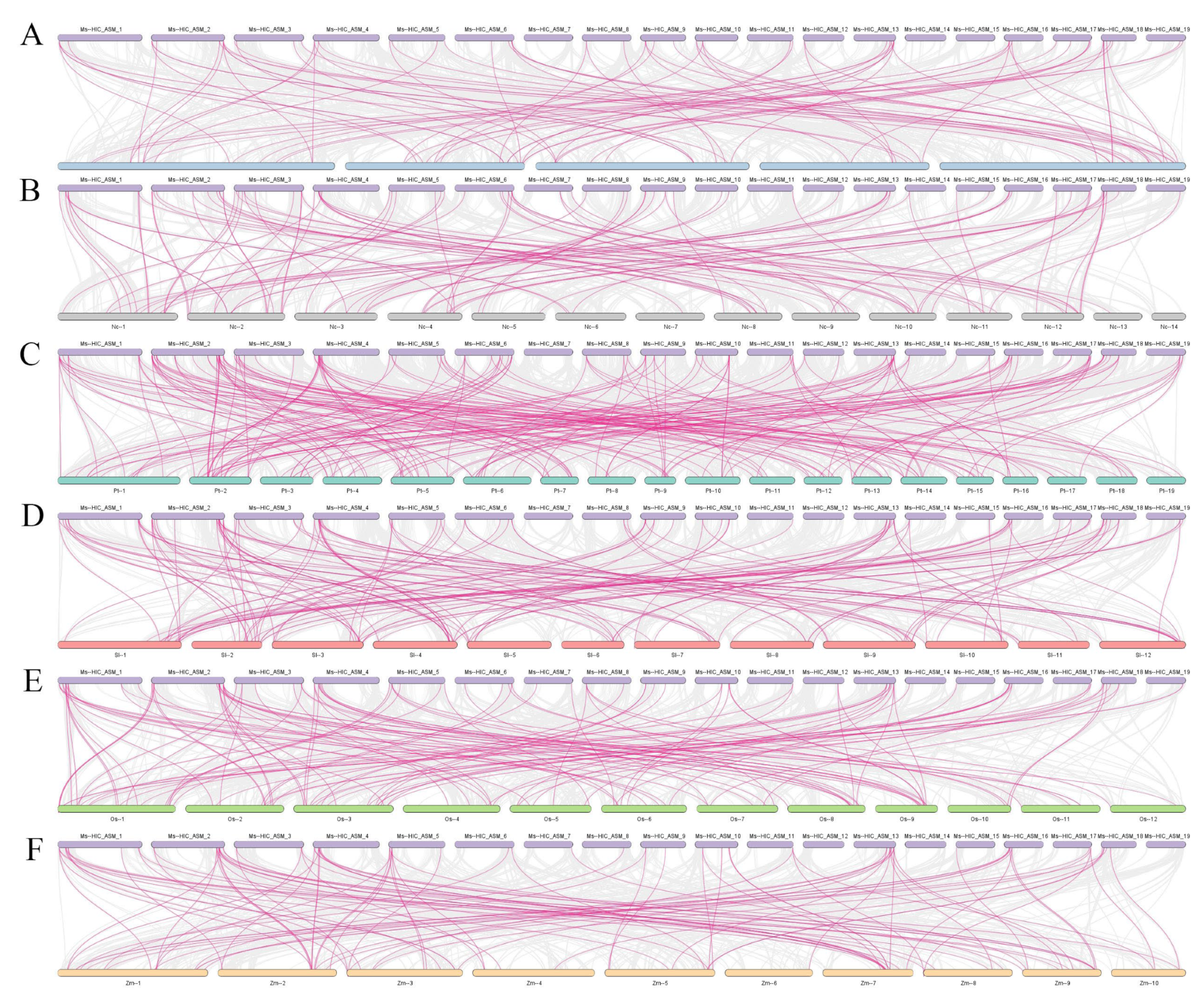
2.4. Intraspecific and Interspecific Collinearity and Gene Duplication Analysis
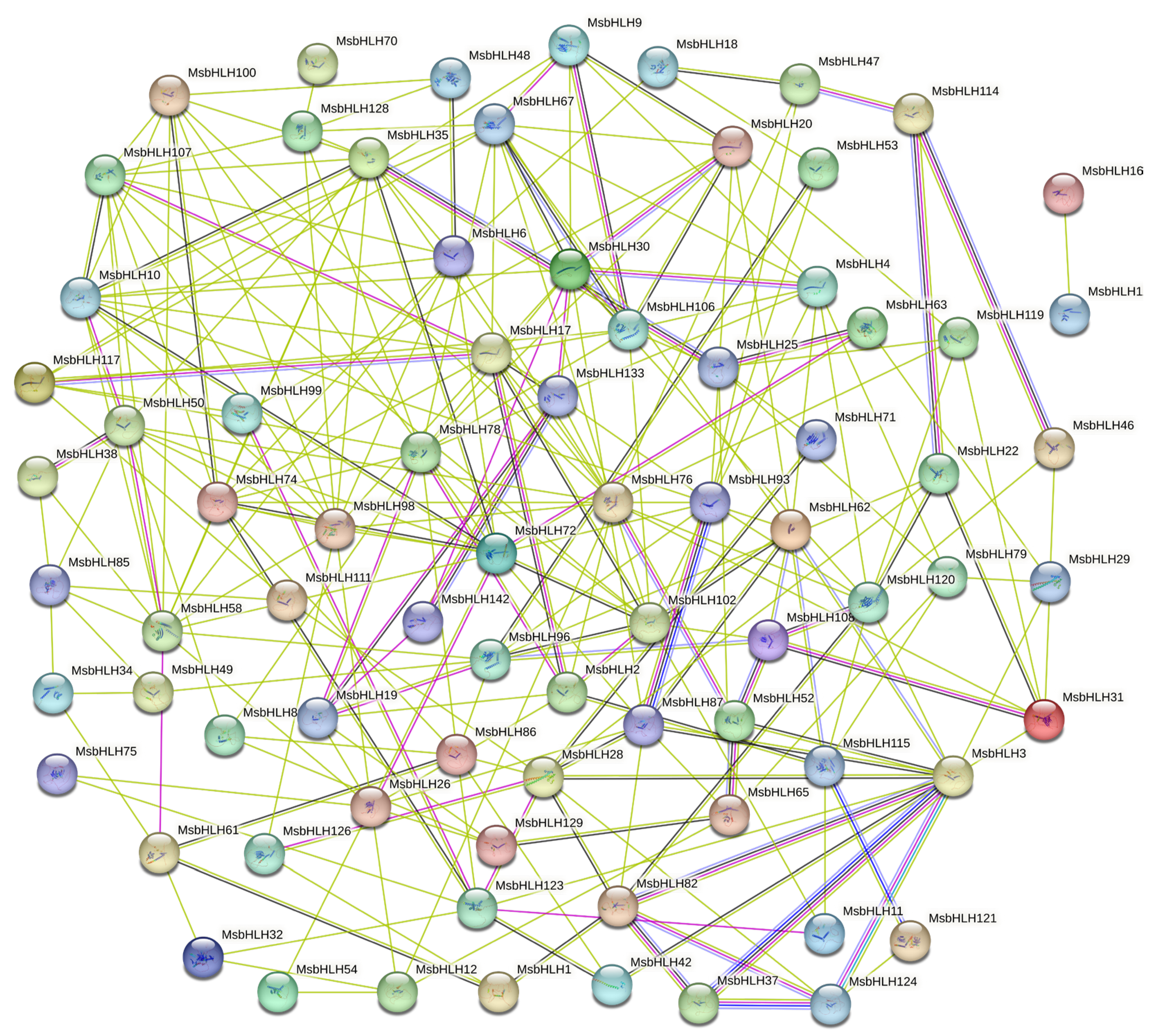
2.5. Protein Interactions Analysis of MsbHLH Protein
2.6. Cis-Regulatory Element Analysis of MsbHLHs
2.7. GO Enrichment Analysis of MsbHLHs
2.8. Expression Pattern of MsbHLHs in Seeds
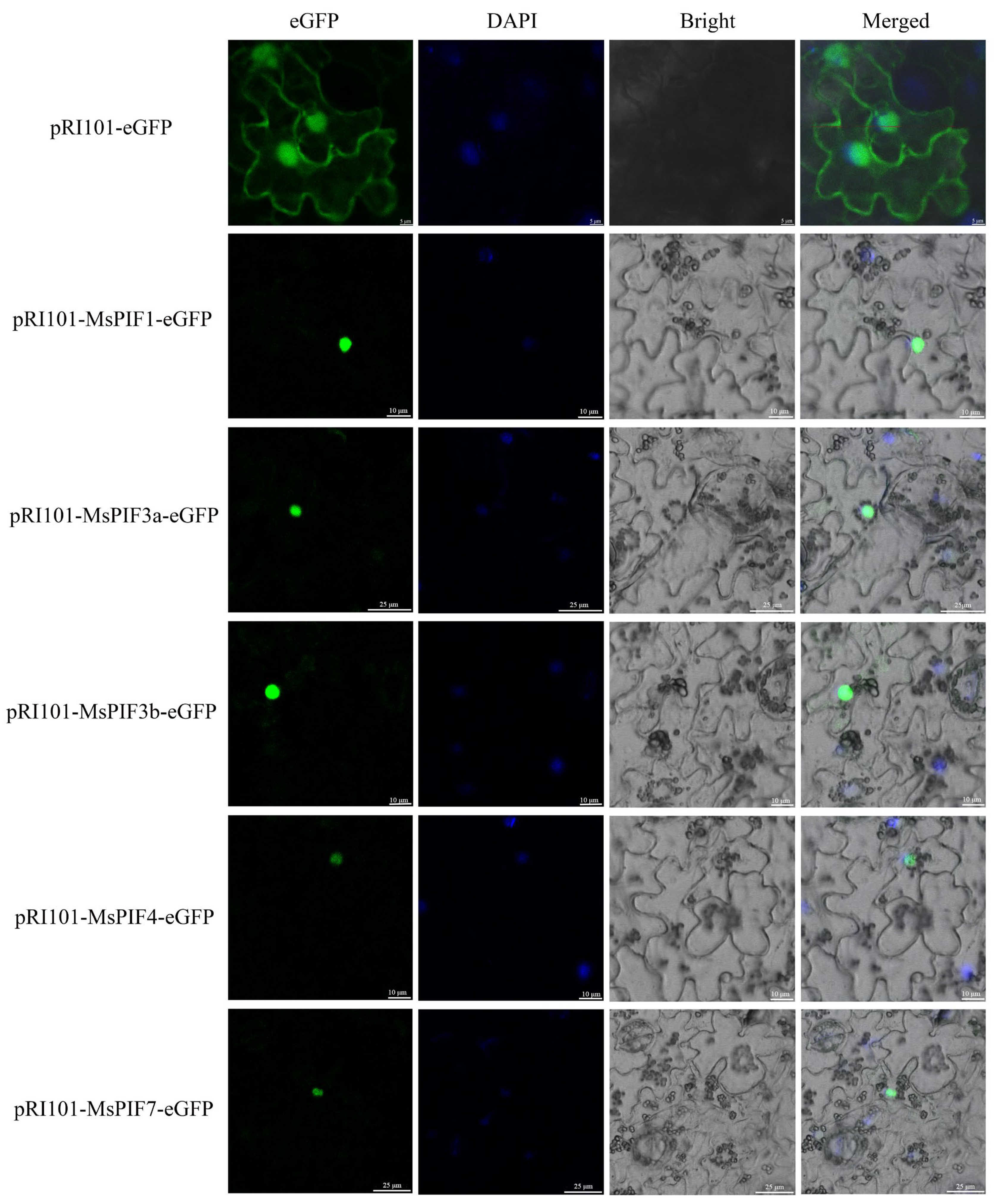
2.9. Subcellular Localization of MsPIFs
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Light Quality Treatments
4.2. Identification of bHLH Genes in the M. sieboldii Genome
4.3. Phylogenetic Analysis of MsbHLHs
4.4. Gene Structure, Conserved Motif, and Cis-Regulatory Elements Analysis
4.5. Duplication and Genome Synteny Analysis
4.6. Protein–Protein Interaction Network Prediction, Homology Modeling, and GO Enrichment Analysis
4.7. Transcriptome and qRT-PCR Analysis
4.8. Subcellular Localization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- He, S. Rare Plants in China; Shanghai Science and Technology Press: Shanghai, China, 1998; 184p, ISBN 7-5323-4934-9. [Google Scholar]
- Du, F.; Diao, S.; Wang, H.; Liu, C.; Yang, D.; Sun, S.; Wang, Z. Phenology and Growth Process of Magnolia sieboldii. J. Northeast. For. Univ. 2006, 34, 39–40. [Google Scholar]
- Mei, M. Functions and Mechanisms of MsARF5 in Embryo Development of Magnolia sieboldii K. Koch During the Seed After-Ripening. Ph.D. Thesis, Shenyang Agricultural University, Shenyang, China, 2021. [Google Scholar]
- Li, P.; Lu, X.; Yao, F.; Guo, R. Preliminary Study on Reasons of Seed Dormancy of Magnolia sieboldii K. Koch. Seed 2006, 25, 36–39. [Google Scholar]
- Lu, X.; Li, T.; Ni, W. Study on the Dormant Characteristic of M. sieboldii K. Koch Seeds. J. Shenyang Agric. Univ. 2006, 37, 703–706. [Google Scholar]
- Murre, C.; McCaw, P.S.; Baltimore, D. A New DNA Binding and Dimerization Motif in Immunoglobulin Enhancer Binding, Daughterless, MyoD, and Myc Proteins. Cell 1989, 56, 777–783. [Google Scholar] [CrossRef]
- Ferré-D’Amaré, A.R.; Prendergast, G.C.; Ziff, E.B.; Burley, S.K. Recognition by Max of Its Cognate DNA through a Dimeric b/HLH/Z Domain. Nature 1993, 363, 38–45. [Google Scholar] [CrossRef]
- Kadesch, T. Consequences of Heteromeric Interactions among Helix-Loop-Helix Proteins. Cell Growth Differ. 1993, 4, 49–55. [Google Scholar]
- Murre, C.; McCaw, P.S.; Vaessin, H.; Caudy, M.; Jan, L.Y.; Jan, Y.N.; Cabrera, C.V.; Buskin, J.N.; Hauschka, S.D.; Lassar, A.B.; et al. Interactions between Heterologous Helix-Loop-Helix Proteins Generate Complexes That Bind Specifically to a Common DNA Sequence. Cell 1989, 58, 537–544. [Google Scholar] [CrossRef]
- Ledent, V.; Vervoort, M. The Basic Helix-Loop-Helix Protein Family: Comparative Genomics and Phylogenetic Analysis. Genome Res. 2001, 11, 754–770. [Google Scholar] [CrossRef]
- Pires, N.; Dolan, L. Early Evolution of bHLH Proteins in Plants. Plant Signal. Behav. 2010, 5, 911–912. [Google Scholar] [CrossRef]
- Groszmann, M.; Bylstra, Y.; Lampugnani, E.R.; Smyth, D.R. Regulation of Tissue-Specific Expression of SPATULA, a bHLH Gene Involved in Carpel Development, Seedling Germination, and Lateral Organ Growth in Arabidopsis. J. Exp. Bot. 2010, 61, 1495. [Google Scholar] [CrossRef]
- Hao, Y.; Zong, X.; Ren, P.; Qian, Y.; Fu, A. Basic Helix-Loop-Helix (bHLH) Transcription Factors Regulate a Wide Range of Functions in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 7152. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Yamaguchi, S.; Kamiya, Y.; Bae, G.; Chung, W.-I.; Choi, G. Light Activates the Degradation of PIL5 Protein to Promote Seed Germination through Gibberellin in Arabidopsis. Plant J. 2006, 47, 124–139. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S.; Josse, E.-M.; Kannangara, R.; Gilday, A.D.; Halliday, K.J.; Graham, I.A. Cold and Light Control Seed Germination through the bHLH Transcription Factor SPATULA. Curr. Biol. 2005, 15, 1998–2006. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Song, Y.H.; Josephson-Day, A.R.; Miller, R.J.; Breton, G.; Olmstead, R.G.; Imaizumi, T. FLOWERING BHLH Transcriptional Activators Control Expression of the Photoperiodic Flowering Regulator CONSTANS in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 3582–3587. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) Function as Transcriptional Activators in Abscisic Acid Signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef]
- Kiribuchi, K.; Sugimori, M.; Takeda, M.; Otani, T.; Okada, K.; Onodera, H.; Ugaki, M.; Tanaka, Y.; Tomiyama-Akimoto, C.; Yamaguchi, T.; et al. RERJ1, a Jasmonic Acid-Responsive Gene from Rice, Encodes a Basic Helix–Loop–Helix Protein. Biochem. Biophys. Res. Commun. 2004, 325, 857–863. [Google Scholar] [CrossRef]
- Seo, J.-S.; Joo, J.; Kim, M.-J.; Kim, Y.-K.; Nahm, B.H.; Song, S.I.; Cheong, J.-J.; Lee, J.S.; Kim, J.-K.; Choi, Y.D. OsbHLH148, a Basic Helix-loop-helix Protein, Interacts with OsJAZ Proteins in a Jasmonate Signaling Pathway Leading to Drought Tolerance in Rice. Plant J. 2011, 65, 907–921. [Google Scholar] [CrossRef]
- Waseem, M.; Rong, X.; Li, Z. Dissecting the Role of a Basic Helix-Loop-Helix Transcription Factor, SlbHLH22, Under Salt and Drought Stresses in Transgenic Solanum lycopersicum L. Front. Plant Sci. 2019, 10, 734. [Google Scholar] [CrossRef]
- Guo, J.; Sun, B.; He, H.; Zhang, Y.; Tian, H.; Wang, B. Current Understanding of bHLH Transcription Factors in Plant Abiotic Stress Tolerance. Int. J. Mol. Sci. 2021, 22, 4921. [Google Scholar] [CrossRef]
- Shen, T.; Wen, X.; Wen, Z.; Qiu, Z.; Hou, Q.; Li, Z.; Mei, L.; Yu, H.; Qiao, G. Genome-Wide Identification and Expression Analysis of bHLH Transcription Factor Family in Response to Cold Stress in Sweet Cherry (Prunus avium L.). Sci. Hortic. 2021, 279, 109905. [Google Scholar] [CrossRef]
- Yang, X.; Wang, R.; Hu, Q.; Li, S.; Mao, X.; Jing, H.; Zhao, J.; Hu, G.; Fu, J.; Liu, C. DlICE1, a Stress-Responsive Gene from Dimocarpus longan, Enhances Cold Tolerance in Transgenic Arabidopsis. Plant Physiol. Biochem. 2019, 142, 490–499. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Fu, X.; Xu, H.; Zhang, Y.; Shi, Z.; Zhou, G.; Bao, W. Comprehensive Analysis of bHLH Transcription Factors in Ipomoea aquatica and Its Response to Anthocyanin Biosynthesis. Int. J. Mol. Sci. 2023, 24, 5652. [Google Scholar] [CrossRef] [PubMed]
- Pires, N.; Dolan, L. Origin and Diversification of Basic-Helix-Loop-Helix Proteins in Plants. Mol. Biol. Evol. 2009, 27, 862. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis Basic/Helix-Loop-Helix Transcription Factor Family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, Y.; Altaf, M.A.; Hao, Y.; Zhou, G.; Li, X.; Zhu, J.; Ma, W.; Wang, Z.; Bao, W. Genome-Wide Identification of Myeloblastosis Gene Family and Its Response to Cadmium Stress in Ipomoea aquatica. Front. Plant Sci. 2022, 13, 979988. [Google Scholar] [CrossRef]
- Liang, J.; Fang, Y.; An, C.; Yao, Y.; Wang, X.; Zhang, W.; Liu, R.; Wang, L.; Aslam, M.; Cheng, Y.; et al. Genome-Wide Identification and Expression Analysis of the bHLH Gene Family in Passion Fruit (Passiflora edulis) and Its Response to Abiotic Stress. Int. J. Biol. Macromol. 2023, 225, 389–403. [Google Scholar]
- Ain-Ali, Q.-U.; Mushtaq, N.; Amir, R.; Gul, A.; Tahir, M.; Munir, F. Genome-Wide Promoter Analysis, Homology Modeling and Protein Interaction Network of Dehydration Responsive Element Binding (DREB) Gene Family in Solanum tuberosum. PLoS ONE 2021, 16, e0261215. [Google Scholar] [CrossRef]
- Wei, K.; Chen, H. Comparative Functional Genomics Analysis of bHLH Gene Family in Rice, Maize and Wheat. BMC Plant Biol. 2018, 18, 309. [Google Scholar] [CrossRef]
- Chen, X.; Yao, C.; Liu, J.; Liu, J.; Fang, J.; Deng, H.; Yao, Q.; Kang, T.; Guo, X. Basic Helix-Loop-Helix (bHLH) Gene Family in Rye (Secale cereale L.): Genome-Wide Identification, Phylogeny, Evolutionary Expansion and Expression Analyses. BMC Genom. 2024, 25, 67. [Google Scholar] [CrossRef]
- Quan, X.; Meng, C.; Zhang, N.; Liang, X.; Li, J.; Li, H.; He, W. Genome-Wide Analysis of Barley bHLH Transcription Factors and the Functional Characterization of HvbHLH56 in Low Nitrogen Tolerance in Arabidopsis. Int. J. Mol. Sci. 2023, 24, 9740. [Google Scholar] [CrossRef]
- Ke, Y.; Wu, Y.; Zhou, H.; Chen, P.; Wang, M.; Liu, M.; Li, P.; Yang, J.; Li, J.; Du, H. Genome-Wide Survey of the bHLH Super Gene Family in Brassica napus. BMC Plant Biol. 2020, 20, 115. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Fan, H.; Ling, H. Genome-Wide Identification and Characterization of the bHLH Gene Family in Tomato. BMC Genom. 2015, 16, 9. [Google Scholar] [CrossRef] [PubMed]
- Bailey, P.C.; Martin, C.; Toledo-Ortiz, G.; Quail, P.H.; Huq, E.; Heim, M.A.; Jakoby, M.; Werber, M.; Weisshaar, B. Update on the Basic Helix-Loop-Helix Transcription Factor Gene Family in Arabidopsis thaliana. Plant Cell 2003, 15, 2497. [Google Scholar] [CrossRef]
- Huang, X.; Su, L.; Xian, B.; Yu, Q.; Zhang, M.; Fan, J.; Zhang, C.; Liu, Y.; He, H.; Zhong, X.; et al. Genome-Wide Identification and Characterization of the Sweet Orange (Citrus sinensis) Basic Helix-Loop-Helix (bHLH) Family Reveals a Role for CsbHLH085 as a Regulator of Citrus Bacterial Canker Resistance. Int. J. Biol. Macromol. 2024, 267, 131442. [Google Scholar] [CrossRef]
- Zhan, H.; Liu, H.; Ai, W.; Han, X.; Wang, Y.; Lu, X. Genome-Wide Identification and Expression Analysis of the bHLH Transcription Factor Family and Its Response to Abiotic Stress in Mongolian Oak (Quercus mongolica). Curr. Issues Mol. Biol. 2023, 45, 1127. [Google Scholar] [CrossRef]
- Xue, G.; Fan, Y.; Zheng, C.; Yang, H.; Feng, L.; Chen, X.; Yang, Y.; Yao, X.; Weng, W.; Kong, L.; et al. bHLH Transcription Factor Family Identification, Phylogeny, and Its Response to Abiotic Stress in Chenopodium quinoa. Front. Plant Sci. 2023, 14, 1171518. [Google Scholar] [CrossRef]
- Zhao, L.; Bi, W.; Jia, Y.; Shi, J.; Chi, Y.; Yu, M.; Wang, C. Genome-Wide Characterization of bHLH Family Genes and Expression Analysis in Response to Osmotic Stress in Betula platyphylla. Plants 2023, 12, 3687. [Google Scholar] [CrossRef]
- Zhou, X.; Liao, Y.; Kim, S.-U.; Chen, Z.; Nie, G.; Cheng, S.; Ye, J.; Xu, F. Genome-Wide Identification and Characterization of bHLH Family Genes from Ginkgo biloba. Sci. Rep. 2020, 10, 13723. [Google Scholar] [CrossRef]
- Tan, C.; Qiao, H.; Ma, M.; Wang, X.; Tian, Y.; Bai, S.; Hasi, A. Genome-Wide Identification and Characterization of Melon bHLH Transcription Factors in Regulation of Fruit Development. Plants 2021, 10, 2721. [Google Scholar] [CrossRef]
- Dong, H.; Chen, Q.; Dai, Y.; Hu, W.; Zhang, S.; Huang, X. Genome-Wide Identification of PbrbHLH Family Genes, and Expression Analysis in Response to Drought and Cold Stresses in Pear (Pyrus bretschneideri). BMC Plant Biol. 2021, 21, 86. [Google Scholar] [CrossRef]
- Liu, W.; Yi, Y.; Zhuang, J.; Ge, C.; Cao, Y.; Zhang, L.; Liu, M. Genome-Wide Identification and Transcriptional Profiling of the Basic Helix-Loop-Helix Gene Family in Tung Tree (Vernicia fordii). PeerJ 2022, 10, e13981. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Lai, D.; Yang, H.; Xue, G.; He, A.; Chen, L.; Feng, L.; Ruan, J.; Xiang, D.; Yan, J.; et al. Genome-Wide Identification and Expression Analysis of the bHLH Transcription Factor Family and Its Response to Abiotic Stress in Foxtail Millet (Setaria italica L.). BMC Genom. 2021, 22, 778. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of Duplicate Genes in Exon–Intron Structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Wang, P.; Su, L.; Gao, H.; Jiang, X.; Wu, X.; Li, Y.; Zhang, Q.; Wang, Y.; Ren, F. Genome-Wide Characterization of bHLH Genes in Grape and Analysis of Their Potential Relevance to Abiotic Stress Tolerance and Secondary Metabolite Biosynthesis. Front. Plant Sci. 2018, 9, 64. [Google Scholar] [CrossRef]
- Yang, M.; Zhou, C.; Yang, H.; Kuang, R.; Huang, B.; Wei, Y. Genome-Wide Analysis of Basic Helix-Loop-Helix Transcription Factors in Papaya (Carica papaya L.). PeerJ 2020, 8, e9319. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, W.; Liu, R.; Shi, B.; Shu, Y.; Zhang, H. Genome-Wide Characterization and Expression Analysis of bHLH Gene Family in Physic Nut (Jatropha curcas L.). PeerJ 2022, 10, e13786. [Google Scholar] [CrossRef]
- Li, X.; Xue, C.; Li, J.; Qiao, X.; Li, L.; Yu, L.; Huang, Y.; Wu, J. Genome-Wide Identification, Evolution and Functional Divergence of MYB Transcription Factors in Chinese White Pear (Pyrus Bretschneideri). Plant Cell Physiol. 2016, 57, 824–847. [Google Scholar] [CrossRef]
- Sun, H.; Pang, B.; Yan, J.; Wang, T.; Wang, L.; Chen, C.; Li, Q.; Ren, Z. Comprehensive Analysis of Cucumber Gibberellin Oxidase Family Genes and Functional Characterization of CsGA20ox1 in Root Development in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 3135. [Google Scholar] [CrossRef]
- Chen, F.; Hu, Y.; Vannozzi, A.; Wu, K.; Cai, H.; Qin, Y.; Mullis, A.; Lin, Z.; Zhang, L. The WRKY Transcription Factor Family in Model Plants and Crops. Crit. Rev. Plant Sci. 2017, 36, 311–335. [Google Scholar] [CrossRef]
- Starr, T.K.; Jameson, S.C.; Hogquist, K.A. Positive and Negative Selection of T Cells. Annu. Rev. Immunol. 2003, 21, 139–176. [Google Scholar] [CrossRef]
- Ding, A.; Ding, A.; Li, P.; Wang, J.; Cheng, T.; Bao, F.; Zhang, Q. Genome-Wide Identification and Low-Temperature Expression Analysis of bHLH Genes in Prunus mume. Front. Genet. 2021, 12, 762135. [Google Scholar] [CrossRef]
- Fu, N.; Wang, L.; Sun, Q.; Wang, Q.; Zhang, Y.; Han, X.; Yang, Q.; Ma, W.; Tong, Z.; Zhang, J. Genome-Wide Identification of the bHLH Transcription Factor Family and the Regulatory Roles of PbbHLH74 in Response to Drought Stress in Phoebe bournei. Int. J. Biol. Macromol. 2024, 283, 137760. [Google Scholar] [CrossRef]
- Luo, X.; Dai, Y.; Xian, B.; Xu, J.; Zhang, R.; Rehmani, M.S.; Zheng, C.; Zhao, X.; Mao, K.; Ren, X.; et al. PIF4 Interacts with ABI4 to Serve as a Transcriptional Activator Complex to Promote Seed Dormancy by Enhancing ABA Biosynthesis and Signaling. J. Integr. Plant Biol. 2024, 66, 909–927. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant Hormone-Mediated Regulation of Stress Responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Shu, K.; Liu, X.; Xie, Q.; He, Z. Two Faces of One Seed: Hormonal Regulation of Dormancy and Germination. Mol. Plant 2016, 9, 34–45. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.; Bressan, R.A.; Song, C.; Zhu, J.; Zhao, Y. Abscisic Acid Dynamics, Signaling, and Functions in Plants. Invit. Expert Rev. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Sano, N.; Marion-Poll, A. ABA Metabolism and Homeostasis in Seed Dormancy and Germination. Int. J. Mol. Sci. 2021, 22, 5069. [Google Scholar] [CrossRef]
- Concha, C.M.; Figueroa, N.E.; Poblete, L.A.; Oñate, F.A.; Schwab, W.; Figueroa, C.R. Methyl Jasmonate Treatment Induces Changes in Fruit Ripening by Modifying the Expression of Several Ripening Genes in Fragaria chiloensis Fruit. Plant Physiol. Biochem. 2013, 70, 433–444. [Google Scholar] [CrossRef]
- Li, H.; Sun, J.; Xu, Y.; Jiang, H.; Wu, X.; Li, C. The bHLH-Type Transcription Factor AtAIB Positively Regulates ABA Response in Arabidopsis. Plant Mol. Biol. 2007, 65, 655–665. [Google Scholar] [CrossRef]
- Mei, S.; Zhang, M.; Ye, J.; Du, J.; Jiang, Y.; Hu, Y. Auxin Contributes to Jasmonate-Mediated Regulation of Abscisic Acid Signaling during Seed Germination in Arabidopsis. Plant Cell 2022, 35, 1110–1133. [Google Scholar] [CrossRef]
- Yang, L.; Liu, S.; Lin, R. The Role of Light in Regulating Seed Dormancy and Germination. J. Integr. Plant Biol. 2020, 62, 1310–1326. [Google Scholar] [CrossRef] [PubMed]
- Godo, T.; Fujiwara, K.; Guan, K.; Miyoshi, K. Effects of Wavelength of LED-Light on in Vitro Asymbiotic Germination and Seedling Growth of Bletilla ochracea Schltr. (Orchidaceae). Plant Biotechnol. 2011, 28, 397–400. [Google Scholar] [CrossRef]
- Katahata, S.-I.; Naramoto, M.; Kakubari, Y.; Mukai, Y. Photosynthetic Capacity and Nitrogen Partitioning in Foliage of the Evergreen Shrub Daphniphyllum humile along a Natural Light Gradient. Tree Physiol. 2007, 27, 199–208. [Google Scholar] [CrossRef]
- Brantley, S.; Young, D. Contribution of Sunflecks Is Minimal in Expanding Shrub Thickets Compared to Temperate Forest. Ecology 2009, 90, 1021–1029. [Google Scholar] [CrossRef]
- Castelain, M.; Hir, R.L.; Bellini, C. The non-DNA-binding bHLH Transcription Factor PRE3/bHLH135/ATBS1/TMO7 Is Involved in the Regulation of Light Signaling Pathway in Arabidopsis. Physiol. Plant. 2012, 145, 450–460. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, H.; Ding, L.; Soppe, W.J.; Xiang, Y. REVERSAL OF RDO5 1, a Homolog of Rice Seed Dormancy4, Interacts with bHLH57 and Controls ABA Biosynthesis and Seed Dormancy in Arabidopsis. Plant Cell 2020, 32, 1933. [Google Scholar] [CrossRef]
- Ni, M.; Tepperman, J.M.; Quail, P.H. PIF3, a Phytochrome-Interacting Factor Necessary for Normal Photoinduced Signal Transduction, Is a Novel Basic Helix-Loop-Helix Protein. Cell 1998, 95, 657–667. [Google Scholar] [CrossRef]
- Oh, E.; Zhu, J.-Y.; Wang, Z.-Y. Interaction between BZR1 and PIF4 Integrates Brassinosteroid and Environmental Responses. Nat. Cell Biol. 2012, 14, 802. [Google Scholar] [CrossRef]
- Penfield, S.; Josse, E.-M.; Halliday, K.J. A Role for an Alternative Splice Variant of PIF6 in the Control of Arabidopsis Primary Seed Dormancy. Plant Mol. Biol. 2010, 73, 89–95. [Google Scholar] [CrossRef]
- Oh, J.; Park, E.; Song, K.; Bae, G.; Choi, G. Phytochrome Interacting Factor8 Inhibits Phytochrome A-Mediated Far-Red Light Responses in Arabidopsis. Plant Cell 2019, 32, 186. [Google Scholar] [CrossRef]
- Zhang, M.; Zhu, J.; Yan, Q. Effects of Light Quality on the Seed Germination of Main Tree Species in a Secondary Forest Ecosystem of Northeast China. Chin. J. Appl. Ecol. 2012, 23, 2625–2631. [Google Scholar]
- Huang, M.; Hu, Y.; Liu, X.; Li, Y.; Hou, X. Arabidopsis Leafy Cotyledon1 Mediates Postembryonic Development via Interacting with Phytochrome-Interacting Factor4. Plant Cell 2015, 27, 3099–3111. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Li, X.; Guo, Y.; Chu, J.; Fang, S.; Yan, C.; Noel, J.P.; Liu, H. Cryptochrome 1 Interacts with PIF4 to Regulate High Temperature-Mediated Hypocotyl Elongation in Response to Blue Light. Proc. Natl. Acad. Sci. USA 2015, 113, 224. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhang, Y.; He, Y.; Wang, Y.; Wang, L. Pseudo Response Regulators Regulate Photoperiodic Hypocotyl Growth by Repressing PIF4/5 Transcription. Plant Physiol. 2020, 183, 686. [Google Scholar] [CrossRef]
- Lu, X.; Mei, M.; Liu, L.; Xu, X.; Ai, W. The Chromosome-Scale Genome of Magnolia sieboldii K. Koch Provides Insight Into the Evolutionary Position of Magnoliids and Seed Germination. Mol. Ecol. Resour. 2025, 25, e14030. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2020, 49, D412–D419. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER Web Server: 2018 Update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The Proteomics Server for in-Depth Protein Knowledge and Analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A Multiple Sequence Alignment Method with Reduced Time and Space Complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; de Peer, Y.V.; Rouzé, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Geourjon, C.; Deléage, G. SOPMA: Significant Improvements in Protein Secondary Structure Prediction by Consensus Prediction from Multiple Alignments. Comput. Appl. Biosci. 1995, 11, 681–684. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A Hierarchical, Functionally and Phylogenetically Annotated Orthology Resource Based on 5090 Organisms and 2502 Viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Ai, W.; Liu, H.; Wang, Y.; Wang, Y.; Wei, J.; Zhang, X.; Lu, X. Identification of Functional Brassinosteroid Receptor Genes in Oaks and Functional Analysis of QmBRI1. Int. J. Mol. Sci. 2023, 24, 16405. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Xu, X.; Yang, X.; Liu, H.; Xu, L.; Ai, W.; Lu, X. Genome-Wide Identification of the bHLH Gene Family in Magnolia sieboldii and Response of MsPIFs to Different Light Qualities. Int. J. Mol. Sci. 2025, 26, 3152. https://doi.org/10.3390/ijms26073152
Liu L, Xu X, Yang X, Liu H, Xu L, Ai W, Lu X. Genome-Wide Identification of the bHLH Gene Family in Magnolia sieboldii and Response of MsPIFs to Different Light Qualities. International Journal of Molecular Sciences. 2025; 26(7):3152. https://doi.org/10.3390/ijms26073152
Chicago/Turabian StyleLiu, Lin, Xin Xu, Xiaohuan Yang, Hanzhang Liu, Lingyi Xu, Wanfeng Ai, and Xiujun Lu. 2025. "Genome-Wide Identification of the bHLH Gene Family in Magnolia sieboldii and Response of MsPIFs to Different Light Qualities" International Journal of Molecular Sciences 26, no. 7: 3152. https://doi.org/10.3390/ijms26073152
APA StyleLiu, L., Xu, X., Yang, X., Liu, H., Xu, L., Ai, W., & Lu, X. (2025). Genome-Wide Identification of the bHLH Gene Family in Magnolia sieboldii and Response of MsPIFs to Different Light Qualities. International Journal of Molecular Sciences, 26(7), 3152. https://doi.org/10.3390/ijms26073152





